Genome-Wide SNP Markers Accelerate Perennial Forest Tree Breeding Rate for Disease Resistance through Marker-Assisted and Genome-Wide Selection
Abstract
1. Introduction
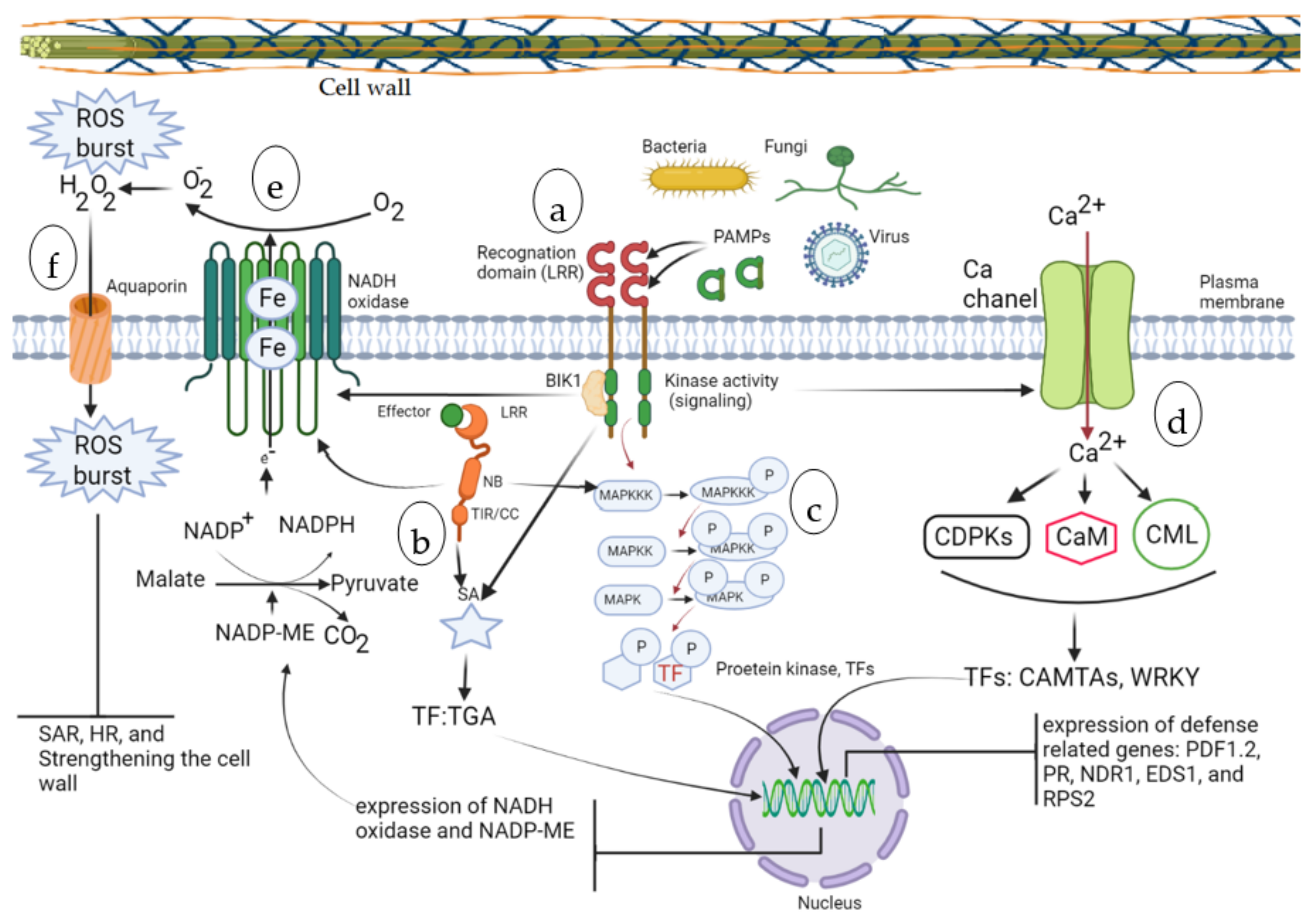
2. Molecular Mechanisms of Tree Responses to Diseases
3. Experimental Approaches to Characterizing Disease Resistance Related Genes
3.1. QTL Mapping
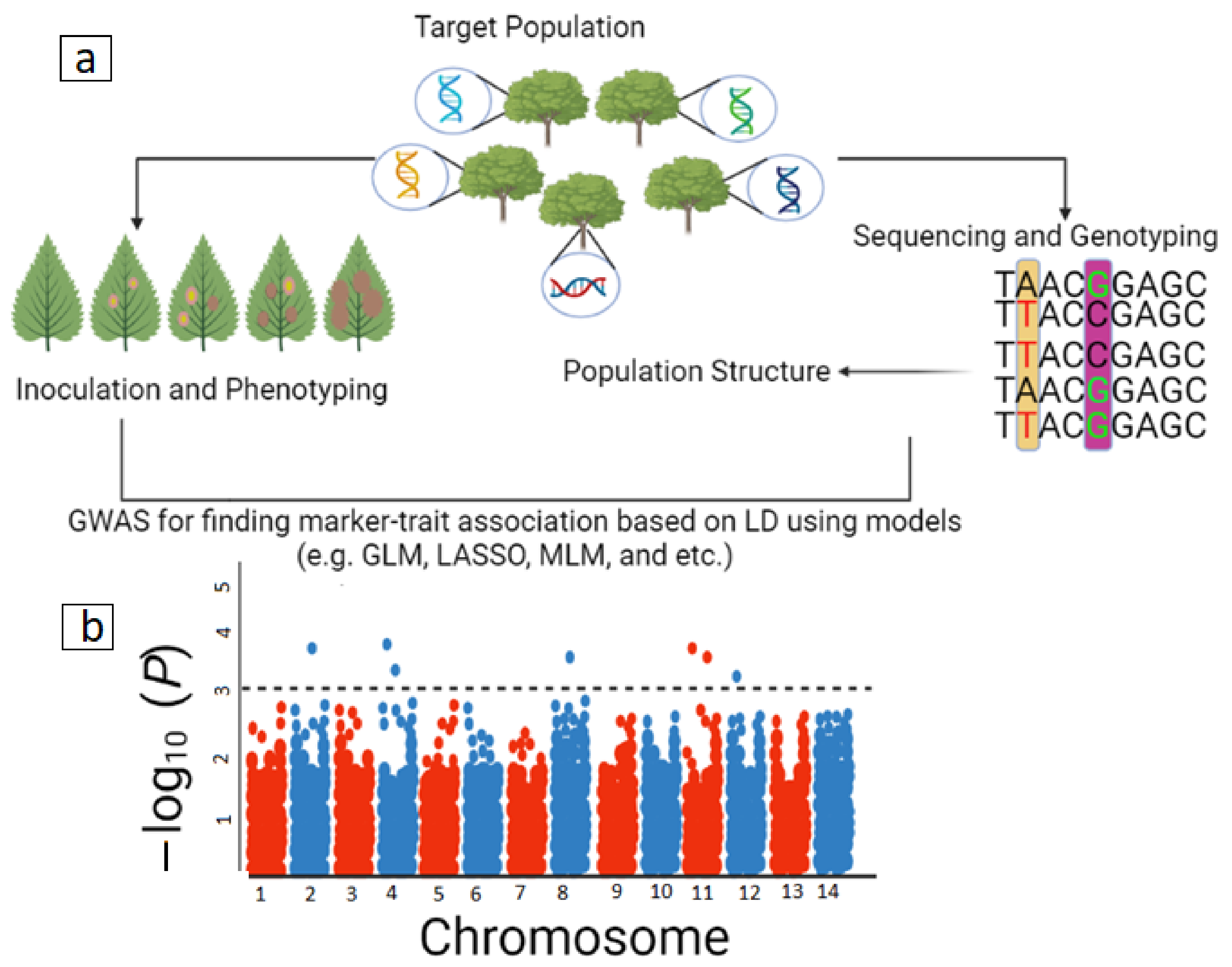
3.2. Association Mapping and Genome-Wide Association Studies (GWAS)
| Species | Target Trait | Number of Samples | Number of Markers | Significant Markers | GWAS Model | References |
|---|---|---|---|---|---|---|
| Cornus florida L. | Resistance to dogwood anthracnose and ecological pressures | 289 | 3134 | 14–29 | Latent factor mixed modeling | [64] |
| Eucalyptus | Growth and wood property traits | 1123 | 37,832 | 78 | FarmCPU | [65] |
| Eucalyptus | DBH, diameter at breast height; HT, total height. | 3373 | 41,320 | 11–3 | linear model-based association (LMA) and mixed linear model-based association (MLMA) | [66] |
| E. globulus | Growth and wood property traits | 303 | 2,364 DArT | 18 | Unified Mixed Model | [67] |
| E. grandis | Terpene traits, 1,8-cineole, c-terpinene, and p-cymene | 416 | 15,387 | 21–32 and 28 | Efficient Mixed-Model Association eXpedited (EMMAX) | [68] |
| Fagus grandifolia Ehrh. | Beech bark disease (BBD) | 514 | 16,000 | 4 | logistic regression model | [69] |
| Hevea brasiliensis | Latex yield | 170 | 21,146 | 4 | MLMA | [70] |
| Picea abies (L.) H. Karst. | Susceptibility to H. parviporum | 533 | 373,384 | 36 | EMMAX | [71] |
| P. abies L. Karst | Heterobasidion rot caused by H. annosum s.s. | 400 | 63,760 | 12 | MLM | [72] |
| P. abies L. Karst | Resistance to H. parviporum | 466 | 197,399 | 11 | LASSO | [73] |
| P. crassifolia Kom. | Earlywood tracheid traits | 106 | 12,275,765 | 96 | MLM | [74] |
| Pinus contorta | Serotiny | 98 | 95,000 | 11 | Bayesian generalized linear model (GLM) | [75] |
| P. taeda | Solid wood and wood chemistry traits | 435 | 58 | 4 | MLM | [76] |
| P. taeda | Carbon isotope discrimination | 961 | 46 | 4 | MLM | [77] |
| P. taeda | Pitch canker resistance | 498 | 7216 | 10 | Bayesian association with missing data (BAMD) | [78] |
| Populus trichocarpa | 16 wood chemistry/ ultrastructure traits | 334 | 29,233 | 141 | regression model | [79] |
| P. trichocarpa | Resistance to Sphaerulina musiva | 3404 | 8,253,066 | 96 | EMMA in EMMAX software | [80] |
| P.trichocarpa | Stomatal traits | 2447 | 34,131 | 25 | GLM | [81] |
3.3. Genomic Selection
| Species | Target Trait | Population Size and Type | Number of Markers | Prediction Accuracy | Prediction Model | References |
|---|---|---|---|---|---|---|
| Pinus taeda L. | Growth, wood quality, and fusiform rust resistance | 921 from 61 full-sib families | 4,853 SNPs | 0.17-0.51 | Bayes A, Cπ Bayesian LASSO and RR–BLUP | [101] |
| P. taeda L. | Fusiform rust resistance | 721 from 5 full-sib families | 16,920 SNPs | 0.71–0.76 | Ridge regression, Bayes B, and Bayes Cπ | [102] |
| Fraxinus excelsior | Resistance to ash dieback disease | 1,250 from 15 native seed zones | 10,000 SNPs | 0.67 | RR–BLUP | [105] |
| Castanea dentata | Mean canker severity | Progeny of 1,230 from BC3F2 | 71,507 SNPs | 0.06-0.30 | HBLUP, ABLUP, and Bayes C | [107] |
| Theobroma cacao | Resistance to black pod rot (BPR), witches’ broom disease (WBD), and frosty pod rot disease (FPRD) diseases | 1,345 accessions from F1 progeny | 9,640 SNPs | 0.065-0.478 | G-BLUP | [109] |
4. Future Application of GWAS and GS
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guevara-Escudero, M.; Osorio, A.N.; Cortés, A.J. Integrative pre-breeding for biotic resistance in forest trees. Plants 2021, 10, 2022. [Google Scholar] [CrossRef]
