Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication
Abstract
1. Introduction
2. Results
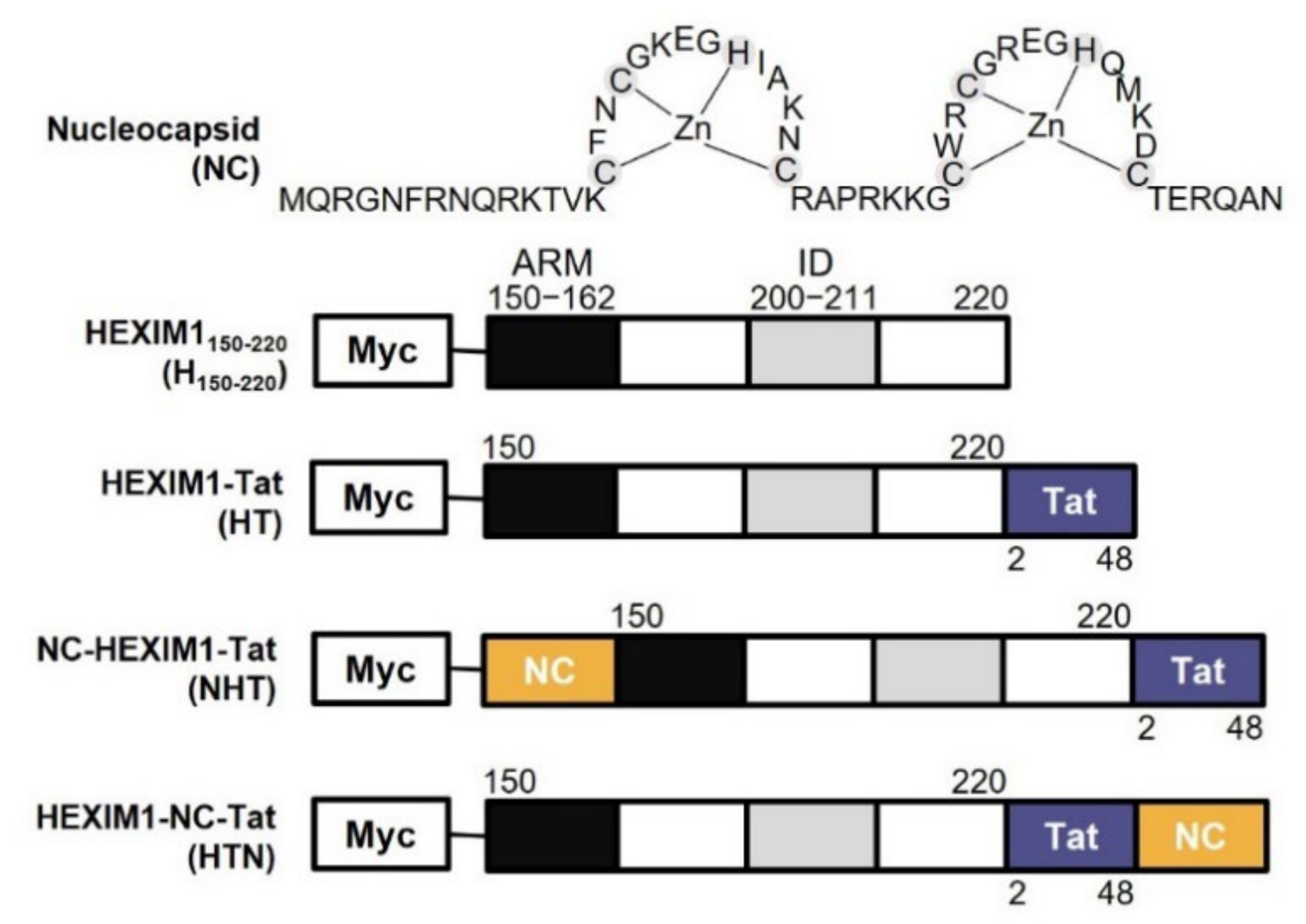
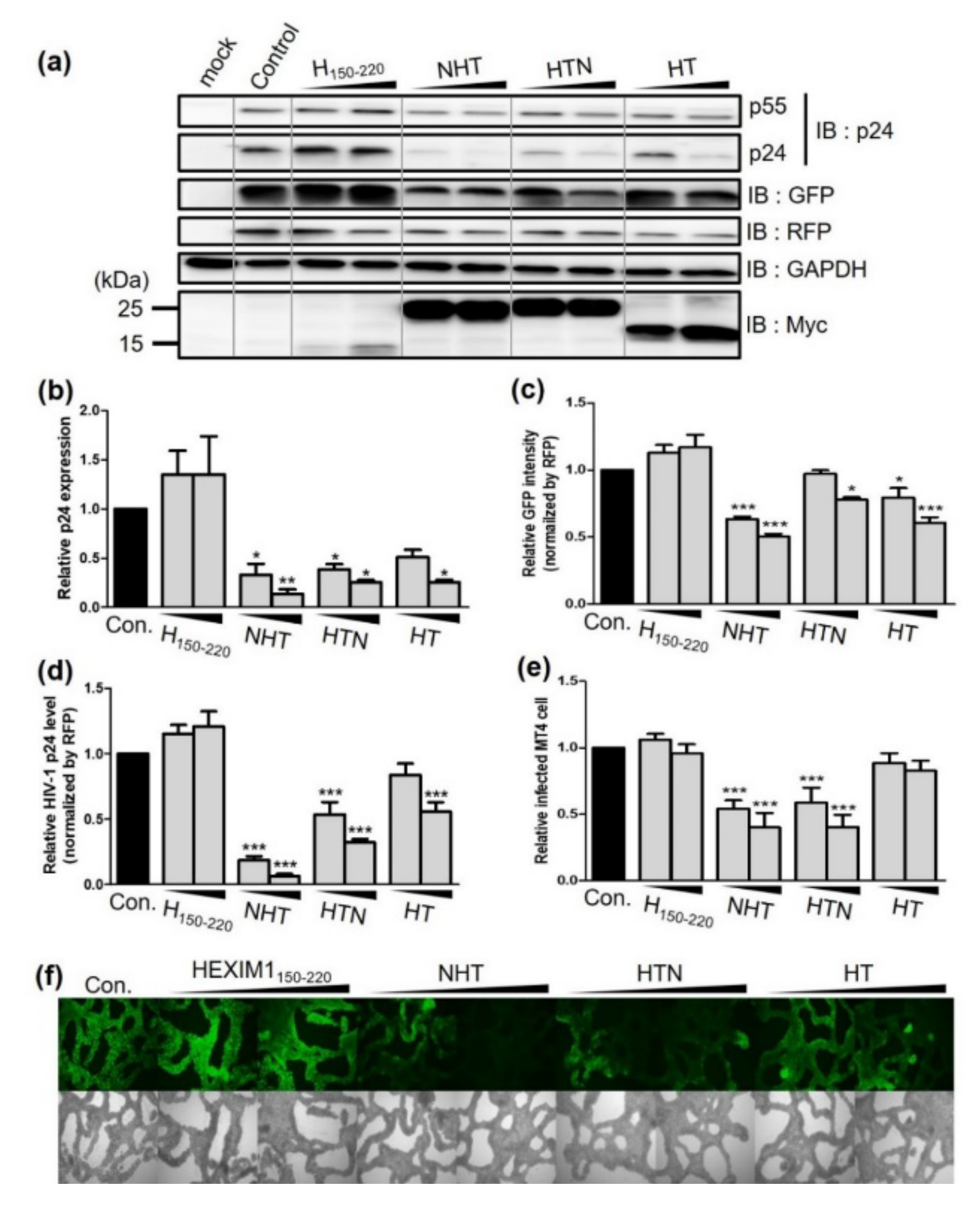
2.1. The Chimeric Proteins NHT and HTN Have Better Antiviral Effects than HT
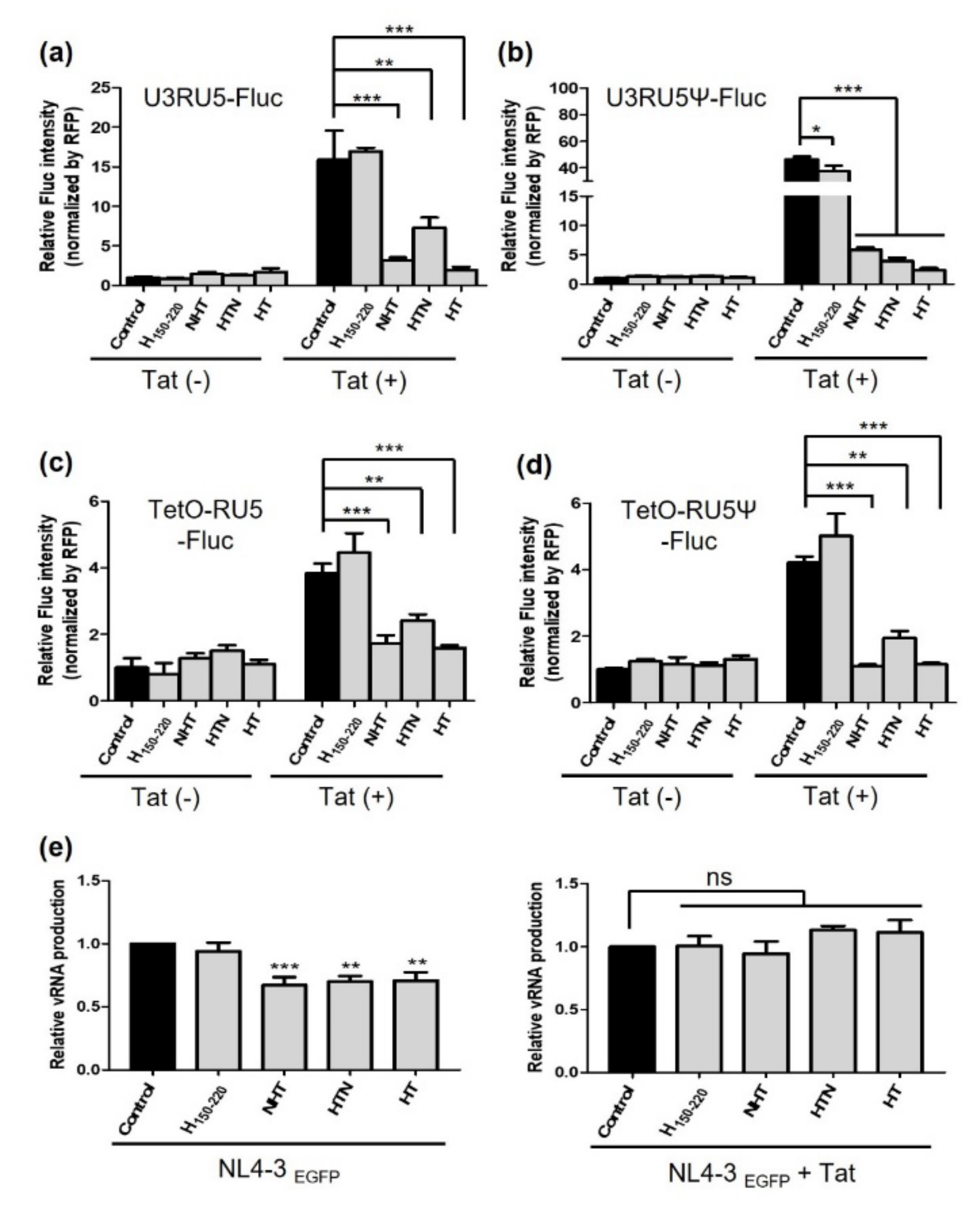
2.2. NHT, HTN, and HT Inhibit the Transcription of the HIV-1 Gene in a Tat-Dependent Manner
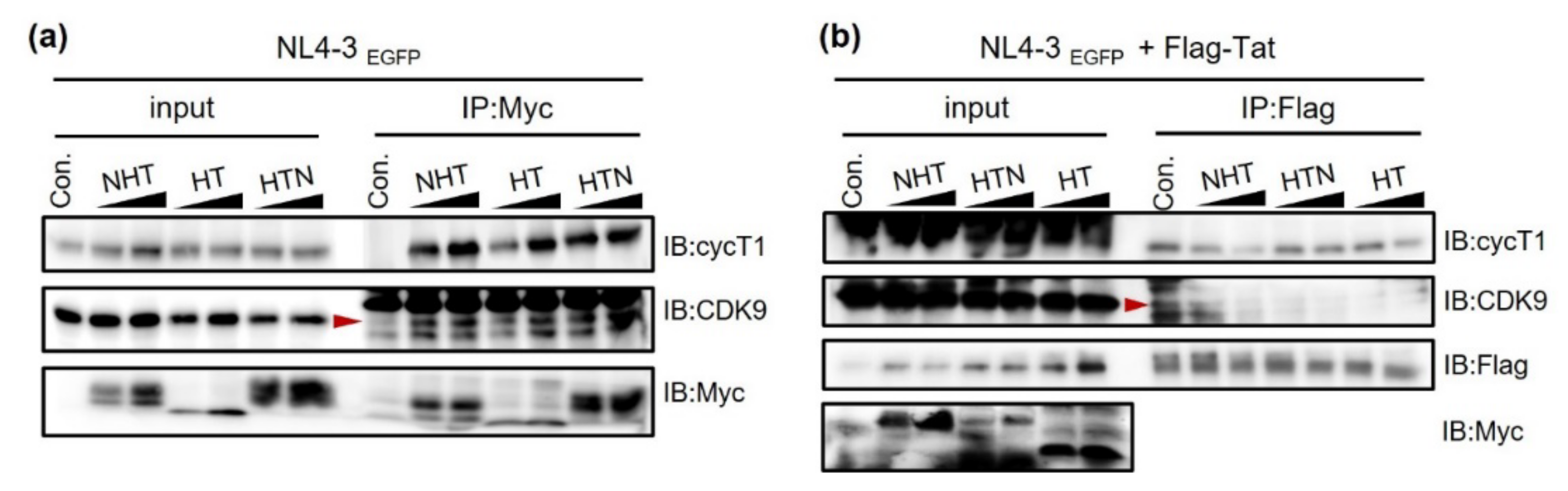
2.3. NTH, HTN, and HT Interact with and Hijack the Host Cell Factor P-TEFb and Viral RNA by Competing with Tat
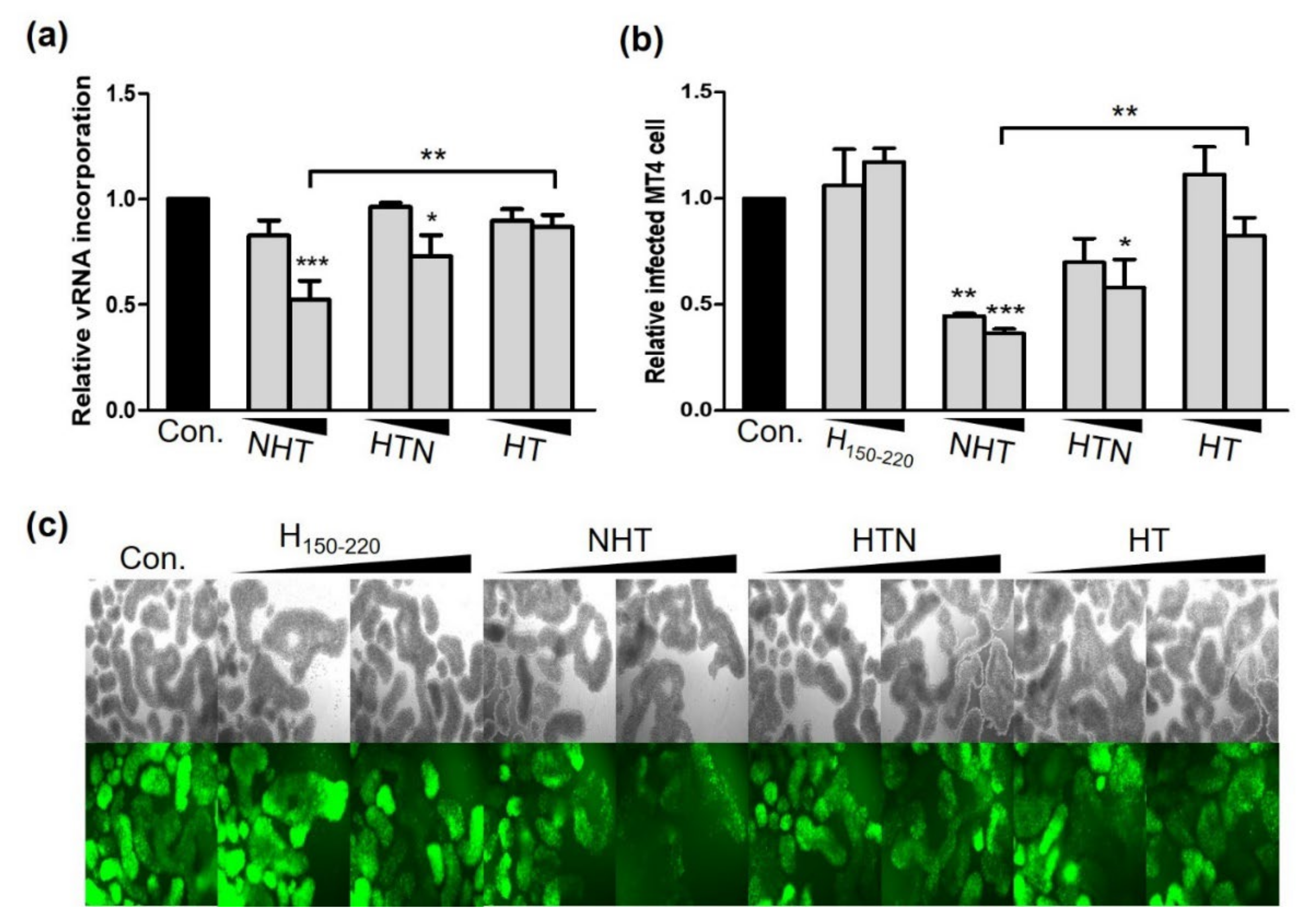
2.4. NTH and HTN Interrupt vRNA Packaging and Reduce the Infectivity of the Progeny Virus
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. Antibodies
4.3. Preparation of the HIV-1 Virus Stock
4.4. HIV-1 p24 ELISA
4.5. MT4 Cell Infection
4.6. Reporter Gene Assay
4.7. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
4.8. RNA Immunoprecipitation
4.9. Co-Immunoprecipitation
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, J.; Marshall, N.F.; Price, D.H. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998, 273, 13855–13860. [Google Scholar] [CrossRef]
- Garber, M.E.; Wei, P.; KewalRamani, V.N.; Mayall, T.P.; Herrmann, C.H.; Rice, A.P.; Littman, D.R.; Jones, K.A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998, 12, 3512–3527. [Google Scholar] [CrossRef] [PubMed]
