Application of the Mutant Libraries for Candida albicans Functional Genomics
Abstract
1. Introduction
2. Methods Used in the Mutant Library Construction in C. albicans
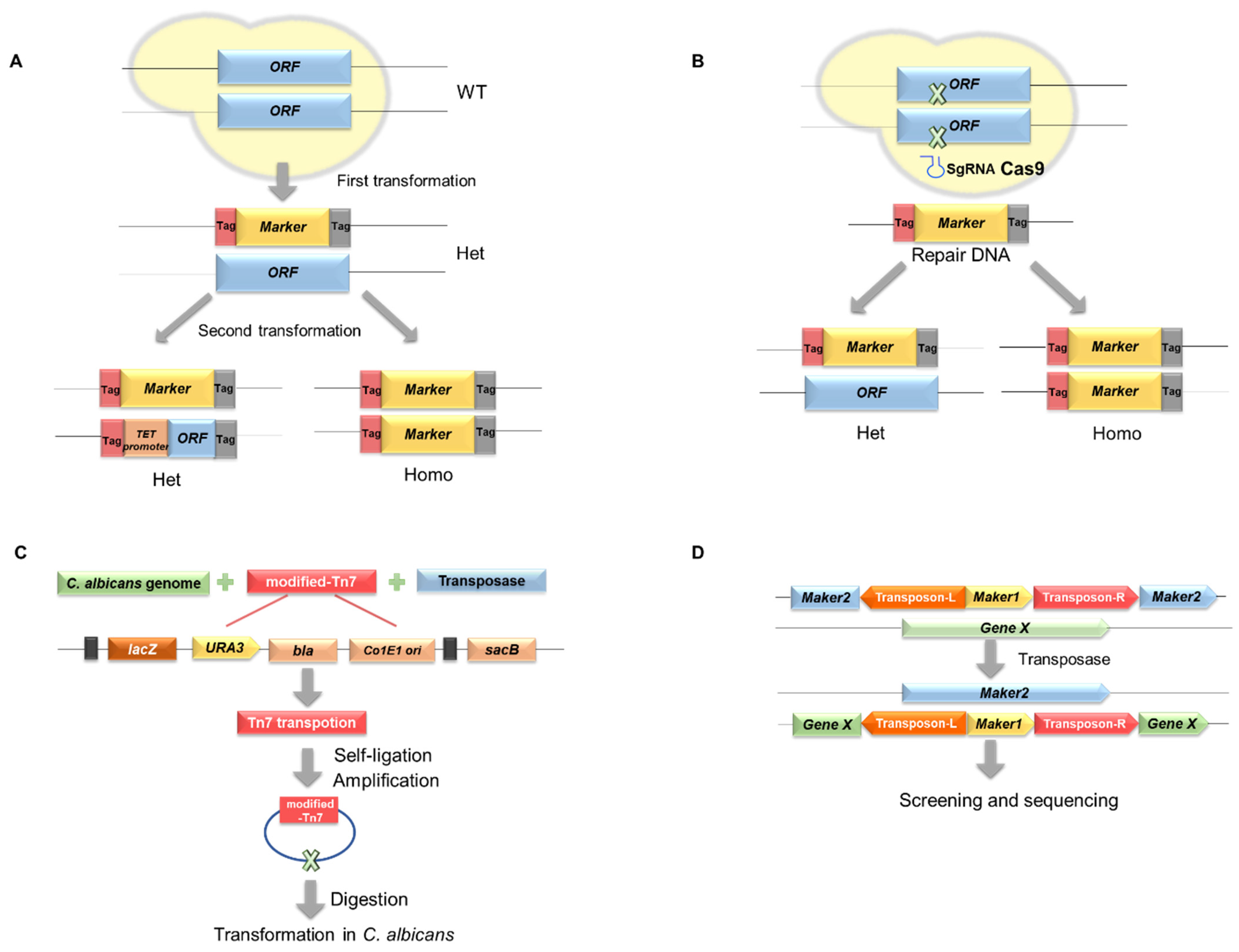
2.1. Gene Disruption
2.2. Transposon Insertion
2.3. Conditional Expression
3. Application of the C. albicans Mutant Library
3.1. Morphological Transformation
3.2. Biofilm Formation
3.3. Fungus–Host Interaction
3.4. Antifungal Targets Identification
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bongomin, F.; Gago, S.; Oladele, R.O.; Denning, D.W. Global and Multi-National Prevalence of Fungal Diseases-Estimate Precision. J. Fungi 2017, 3, 57. [Google Scholar] [CrossRef]
- Mota Fernandes, C.; Dasilva, D.; Haranahalli, K.; McCarthy, J.B.; Mallamo, J.; Ojima, I.; Del Poeta, M. The Future of Antifungal Drug Therapy: Novel Compounds and Targets. Antimicrob. Agents Chemother. 2021, 65, e01719-20. [Google Scholar] [CrossRef]
- Whiteway, M.; Bachewich, C. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 2007, 61, 529–553. [Google Scholar] [CrossRef]
- Ramirez-Zavala, B.; Reuss, O.; Park, Y.N.; Ohlsen, K.; Morschhauser, J. Environmental induction of white-opaque switching in Candida albicans. PLoS Pathog. 2008, 4, e1000089. [Google Scholar] [CrossRef]
- Ramage, G.; Saville, S.P.; Thomas, D.P.; López-Ribot, J.L. Candida biofilms: An update. Eukaryot. Cell 2005, 4, 633–638. [Google Scholar] [CrossRef]
- Homann, O.R.; Dea, J.; Noble, S.M.; Johnson, A.D. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009, 5, e1000783. [Google Scholar] [CrossRef]
- Noble, S.M.; French, S.; Kohn, L.A.; Chen, V.; Johnson, A.D. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010, 42, 590–598. [Google Scholar] [CrossRef]
- Shapiro, R.S.; Chavez, A.; Porter, C.B.M.; Hamblin, M.; Kaas, C.S.; DiCarlo, J.E.; Collins, J.J. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nat. Microbiol. 2018, 3, 73–82. [Google Scholar] [CrossRef]
- Xu, D.; Jiang, B.; Ketela, T.; Lemieux, S.; Veillette, K.; Martel, N.; Roemer, T. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 2007, 3, e92. [Google Scholar] [CrossRef]
- Bharucha, N.; Chabrier-Rosello, Y.; Xu, T.; Johnson, C.; Sobczynski, S.; Song, Q.; Krysan, D.J. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: Analysis of the RAM network during morphogenesis. PLoS Genet. 2011, 7, e1002058. [Google Scholar] [CrossRef]
- Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Mitchell, A.P. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010, 6, e1000752. [Google Scholar] [CrossRef] [PubMed]