- Ghanbary, E.; Fathizadeh, O.; Pazhouhan, I.; Zarafshar, M.; Kouchaksaraei, M.; Jafarnia, S.; Parad, G.; Bader, M. Drought and pathogen effects on survival, leaf physiology, oxidative damage, and defense in two Middle Eastern oak species. Forests 2021, 12, 247. [Google Scholar] [CrossRef]
- Naidoo, S.; Slippers, B.; Plett, J.M.; Coles, D.; Oates, C.N. The road to resistance in forest trees. Front. Plant Sci. 2019, 10, 273. [Google Scholar] [CrossRef]
- Coker, T.L.R.; Rozsypálek, J.; Edwards, A.; Harwood, T.P.; Butfoy, L.; Buggs, R.J.A. Estimating mortality rates of European ash (Fraxinus excelsior) under the ash dieback (Hymenoscyphus fraxineus) epidemic. Plants People Planet 2019, 1, 48–58. [Google Scholar] [CrossRef]
- Mirabolfathy, M.; Ju, Y.-M.; Hsieh, H.-M.; Rogers, J.D. Obolarina persica sp. nov., associated with dying Quercus in Iran. Mycoscience 2013, 54, 315–320. [Google Scholar] [CrossRef]
- Safaee, D.; Khodaparast, S.A.; Mirabolfathy, M.; Mousanejad, S. A multiplex PCR-based technique for identification of Biscogniauxia mediterranea and Obolarina persica causing charcoal disease of oak trees in Zagros forests. For. Pathol. 2017, 47, e12330. [Google Scholar] [CrossRef]
- Şimşek, S.A.; Akyüz, B.; Katircioğlu, Y.Z.; Serdar, U.; Maden, S. Effectiveness and efficacy of superficial disinfectants to prevent mechanical transmission of Cryphonectria parasitica from chestnut scion woods. Eur. J. Plant Pathol. 2021, 159, 131–138. [Google Scholar] [CrossRef]
- Newhouse, A.E.; Polin-McGuigan, L.D.; Baier, K.A.; Valletta, K.E.; Rottmann, W.H.; Tschaplinski, T.J.; Maynard, C.A.; Powell, W.A. Transgenic American chestnuts show enhanced blight resistance and transmit the trait to T1 progeny. Plant Sci. 2014, 228, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.G.; de Jesus Junior, W.C.; Pezzopane, J.E.M.; Alves, F.R.; Moraes, W.B.; Pirovani, D.B.; Mafia, R.G.; do Amaral, J.F.T.; Guerra Filho, P.A.; Curty, T.A.; et al. Evasive planning for the management of Eucalyptus rust Austropuccinia psidii for Espírito Santo State, Brazil. Forests 2022, 13, 646. [Google Scholar] [CrossRef]
- Kimic, K.; Mirzwa-Mróz, E.; Szyndel, M.S. Diagnosis and recommendations for management of trees and shrubs in green squares in Warsaw based on research on fungal diseases. Trees 2022, 1–15. [Google Scholar] [CrossRef]
- Eyles, A.; Bonello, P.; Ganley, R.; Mohammed, C. Induced resistance to pests and pathogens in trees. New Phytol. 2010, 185, 893–908. [Google Scholar] [CrossRef]
- Stępniewska, H.; Jankowiak, R.; Bilański, P.; Hausner, G. Structure and abundance of Fusarium communities inhabiting the litter of beech forests in central Europe. Forests 2021, 12, 811. [Google Scholar] [CrossRef]
- Hennon, P.E.; Frankel, S.J.; Woods, A.J.; Worrall, J.J.; Ramsfield, T.D.; Zambino, P.J.; Shaw, D.C.; Ritóková, G.; Warwell, M.V.; Norlander, D.; et al. Applications of a conceptual framework to assess climate controls of forest tree diseases. For. Pathol. 2021, 51, e12719. [Google Scholar] [CrossRef]
- Basak, A. Chapter-5 Breeding Techniques of Forest Trees. For. Sci. 2022, 3, 75–105. [Google Scholar]
- Harper, A.L.; McKinney, L.V.; Nielsen, L.; Havlickova, L.; Li, Y.; Trick, M.; Fraser, F.; Wang, L.; Fellgett, A.; Sollars, E.S.A.; et al. Molecular markers for tolerance of European ash (Fraxinus excelsior) to dieback disease identified using Associative Transcriptomics. Sci. Rep. 2016, 6, 19335. [Google Scholar] [CrossRef] [PubMed]
- Neale, D.B.; Kremer, A. Forest tree genomics: Growing resources and applications. Nat. Rev. Genet. 2011, 12, 111–122. [Google Scholar] [CrossRef]
- Sollars, E.S.A.; Harper, A.L.; Kelly, L.J.; Sambles, C.M.; Ramirez-Gonzalez, R.H.; Swarbreck, D.; Kaithakottil, G.; Cooper, E.D.; Uauy, C.; Havlickova, L.; et al. Genome sequence and genetic diversity of European ash trees. Nature 2017, 541, 212–216. [Google Scholar] [CrossRef] [PubMed]