- Wei, P.; Garber, M.E.; Fang, S.-M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Marshall, N.F.; Peng, J.; Xie, Z.; Price, D.H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996, 271, 27176–27183. [Google Scholar] [CrossRef] [PubMed]
- Taube, R.; Lin, X.; Irwin, D.; Fujinaga, K.; Peterlin, B.M. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 2002, 22, 321–331. [Google Scholar] [CrossRef]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Ping, Y.-H.; Rana, T.M. Tat-associated kinase (P-TEFb): A component of transcription preinitiation and elongation complexes. J. Biol. Chem. 1999, 274, 7399–7404. [Google Scholar] [CrossRef] [PubMed]
- Isel, C.; Karn, J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 1999, 290, 929–941. [Google Scholar] [CrossRef] [PubMed]
- Dahmus, M.E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 1996, 271, 19009–19012. [Google Scholar] [CrossRef] [PubMed]
- Fujinaga, K.; Luo, Z.; Peterlin, B.M. Genetic analysis of the structure and function of 7SK small nuclear ribonucleoprotein (snRNP) in cells. J. Biol. Chem. 2014, 289, 21181–21190. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Brogie, J.E.; Price, D.H. 7SK snRNA: A noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley Interdiscip. Rev. RNA 2012, 3, 92–103. [Google Scholar] [CrossRef]
- Yik, J.H.; Chen, R.; Pezda, A.C.; Samford, C.S.; Zhou, Q. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 2004, 24, 5094–5105. [Google Scholar] [CrossRef]