- Davis, D.A.; Bruno, V.M.; Loza, L.; Filler, S.G.; Mitchell, A.P. Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 2002, 162, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Epp, E.; Walther, A.; Lépine, G.; Leon, Z.; Mullick, A.; Raymond, M.; Whiteway, M. Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol. Microbiol. 2010, 75, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, H.; Li, Z.; Wong, A.H.H.; Wang, Y.Z.; Guo, Y.; Wang, J. Candida albicans gains azole resistance by altering sphingolipid composition. Nat. Commun. 2018, 9, 4495. [Google Scholar] [CrossRef]
- Mielich, K.; Shtifman-Segal, E.; Golz, J.C.; Zeng, G.; Wang, Y.; Berman, J.; Kunze, R. Maize Transposable Elements Ac/Ds as Insertion Mutagenesis Tools in Candida albicans. G3 2018, 8, 1139–1145. [Google Scholar] [CrossRef]
- Nobile, C.J.; Mitchell, A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005, 15, 1150–1155. [Google Scholar] [CrossRef]
- Norice, C.T.; Smith, F.J., Jr.; Solis, N.; Filler, S.G.; Mitchell, A.P. Requirement for Candida albicans Sun41 in biofilm formation and virulence. Eukaryot. Cell 2007, 6, 2046–2055. [Google Scholar] [CrossRef]
- Oh, J.; Fung, E.; Schlecht, U.; Davis, R.W.; Giaever, G.; St Onge, R.P.; Nislow, C. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog. 2010, 6, e1001140. [Google Scholar] [CrossRef]
- Uhl, M.A.; Biery, M.; Craig, N.; Johnson, A.D. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C.albicans. EMBO J. 2003, 22, 2668–2678. [Google Scholar] [CrossRef]
- Chauvel, M.; Nesseir, A.; Cabral, V.; Znaidi, S.; Goyard, S.; Bachellier-Bassi, S.; d’Enfert, C. A versatile overexpression strategy in the pathogenic yeast Candida albicans: Identification of regulators of morphogenesis and fitness. PLoS ONE 2012, 7, e45912. [Google Scholar] [CrossRef]
- O’Meara, T.R.; Veri, A.O.; Ketela, T.; Jiang, B.; Roemer, T.; Cowen, L.E. Global analysis of fungal morphology exposes mechanisms of host cell escape. Nat. Commun. 2015, 6, 6741. [Google Scholar] [CrossRef] [PubMed]
- Roemer, T.; Jiang, B.; Davison, J.; Ketela, T.; Veillette, K.; Breton, A.; Bussey, H. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003, 50, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Sahni, N.; Yi, S.; Daniels, K.J.; Huang, G.; Srikantha, T.; Soll, D.R. Tec1 mediates the pheromone response of the white phenotype of Candida albicans: Insights into the evolution of new signal transduction pathways. PLoS Biol. 2010, 8, e1000363. [Google Scholar] [CrossRef] [PubMed]
- Schillig, R.; Morschhauser, J. Analysis of a fungus-specific transcription factor family, the Candida albicans zinc cluster proteins, by artificial activation. Mol. Microbiol. 2013, 89, 1003–1017. [Google Scholar] [CrossRef] [PubMed]