- McMullan, M.; Rafiqi, M.; Kaithakottil, G.; Clavijo, B.J.; Bilham, L.; Orton, E.; Percival-Alwyn, L.; Ward, B.J.; Edwards, A.; Saunders, D.G.O.; et al. The ash dieback invasion of Europe was founded by two genetically divergent individuals. Nat. Ecol. Evol. 2018, 2, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Harfouche, A.; Meilan, R.; Kirst, M.; Morgante, M.; Boerjan, W.; Sabatti, M.; Mugnozza, G.S. Accelerating the domestication of forest trees in a changing world. Trends Plant Sci. 2012, 17, 64–72. [Google Scholar] [CrossRef]
- Telfer, E.; Graham, N.; Macdonald, L.; Sturrock, S.; Wilcox, P.; Stanbra, L. Approaches to variant discovery for conifer transcriptome sequencing. PLoS ONE 2018, 13, e0205835. [Google Scholar] [CrossRef] [PubMed]
- Büchel, K.; Fenning, T.; Gershenzon, J.; Hilker, M.; Meiners, T. Elm defence against herbivores and pathogens: Morphological, chemical and molecular regulation aspects. Phytochem. Rev. 2016, 15, 961–983. [Google Scholar] [CrossRef]
- Kovalchuk, A.; Keriö, S.; Oghenekaro, A.O.; Jaber, E.; Raffaello, T.; Asiegbu, F.O. Antimicrobial defenses and resistance in forest trees: Challenges and perspectives in a genomic era. Annu. Rev. Phytopathol. 2013, 51, 221–244. [Google Scholar] [CrossRef] [PubMed]
- Veluthakkal, R.; Dasgupta, M.G. Dasgupta, Pathogenesis-related genes and proteins in forest tree species. Trees 2010, 24, 993–1006. [Google Scholar] [CrossRef]
- Iqbal, Z.; Iqbal, M.S.; Hashem, A.; Abd_Allah, E.F.; Ansari, M.I. Plant defense responses to biotic stress and its interplay with fluctuating dark/light conditions. Front. Plant Sci. 2021, 12, 631810. [Google Scholar] [CrossRef]
- Andersen, E.J.; Ali, S.; Byamukama, E.; Yen, Y.; Nepal, M.P. Disease resistance mechanisms in plants. Genes 2018, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Barragan, A.C.; Weigel, D. Plant NLR diversity: The known unknowns of pan-NLRomes. Plant Cell 2021, 33, 814–831. [Google Scholar] [CrossRef]
- Bonardi, V.; Dangl, J.L. How complex are intracellular immune receptor signaling complexes? Front. Plant Sci. 2012, 3, 237. [Google Scholar] [CrossRef]
- Danquah, A.; de Zelicourt, A.; Colcombet, J.; Hirt, H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 2014, 32, 40–52. [Google Scholar] [CrossRef]
- Ghozlan, M.H.; El-Argawy, E.; Tokgöz, S.; Lakshman, D.K.; Mitra, A. Plant defense against necrotrophic pathogens. Am. J. Plant Sci. 2020, 11, 2122–2138. [Google Scholar] [CrossRef]
- Aftab, T.; Roychoudhury, A. Crosstalk among Plant Growth Regulators and Signaling Molecules during Biotic and Abiotic Stresses: Molecular Responses and Signaling Pathways; Springer: Berlin/Heidelberg, Germany, 2021; pp. 2017–2019. [Google Scholar]
- Li, G.; Chen, T.; Zhang, Z.; Li, B.; Tian, S. Roles of aquaporins in plant-pathogen interaction. Plants 2020, 9, 1134. [Google Scholar] [CrossRef]
- Marshall, R.; Kombrink, A.; Motteram, J.; Loza-Reyes, E.; Lucas, J.; Hammond-Kosack, K.E.; Thomma, B.P.; Rudd, J.J. Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 2011, 156, 756–769. [Google Scholar] [CrossRef]
- Jwa, N.-S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 8, 1687. [Google Scholar] [CrossRef] [PubMed]
- Younessi-Hamzekhanlu, M.; Ozturk, M.; Jafarpour, P.; Mahna, N. Exploitation of next generation sequencing technologies for unraveling metabolic pathways in medicinal plants: A concise review. Ind. Crops Prod. 2022, 178, 114669. [Google Scholar] [CrossRef]
- Chang, S.; Mahon, E.L.; MacKay, H.A.; Rottmann, W.H.; Strauss, S.H.; Pijut, P.M.; Powell, W.A.; Coffey, V.; Lu, H.; Mansfield, S.D.; et al. Genetic engineering of trees: Progress and new horizons. Vitr. Cell. Dev. Biol. Plant 2018, 54, 341–376. [Google Scholar] [CrossRef]
- Sánchez, A.R.; Pineda, J.E.M.; Casas, X.M.; Calderón, J.H.M. Influence of edaphic salinity on -leaf morphoanatomical functional traits on juvenile and adult trees of red mangrove (Rhizophora mangle): Implications with Relation to Climate Change. Forests 2021, 12, 1586. [Google Scholar] [CrossRef]