- Schulte, A.; Czudnochowski, N.; Barboric, M.; Schönichen, A.; Blazek, D.; Peterlin, B.M.; Geyer, M. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J. Biol. Chem. 2005, 280, 24968–24977. [Google Scholar] [CrossRef] [PubMed]
- Dames, S.A.; Schönichen, A.; Schulte, A.; Barboric, M.; Peterlin, B.M.; Grzesiek, S.; Geyer, M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc. Natl. Acad. Sci. USA 2007, 104, 14312–14317. [Google Scholar] [CrossRef]
- Kobbi, L.; Demey-Thomas, E.; Braye, F.; Proux, F.; Kolesnikova, O.; Vinh, J.; Poterszman, A.; Bensaude, O. An evolutionary conserved Hexim1 peptide binds to the Cdk9 catalytic site to inhibit P-TEFb. Proc. Natl. Acad. Sci. USA 2016, 113, 12721–12726. [Google Scholar] [CrossRef] [PubMed]
- Czudnochowski, N.; Bösken, C.A.; Geyer, M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat. Commun. 2012, 3, 842. [Google Scholar] [CrossRef] [PubMed]
- Muniz, L.; Egloff, S.; Ughy, B.; Jády, B.E.; Kiss, T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog. 2010, 6, e1001152. [Google Scholar] [CrossRef]
- Fujinaga, K.; Taube, R.; Wimmer, J.; Cujec, T.P.; Peterlin, B.M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 1999, 96, 1285–1290. [Google Scholar] [CrossRef]
- Barboric, M.; Yik, J.H.; Czudnochowski, N.; Yang, Z.; Chen, R.; Contreras, X.; Geyer, M.; Matija Peterlin, B.; Zhou, Q. Tat competes with HEXIM1 to increase the active pool of P-TEFb for HIV-1 transcription. Nucleic Acids Res. 2007, 35, 2003–2012. [Google Scholar] [CrossRef]