- Znaidi, S.; van Wijlick, L.; Hernandez-Cervantes, A.; Sertour, N.; Desseyn, J.L.; Vincent, F.; d’Enfert, C. Systematic gene overexpression in Candida albicans identifies a regulator of early adaptation to the mammalian gut. Cell Microbiol. 2018, 20, e12890. [Google Scholar] [CrossRef]
- Chen, Y.; Mallick, J.; Maqnas, A.; Sun, Y.; Choudhury, B.I.; Côte PWhiteway, M. Chemogenomic Profiling of the Fungal Pathogen Candida albicans. Antimicrob. Agents Chemother. 2018, 62, e02365-17. [Google Scholar] [CrossRef]
- Fu, C.; Zhang, X.; Veri, A.O.; Iyer, K.R.; Lash, E.; Xue, A.; Cowen, L.E. Leveraging machine learning essentiality predictions and chemogenomic interactions to identify antifungal targets. Nat. Commun. 2021, 12, 6497. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Guo, W.; Köhler, J.R. CaNAT1, a heterologous dominant selectable marker for transformation of Candida albicans and other pathogenic Candida species. Infect. Immun. 2005, 73, 1239–1242. [Google Scholar] [CrossRef]
- Min, K.; Ichikawa, Y.; Woolford, C.A.; Mitchell, A.P. Candida albicans Gene Deletion with a Transient CRISPR-Cas9 System. mSphere 2016, 1, e00130-e16. [Google Scholar] [CrossRef]
- Vyas, V.K.; Barrasa, M.I.; Fink, G.R. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 2015, 1, e1500248. [Google Scholar] [CrossRef]
- Wensing, L.; Sharma, J.; Uthayakumar, D.; Proteau, Y.; Chavez, A.; Shapiro, R.S. A CRISPR Interference Platform for Efficient Genetic Repression in Candida albicans. mSphere 2019, 4, e00002-19. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhu, S.; Cai, C.; Yuan, P.; Li, C.; Huang, Y.; Wei, W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 2014, 509, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.B.; Wang, M.; Zhang, W.B.; Xiao, T.; Chen, C.H.; Wu, A.; Liu, X.S. Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat Protoc. 2019, 14, 756–780. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Kim, K.S. RELATe enables genome-scale engineering in fungal genomics. Sci. Adv. 2020, 6, eabb8783. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Davis, R.W. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef]
- Mitra, R.; Fain-Thornton, J.; Craig, N.L. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008, 27, 1097–1109. [Google Scholar] [CrossRef]
- Elick, T.A.; Bauser, C.A.; Fraser, M.J. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica 1996, 98, 33–41. [Google Scholar] [CrossRef]
- Segal, E.S.; Gritsenko, V.; Levitan, A.; Yadav, B.; Dror, N.; Steenwyk, J.L.; Berman, J. Gene Essentiality Analyzed by In Vivo Transposon Mutagenesis and Machine Learning in a Stable Haploid Isolate of Candida albicans. mBio 2018, 9, e02048-18. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, G.; Spellberg, B.J.; Edwards, J.E., Jr.; Ibrahim, A.S. Gene overexpression/suppression analysis of candidate virulence factors of Candida albicans. Eukaryot Cell 2008, 7, 483–492. [Google Scholar] [CrossRef]