- Ji, F.; Wei, W.; Liu, Y.; Wang, G.; Zhang, Q.; Xing, Y.; Zhang, S.; Liu, Z.; Cao, Q.; Qin, L. Construction of a SNP-based high-density genetic map using genotyping by sequencing (GBS) and QTL analysis of nut traits in Chinese chestnut (Castanea mollissima Blume). Front. Plant Sci. 2018, 9, 816. [Google Scholar] [CrossRef]
- Singh, B.D.; Singh, A.K. Marker-Assisted Plant Breeding: Principles and Practices; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Grattapaglia, D. Pseudo-testcross mapping strategy using RAPD markers. In Fingerprinting Methods Based on Arbitrarily Primed Pcr; Springer: Berlin/Heidelberg, Germany, 1997; pp. 201–218. [Google Scholar]
- Zhou, W.; Tang, Z.; Hou, J.; Hu, N.; Yin, T. Genetic map construction and detection of genetic loci underlying segregation distortion in an intraspecific cross of Populus deltoides. PLoS ONE 2015, 10, e0126077. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137. [Google Scholar] [CrossRef]
- Butler, J.B.; Freeman, J.S.; Vaillancourt, R.E.; Potts, B.M.; Glen, M.; Lee, D.J.; Pegg, G.S. Evidence for different QTL underlying the immune and hypersensitive responses of Eucalyptus globulus to the rust pathogen Puccinia psidii. Tree Genet. Genomes 2016, 12, 39. [Google Scholar] [CrossRef]
- Rosado, C.C.G.; Guimarães, L.M.D.S.; Faria, D.A.; de Resende, M.D.V.; Cruz, C.D.; Grattapaglia, D.; Alfenas, A.C. QTL mapping for resistance to Ceratocystis wilt in Eucalyptus. Tree Genet. Genomes 2016, 12, 72. [Google Scholar] [CrossRef]
- Hirao, T.; Matsunaga, K.; Shirasawa, K. Quantitative trait loci analysis based on high-density mapping of single-nucleotide polymorphisms by genotyping-by-sequencing against pine wilt disease in japanese black pine (Pinus thunbergii). Front. Plant Sci. 2022, 13, 850660. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J. JoinMap 4. Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma BV: Wageningen, The Netherlands, 2006; p. 33. [Google Scholar]
- Lander, E.S.; Green, P.; Abrahamson, J.; Barlow, A.; Daly, M.J.; Lincoln, S.E.; Newburg, L. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1987, 1, 174–181. [Google Scholar] [CrossRef]
- Van Ooijen, J. MapQTL® 6, Software for the Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2009; p. 64. [Google Scholar]
- Zarpelon, T.G.; Guimarães, L.M.D.S.; Faria, D.A.; Coutinho, M.M.; Neto, B.C.; Teixeira, R.U.; Grattapaglia, D.; Alfenas, A.C. Genetic mapping and validation of QTLs associated with resistance to Calonectria leaf blight caused by Calonectria pteridis in Eucalyptus. Tree Genet. Genomes 2015, 11, 803. [Google Scholar] [CrossRef]
- Zhebentyayeva, T.N.; Sisco, P.H.; Georgi, L.L.; Jeffers, S.N.; Perkins, M.T.; James, J.B.; Hebard, F.V.; Saski, C.; Nelson, C.D.; Abbott, A.G. Dissecting resistance to Phytophthora cinnamomi in interspecific hybrid chestnut crosses using sequence-based genotyping and QTL mapping. Phytopathology 2019, 109, 1594–1604. [Google Scholar] [CrossRef]
- Kushalappa, A.C.; Yogendra, K.N.; Karre, S. Plant innate immune response: Qualitative and quantitative resistance. Crit. Rev. Plant Sci. 2016, 35, 38–55. [Google Scholar] [CrossRef]
- Butler, J.B.; Potts, B.M.; Vaillancourt, R.E.; Lee, D.J.; Pegg, G.S.; Freeman, J.S. Independent QTL underlie resistance to the native pathogen Quambalaria pitereka and the exotic pathogen Austropuccinia psidii in Corymbia. Tree Genet. Genomes 2019, 15, 72. [Google Scholar] [CrossRef]
- Freeman, J.S.; Potts, B.M.; Vaillancourt, R.E. Few Mendelian genes underlie the quantitative response of a forest tree, Eucalyptus globulus, to a natural fungal epidemic. Genetics 2008, 178, 563–571. [Google Scholar] [CrossRef]
- Dorrance, A.E. Oomycete and fungal pathogens of soybean. In Achieving Sustainable Cultivation of Soybeans; Burleigh Dodds Science Publishing: Cambridge, UK, 2018; Volume 2, pp. 3–25. [Google Scholar]