- Zhu, Y.; Pe’ery, T.; Peng, J.; Ramanathan, Y.; Marshall, N.; Marshall, T.; Amendt, B.; Mathews, M.B.; Price, D.H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997, 11, 2622–2632. [Google Scholar] [CrossRef] [PubMed]
- Mancebo, H.S.; Lee, G.; Flygare, J.; Tomassini, J.; Luu, P.; Zhu, Y.; Peng, J.; Blau, C.; Hazuda, D.; Price, D. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997, 11, 2633–2644. [Google Scholar] [CrossRef] [PubMed]
- Bieniasz, P.D.; Grdina, T.A.; Bogerd, H.P.; Cullen, B.R. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 7791–7796. [Google Scholar] [CrossRef]
- Leoz, M.; Kukanja, P.; Luo, Z.; Huang, F.; Cary, D.C.; Peterlin, B.M.; Fujinaga, K. HEXIM1-Tat chimera inhibits HIV-1 replication. PLoS Pathog. 2018, 14, e1007402. [Google Scholar] [CrossRef] [PubMed]
- Green, L.M.; Berg, J.M. Retroviral nucleocapsid protein-metal ion interactions: Folding and sequence variants. Proc. Natl. Acad. Sci. USA 1990, 87, 6403–6407. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.M.; Sargent, T.D. Localized and inducible expression of Xenopus-posterior (Xpo), a novel gene active in early frog embryos, encoding a protein with a ‘CCHC’finger domain. Development 1991, 112, 747–753. [Google Scholar] [CrossRef]
- Berkowitz, R.D.; Goff, S.P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology 1994, 202, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Lapadat-Tapolsky, M.; Rocquigny, H.D.; Gent, D.V.; Roques, B.; Plasterk, R.; Darlix, J.-L. Interactions between HIV-1 nucleocapsid protein and viral DNA may have important functions in the viral life cycle. Nucleic Acids Res. 1993, 21, 831–839. [Google Scholar] [CrossRef]