- Flanagan, P.R.; Liu, N.N.; Fitzpatrick, D.J.; Hokamp, K.; Kohler, J.R.; Moran, G.P. The Candida albicans TOR-Activating GTPases Gtr1 and Rhb1 Coregulate Starvation Responses and Biofilm Formation. mSphere 2017, 2, e00477-17. [Google Scholar] [CrossRef]
- Basso, L.R., Jr.; Gast, C.E.; Mao, Y.; Wong, B. Fluconazole transport into Candida albicans secretory vesicles by the membrane proteins Cdr1p, Cdr2p, and Mdr1p. Eukaryot Cell 2010, 9, 960–970. [Google Scholar] [CrossRef] [PubMed]
- Hickman, M.A.; Zeng, G.S.; Forche, A.; Hirakawa, M.P.; Abbey, D.; Harrison, B.D.; Berman, J. The ‘obligate diploid’ Candida albicans forms mating-competent haploids. Nature 2013, 494, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Huang, G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence 2012, 3, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Veri, A.O.; Liu, Z.; Iyer, K.R.; O’Meara, T.R.; Robbins, N.; Cowen, L.E. Mitochondrial perturbation reduces susceptibility to xenobiotics through altered efflux in Candida albicans. Genetics 2021, 219, iyab095. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; Zeng, G.; Chan, F.Y.; Wang, Y.M.; Yang, D.; Wang, Y. Sac7 and Rho1 regulate the white-to-opaque switching in Candida albicans. Sci. Rep. 2018, 8, 875. [Google Scholar] [CrossRef] [PubMed]
- Desai, J.V.; Mitchell, A.P. Candida albicans Biofilm Development and Its Genetic Control. Microbiol. Spectr. 2015, 3, 99–114. [Google Scholar] [CrossRef]
- Nobile, C.J.; Fox, E.P.; Nett, J.E.; Sorrells, T.R.; Mitrovich, Q.M.; Hernday, A.D.; Johnson, A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 2012, 148, 126–138. [Google Scholar] [CrossRef]
- Finkel, J.S.; Xu, W.; Huang, D.; Hill, E.M.; Desai, J.V.; Woolford, C.A.; Mitchell, A.P. Portrait of Candida albicans adherence regulators. PLoS Pathog. 2012, 8, e1002525. [Google Scholar] [CrossRef]
- Lee, J.A.; Robbins, N.; Xie, J.L.; Ketela, T.; Cowen, L.E. Functional Genomic Analysis of Candida albicans Adherence Reveals a Key Role for the Arp2/3 Complex in Cell Wall Remodelling and Biofilm Formation. PLoS Genet. 2016, 12, e1006452. [Google Scholar] [CrossRef]
- Costa, A.; Omran, R.P.; Correia-Mesquita, T.O.; Dumeaux, V.; Whiteway, M. Screening of Candida albicans GRACE library revealed a unique pattern of biofilm formation under repression of the essential gene ILS1. Sci. Rep. 2019, 9, 9187. [Google Scholar] [CrossRef]