- Du, Q.; Lu, W.; Quan, M.; Xiao, L.; Song, F.; Li, P.; Zhou, D.; Xie, J.; Wang, L.; Zhang, D. Genome-wide association studies to improve wood properties: Challenges and prospects. Front. Plant Sci. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.; Baenziger, P.S.; Börner, A. GWAS: Fast-forwarding gene identification and characterization in temperate cereals: Lessons from barley–a review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef]
- Ibrahim, A.K.; Zhang, L.; Niyitanga, S.; Afzal, M.Z.; Xu, Y.; Zhang, L.; Zhang, L.; Qi, J. Principles and approaches of association mapping in plant breeding. Trop. Plant Biol. 2020, 13, 212–224. [Google Scholar] [CrossRef]
- Zahid, G.; Kaçar, Y.A.; Dönmez, D.; Küden, A.; Giordani, T. Perspectives and recent progress of genome-wide association studies (GWAS) in fruits. Mol. Biol. Rep. 2022, 49, 5341–5352. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.T.L.; Ades, P.K.; Runa, F.A.; Bossinger, G.; Sandhu, K.S.; Potts, B.M.; Tibbits, J.F. Genome-wide association study of myrtle rust (Austropuccinia psidii) resistance in Eucalyptus obliqua (subgenus Eucalyptus). Tree Genet. Genomes 2021, 17, 31. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Youssef, H.M.; Eggert, K.; Koppolu, R.; Alqudah, A.M.; Poursarebani, N.; Fazeli, A.; Sakuma, S.; Tagiri, A.; Rutten, T.; Govind, G.; et al. VRS2 regulates hormone-mediated inflorescence patterning in barley. Nat. Genet. 2017, 49, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Charles, T.M.; Newton, R.J. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol. Biol. 2005, 59, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Muqaddasi, Q.H.; Lohwasser, U.; Nagel, M.; Börner, A.; Pillen, K.; Röder, M.S. Genome-wide association mapping of anther extrusion in hexaploid spring wheat. PLoS ONE 2016, 11, e0155494. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Vroh Bi, I.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Pais, A.L.; Whetten, R.W.; Xiang, Q.Y. Population structure, landscape genomics, and genetic signatures of adaptation to exotic disease pressure in Cornus florida L.—Insights from GWAS and GBS data. J. Syst. Evol. 2020, 58, 546–570. [Google Scholar] [CrossRef]
- Tan, B.; Ingvarsson, P.K. Integrating genome-wide association mapping of additive and dominance genetic effects to improve genomic prediction accuracy in Eucalyptus. Plant Genome 2022, 15, e20208. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Silva-Junior, O.B.; Resende, R.T.; Cappa, E.P.; Müller, B.; Tan, B.; Isik, F.; Ratcliffe, B.; El-Kassaby, Y.A. Quantitative genetics and genomics converge to accelerate forest tree breeding. Front. Plant Sci. 2018, 9, 1693. [Google Scholar] [CrossRef]
- Cappa, E.P.; El-Kassaby, Y.A.; Garcia, M.N.; Acuña, C.; Borralho, N.M.G.; Grattapaglia, D.; Poltri, S.N.M. Impacts of population structure and analytical models in genome-wide association studies of complex traits in forest trees: A case study in Eucalyptus globulus. PLoS ONE 2013, 8, e81267. [Google Scholar] [CrossRef] [PubMed]
- Mhoswa, L.; Myburg, A.A.; Slippers, B.; Külheim, C.; Naidoo, S. Genome-wide association study identifies SNP markers and putative candidate genes for terpene traits important for Leptocybe invasa resistance in Eucalyptus grandis. G3 Genes Genomes Genet. 2022, 12, jkac004. [Google Scholar] [CrossRef] [PubMed]
- Ćalić, I.; Koch, J.; Carey, D.; Addo-Quaye, C.; Carlson, J.E.; Neale, D.B. Genome-wide association study identifies a major gene for beech bark disease resistance in American beech (Fagus grandifolia Ehrh.). BMC Genom. 2017, 18, 547. [Google Scholar] [CrossRef]
- Chanroj, V.; Rattanawong, R.; Phumichai, T.; Tangphatsornruang, S.; Ukoskit, K. Genome-wide association mapping of latex yield and girth in Amazonian accessions of Hevea brasiliensis grown in a suboptimal climate zone. Genomics 2017, 109, 475–484. [Google Scholar] [CrossRef]