- Purcell, D.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [CrossRef]
- De Guzman, R.N.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Summers, M.F.; Henderson, L.E.; Chance, M.R.; South, T.L.; Blake, P.R.; Perez-Alvarado, G.; Bess, J.W., Jr.; Sowder III, R.C.; Arthur, L.O.; Sagi, I. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1992, 1, 563–574. [Google Scholar] [CrossRef] [PubMed]
- South, T.L.; Summers, M.F. Zinc-and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Sci. 1993, 2, 3–19. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.S.; Panganiban, A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [CrossRef]
- Muriaux, D.; Darlix, J.-L. Properties and functions of the nucleocapsid protein in virus assembly. RNA Biol. 2010, 7, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Kim, M.Y.; Lee, J.H.; You, J.C.; Jeong, S. Selection and stabilization of the RNA aptamers against the human immunodeficiency virus type-1 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2002, 291, 925–931. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.I.; Kim, Y.H.; Paik, S.Y.; You, J.C. Development of a cell-based assay probing the specific interaction between the human immunodeficiency virus type 1 nucleocapsid and psi RNA in vivo. J. Virol. 2007, 81, 6151–6155. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.-Y.; Kim, S.-H.; Jang, S.-I.; You, J.-C. Examination of specific binding activity of aptamer RNAs to the HIV-NC by using a cell-based in vivo assay for protein-RNA interaction. BMB Rep. 2008, 41, 511–515. [Google Scholar] [CrossRef]
- Berkhout, B.; Vastenhouw, N.L.; Klasens, B.I.; Huthoff, H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. Rna 2001, 7, 1097–1114. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.; Berkhout, B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Muesing, M.A.; Smith, D.H.; Cabradilla, C.D.; Benton, C.V.; Lasky, L.A.; Capon, D.J. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 1985, 313, 450–458. [Google Scholar] [CrossRef]
- Masuda, T.; Sato, Y.; Huang, Y.-L.; Takahata, T.; Hasegawa, A.; Kawai, G.; Kannagi, M. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci. Rep. 2016, 5, 17680. [Google Scholar] [CrossRef] [PubMed]
- Didierlaurent, L.; Houzet, L.; Morichaud, Z.; Darlix, J.-L.; Mougel, M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008, 36, 4745–4753. [Google Scholar] [CrossRef] [PubMed]
- Chamontin, C.; Rassam, P.; Ferrer, M.; Racine, P.-J.; Neyret, A.; Lainé, S.; Milhiet, P.-E.; Mougel, M. HIV-1 nucleocapsid and ESCRT-component Tsg101 interplay prevents HIV from turning into a DNA-containing virus. Nucleic Acids Res. 2015, 43, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Berthoux, L.; Péchoux, C.; Ottmann, M.; Morel, G.; Darlix, J.-L. Mutations in the N-terminal domain of human immunodeficiency virus type 1 nucleocapsid protein affect virion core structure and proviral DNA synthesis. J. Virol. 1997, 71, 6973–6981. [Google Scholar] [CrossRef]