- Mukrimin, M.; Kovalchuk, A.; Neves, L.G.; Jaber, E.H.A.; Haapanen, M.; Kirst, M.; Asiegbu, F.O. Genome-wide exon-capture approach identifies genetic variants of Norway spruce genes associated with susceptibility to Heterobasidion parviporum infection. Front. Plant Sci. 2018, 9, 793. [Google Scholar] [CrossRef] [PubMed]
- Capador-Barreto, H.D.; Bernhardsson, C.; Milesi, P.; Vos, I.; Lundén, K.; Wu, H.X.; Karlsson, B.; Ingvarsson, P.K.; Stenlid, J.; Elfstrand, M. Killing two enemies with one stone? Genomics of resistance to two sympatric pathogens in Norway spruce. Mol. Ecol. 2021, 30, 4433–4447. [Google Scholar] [CrossRef]
- Elfstrand, M.; Baison, J.; Lundén, K.; Zhou, L.; Vos, I.; Capador, H.D.; Åslund, M.S.; Chen, Z.; Chaudhary, R.; Olson, Å.; et al. Association genetics identifies a specifically regulated Norway spruce laccase gene, PaLAC5, linked to Heterobasidion parviporum resistance. Plant Cell Environ. 2020, 43, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Guo, Y.; Chen, Y.; Zhang, H.; El-Kassaby, Y.A.; Li, W. Genome wide association study identifies candidate genes related to the earlywood tracheid properties in Picea crassifolia Kom. Forests 2022, 13, 332. [Google Scholar] [CrossRef]
- Parchman, T.L.; Gompert, Z.; Mudge, J.; Schilkey, F.D.; Benkman, C.W.; Buerkle, C.A. Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol. Ecol. 2012, 21, 2991–3005. [Google Scholar] [CrossRef] [PubMed]
- González-Martínez, S.C.; Wheeler, N.C.; Ersoz, E.; Nelson, C.D.; Neale, D.B. Association genetics in Pinus taeda LI Wood property traits. Genetics 2007, 175, 399–409. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, S.C.; Huber, D.; Ersoz, E.; Davis, J.M.; Neale, D.B. Association genetics in Pinus taeda L. II. Carbon isotope discrimination. Heredity 2008, 101, 19–26. [Google Scholar] [CrossRef]
- Quesada, T.; Gopal, V.; Cumbie, W.P.; Eckert, A.J.; Wegrzyn, J.L.; Neale, D.B.; Goldfarb, B.; Huber, D.A.; Casella, G.; Davis, J.M. Association mapping of quantitative disease resistance in a natural population of loblolly pine (Pinus taeda L.). Genetics 2010, 186, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Porth, I.; Klapšte, J.; Skyba, O.; Hannemann, J.; McKown, A.D.; Guy, R.D.; DiFazio, S.P.; Muchero, W.; Ranjan, P.; Tuskan, G.A.; et al. Genome-wide association mapping for wood characteristics in Populus identifies an array of candidate single nucleotide polymorphisms. New Phytol. 2013, 200, 710–726. [Google Scholar] [CrossRef] [PubMed]
- Muchero, W.; Sondreli, K.L.; Chen, J.-G.; LeBoldus, J.M. Association mapping, transcriptomics, and transient expression identify candidate genes mediating plant–pathogen interactions in a tree. Proc. Natl. Acad. Sci. USA 2018, 115, 11573–11578. [Google Scholar] [CrossRef] [PubMed]
- McKown, A.D.; Guy, R.D.; Quamme, L.; Klápště, J.; La Mantia, J.; Constabel, C.P.; El-Kassaby, Y.A.; Hamelin, R.C.; Zifkin, M.; Azam, M.S. Association genetics, geography and ecophysiology link stomatal patterning in Populus trichocarpa with carbon gain and disease resistance trade-offs. Mol. Ecol. 2014, 23, 5771–5790. [Google Scholar] [CrossRef] [PubMed]
- Resende, R.T.; de Resende, M.D.V.; Azevedo, C.F.; e Silva, F.F.; Melo, L.C.; Pereira, H.S.; Souza, T.L.P.O.; Valdisser, P.A.M.R.; Brondani, C.; Vianello, R.P. Genome-wide association and regional heritability mapping of plant architecture, lodging and productivity in Phaseolus vulgaris. G3 Genes Genomes Genet. 2018, 8, 2841–2854. [Google Scholar]
- Resende, R.T.; Resende, M.D.V.; Silva, F.F.; Azevedo, C.F.; Takahashi, E.K.; Silva-Junior, O.B.; Grattapaglia, D. Regional heritability mapping and genome-wide association identify loci for complex growth, wood and disease resistance traits in Eucalyptus. New Phytol. 2017, 213, 1287–1300. [Google Scholar] [CrossRef]