- Druillennec, S.; Dong, C.; Escaich, S.; Gresh, N.; Bousseau, A.; Roques, B.; Fournie-Zaluski, M. A mimic of HIV-1 nucleocapsid protein impairs reverse transcription and displays antiviral activity. Proc. Natl. Acad. Sci. USA 1999, 96, 4886–4891. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef]
- Morales, J.O.; Fathe, K.R.; Brunaugh, A.; Ferrati, S.; Li, S.; Montenegro-Nicolini, M.; Mousavikhamene, Z.; McConville, J.T.; Prausnitz, M.R.; Smyth, H.D. Challenges and future prospects for the delivery of biologics: Oral mucosal, pulmonary, and transdermal routes. AAPS J. 2017, 19, 652–668. [Google Scholar] [CrossRef]
- Lewin, S.R.; Deeks, S.G.; Barré-Sinoussi, F. Towards a cure for HIV—Are we making progress? Lancet 2014, 384, 209–211. [Google Scholar] [CrossRef]
- Peritz, T.; Zeng, F.; Kannanayakal, T.J.; Kilk, K.; Eiríksdóttir, E.; Langel, U.; Eberwine, J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006, 1, 577–580. [Google Scholar] [CrossRef]

| LTR Site | Sequence |
|---|---|
| U3 | tggaagggctaatttggtcccaaaaaagacaagagatccttgatctgtggatctaccacacacaaggctacttccctgattggcagaactacacaccagggccagggatcagatatccactgacctttggatggtgcttcaagttagtaccagttgaaccagagcaagtagaagaggccaaataaggagagaagaacagcttgttacaccctatgagccagcatgggatggaggacccggagggagaagtattagtgtggaagtttgacagcctcctagcatttcgtcacatggcccgagagctgcatccggagtactacaaagactgctgacatcgagctttctacaagggactttccgctggggactttccagggaggtgtggcctgggcgggactggggagtggcgagccctcagatgctacatataagcagctgctttttgcctgtact |