- Junghans, D.T.; Alfenas, A.C.; Brommonschenkel, S.H.; Oda, S.; Mello, E.J.; Grattapaglia, D. Resistance to rust (Puccinia psidii Winter) in Eucalyptus: Mode of inheritance and mapping of a major gene with RAPD markers. Theor. Appl. Genet. 2003, 108, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Mamani, E.M.C.; Bueno, N.W.; Faria, D.A.; Guimarães, L.M.S.; Lau, D.; Alfenas, A.C.; Grattapaglia, D. Positioning of the major locus for Puccinia psidii rust resistance (Ppr1) on the Eucalyptus reference map and its validation across unrelated pedigrees. Tree Genet. Genomes 2010, 6, 953–962. [Google Scholar] [CrossRef]
- Alves, A.A.; Rosado, C.C.G.; Faria, D.A.; da Silva Guimarães, L.M.; Lau, D.; Brommomschenkel, S.H.; Grattapaglia, D.; Couto, A. Genetic mapping provides evidence for the role of additive and non-additive QTLs in the response of inter-specific hybrids of Eucalyptus to Puccinia psidii rust infection. Euphytica 2012, 183, 27–38. [Google Scholar] [CrossRef]
- Christie, N.; Tobias, P.A.; Naidoo, S.; Kulheim, C. The Eucalyptus grandis NBS-LRR gene family: Physical clustering and expression hotspots. Front. Plant Sci. 2016, 6, 1238. [Google Scholar] [CrossRef]
- Barbosa-Da-Silva, A.; Wanderley-Nogueira, A.C.; Silva, R.R.; Berlarmino, L.C.; Soares-Cavalcanti, N.M.; Benko-Iseppon, A.M. In silico survey of resistance (R) genes in Eucalyptus transcriptome. Genet. Mol. Biol. 2005, 28, 562–574. [Google Scholar] [CrossRef]
- Santos, S.A.; Vidigal, P.M.P.; Guimarães, L.M.S.; Mafia, R.G.; Templeton, M.D.; Alfenas, A.C. Transcriptome analysis of Eucalyptus grandis genotypes reveals constitutive overexpression of genes related to rust (Austropuccinia psidii) resistance. Plant Mol. Biol. 2020, 104, 339–357. [Google Scholar] [CrossRef]
- Rosado, T.B.; Tomaz, R.S.; Junior, M.F.R.; Rosado, A.M.; Guimarães, L.M.D.S.; de Araújo, E.F.; Alfenas, A.C.; Cruz, C.D. Detection of QTL associated with rust resistance using IBD-based methodologies in exogamic Eucalyptus spp. populations. Crop Breed. Appl. Biotechnol. 2010, 10, 321–328. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Resende, M.D. Genomic selection in forest tree breeding. Tree Genet. Genomes 2011, 7, 241–255. [Google Scholar] [CrossRef]
- Ahmar, S.; Ballesta, P.; Ali, M.; Mora-Poblete, F. Achievements and challenges of genomics-assisted breeding in forest trees: From marker-assisted selection to genome editing. Int. J. Mol. Sci. 2021, 22, 10583. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef]
- O’Connor, K.M.; Hayes, B.J.; Hardner, C.M.; Alam, M.; Henry, R.J.; Topp, B.L. Genomic selection and genetic gain for nut yield in an Australian Macadamia breeding population. BMC Genom. 2021, 22, 370. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.S.F.; Neves, L.G.; Filho, J.E.D.A.; ResendeJr, M.F.R.; Muñoz, P.R.; dos Santos, P.E.T.; Filho, E.P.; Kirst, M.; Grattapaglia, D. Genomic prediction in contrast to a genome-wide association study in explaining heritable variation of complex growth traits in breeding populations of Eucalyptus. BMC Genom. 2017, 18, 524. [Google Scholar] [CrossRef]
- Beaulieu, J.; Doerksen, T.; Clément, S.; MacKay, J.; Bousquet, J. Accuracy of genomic selection models in a large population of open-pollinated families in white spruce. Heredity 2014, 113, 343–352. [Google Scholar] [CrossRef]
- Isik, F.; Bartholomé, J.; Farjat, A.; Chancerel, E.; Raffin, A.; Sanchez, L.; Plomion, C.; Bouffier, L. Genomic selection in maritime pine. Plant Sci. 2016, 242, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Krutovsky, K.V.; Nelson, C.D.; Koralewski, T.E.; Byram, T.D.; Loopstra, C.A. Exome genotyping, linkage disequilibrium and population structure in loblolly pine (Pinus taeda L.). BMC Genom. 2016, 17, 730. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Grattapaglia, D.; Martins, G.S.; Ferreira, K.Z.; Sundberg, B.; Ingvarsson, P.K. Evaluating the accuracy of genomic prediction of growth and wood traits in two Eucalyptus species and their F1 hybrids. BMC Plant Biol. 2017, 17, 110. [Google Scholar] [CrossRef]