| R | gggtctctctggttagaccagatctgagcctgggagctctctggctaactagggaacccactgcttaagcctcaataaagcttgccttgagtgctc |
| U5 | aaagtagtgtgtgcccgtctgttgtgtgactctggtaactagagatccctcagacccttttagtcagtgtggaaaatctctagca |
| Ψ | gcaggactcggcttgctgaagcgcgcacggcaagaggcgaggggcggcgactggtgagtacgccaaaaattttgactagcggaggctagaaggagagagatgggtgcgagagcgtcggtattaagcg |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.-I.; Kim, G.-N.; Yu, K.-L.; Park, S.-H.; You, J.C. Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication. Int. J. Mol. Sci. 2022, 23, 12340. https://doi.org/10.3390/ijms232012340
Kim H-I, Kim G-N, Yu K-L, Park S-H, You JC. Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication. International Journal of Molecular Sciences. 2022; 23(20):12340. https://doi.org/10.3390/ijms232012340
Chicago/Turabian StyleKim, Hae-In, Ga-Na Kim, Kyung-Lee Yu, Seong-Hyun Park, and Ji Chang You. 2022. "Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication" International Journal of Molecular Sciences 23, no. 20: 12340. https://doi.org/10.3390/ijms232012340
APA StyleKim, H.-I., Kim, G.-N., Yu, K.-L., Park, S.-H., & You, J. C. (2022). Identification of Novel Nucleocapsid Chimeric Proteins Inhibiting HIV-1 Replication. International Journal of Molecular Sciences, 23(20), 12340. https://doi.org/10.3390/ijms232012340