- Lebedev, V.G.; Lebedeva, T.N.; Chernodubov, A.I.; Shestibratov, K.A. Genomic selection for forest tree improvement: Methods, achievements and perspectives. Forests 2020, 11, 1190. [Google Scholar] [CrossRef]
- Resende, M.F.R., Jr.; Muñoz, P.; Resende, M.D.V.; Garrick, D.J.; Fernando, R.L.; Davis, J.M.; Jokela, E.J.; Martin, T.A.; Peter, G.F.; Kirst, M. Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 2012, 190, 1503–1510. [Google Scholar] [CrossRef]
- Shalizi, M.N.; Cumbie, W.P.; Isik, F. Genomic prediction for fusiform rust disease incidence in a large cloned population of Pinus taeda. G3 Genes Genomes Genet. 2021, 11, jkab235. [Google Scholar] [CrossRef]
- De Almeida Filho, J.E.; Guimarães, J.F.R.; e Silva, F.F.; de Resende, M.D.V.; Muñoz, P.; Kirst, M. The contribution of dominance to phenotype prediction in a pine breeding and simulated population. Heredity 2016, 117, 33–41. [Google Scholar] [CrossRef]
- Toro, M.A.; Varona, L. A note on mate allocation for dominance handling in genomic selection. Genet. Sel. Evol. 2010, 42, 33. [Google Scholar] [CrossRef]
- Stocks, J.J.; Metheringham, C.L.; Plumb, W.J.; Lee, S.J.; Kelly, L.J.; Nichols, R.A.; Buggs, R.J.A. Genomic basis of European ash tree resistance to ash dieback fungus. Nat. Ecol. Evol. 2019, 3, 1686–1696. [Google Scholar] [CrossRef]
- Anagnostakis, S.L. Chestnut breeding in the United States for disease and insect resistance. Plant Dis. 2012, 96, 1392–1403. [Google Scholar] [CrossRef]
- Westbrook, J.W.; Zhang, Q.; Mandal, M.K.; Jenkins, E.V.; Barth, L.E.; Jenkins, J.W.; Grimwood, J.; Schmutz, J.; Holliday, J.A. Optimizing genomic selection for blight resistance in American chestnut backcross populations: A trade-off with American chestnut ancestry implies resistance is polygenic. Evol. Appl. 2020, 13, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Isik, F. Genomic Prediction of Complex Traits in Perennial Plants: A Case for Forest Trees. In Complex Trait Prediction; Springer: Berlin/Heidelberg, Germany, 2022; pp. 493–520. [Google Scholar]
- McElroy, M.S.; Navarro, A.J.R.; Mustiga, G.; Stack, C.; Gezan, S.; Peña, G.; Sarabia, W.; Saquicela, D.; Sotomayor, I.; Douglas, G.M.; et al. Prediction of cacao (Theobroma cacao) resistance to Moniliophthora spp. diseases via genome-wide association analysis and genomic selection. Front. Plant Sci. 2018, 9, 343. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Younessi-Hamzekhanlu, M.; Gailing, O. Genome-Wide SNP Markers Accelerate Perennial Forest Tree Breeding Rate for Disease Resistance through Marker-Assisted and Genome-Wide Selection. Int. J. Mol. Sci. 2022, 23, 12315. https://doi.org/10.3390/ijms232012315
Younessi-Hamzekhanlu M, Gailing O. Genome-Wide SNP Markers Accelerate Perennial Forest Tree Breeding Rate for Disease Resistance through Marker-Assisted and Genome-Wide Selection. International Journal of Molecular Sciences. 2022; 23(20):12315. https://doi.org/10.3390/ijms232012315
Chicago/Turabian StyleYounessi-Hamzekhanlu, Mehdi, and Oliver Gailing. 2022. "Genome-Wide SNP Markers Accelerate Perennial Forest Tree Breeding Rate for Disease Resistance through Marker-Assisted and Genome-Wide Selection" International Journal of Molecular Sciences 23, no. 20: 12315. https://doi.org/10.3390/ijms232012315
APA StyleYounessi-Hamzekhanlu, M., & Gailing, O. (2022). Genome-Wide SNP Markers Accelerate Perennial Forest Tree Breeding Rate for Disease Resistance through Marker-Assisted and Genome-Wide Selection. International Journal of Molecular Sciences, 23(20), 12315. https://doi.org/10.3390/ijms232012315