- Rosiana, S.; Zhang, L.; Kim, G.H.; Revtovich, A.V.; Uthayakumar, D.; Sukumaran, A.; Shapiro, R.S. Comprehensive genetic analysis of adhesin proteins and their role in virulence of Candida albicans. Genetics 2021, 217, iyab003. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.J.; Oh, S.; Underhill, D.M. Host-microbe interactions: Commensal fungi in the gut. Curr. Opin. Microbiol. 2017, 40, 131–137. [Google Scholar] [CrossRef] [PubMed]
- Wellington, M.; Koselny, K.; Krysan, D.J. Candida albicans morphogenesis is not required for macrophage interleukin 1beta production. mBio 2012, 4, e00433-00412. [Google Scholar]
- O’Meara, T.R.; Duah, K.; Guo, C.X.; Maxson, M.E.; Gaudet, R.G.; Koselny, K.; Cowen, L.E. High-Throughput Screening Identifies Genes Required for Candida albicans Induction of Macrophage Pyroptosis. mBio 2018, 9, e01581-18. [Google Scholar] [CrossRef]
- Witchley, J.N.; Penumetcha, P.; Abon, N.V.; Woolford, C.A.; Mitchell, A.P.; Noble, S.M. Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection. Cell Host Microbe 2019, 25, 432–443.e436. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.P.; White, C.M.; Pappas, P.G. Candidemia and Invasive Candidiasis. Infectious Dis. Clin. N. Am. 2021, 35, 389–413. [Google Scholar] [CrossRef]
- Rauceo, J.M.; Blankenship, J.R.; Fanning, S.; Hamaker, J.J.; Deneault, J.S.; Smith, F.J.; Mitchell, A.P. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Mol. Biol. Cell 2008, 19, 2741–2751. [Google Scholar] [CrossRef]
- Richard, M.L.; Nobile, C.J.; Bruno, V.M.; Mitchell, A.P. Candida albicans biofilm-defective mutants. Eukaryot Cell 2005, 4, 493–1502. [Google Scholar] [CrossRef]
- Zeng, G.; Xu, X.; Gao, J.; da Silva Dantas, A.; Gow, N.A.R.; Wang, Y. Inactivating the mannose-ethanolamine phosphotransferase Gpi7 confers caspofungin resistance in the human fungal pathogen Candida albicans. Cell Surf. 2021, 7, 100057. [Google Scholar] [CrossRef]
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Bugni, T.S. A marine microbiome antifungal targets urgent-threat drug-resistant fungi. Science 2020, 370, 974–978. [Google Scholar] [CrossRef]
- Gervais, N.C.; Halder, V.; Shapiro, R.S. A data library of Candida albicans functional genomic screens. FEMS Yeast Res. 2021, 21, foab060. [Google Scholar] [CrossRef] [PubMed]

| Type of Mutation | Libraries and References | Methods | Parental Strains | Mutant Strains Generated | Genes Affected | Numbers of Alleles Affected | Application of Library |
|---|---|---|---|---|---|---|---|
| Gene disruption | Xu et al., 2007 [9] | gene knockout (HIS1 and LEU3 marker) | unreported | 2868 | 69 | Monoallelic | Identification of the mechanism of action of novel antifungal agents |
| Homann et al., 2009 [6] | gene knockout (HIS1, LEU2, and ARG4 marker) | SN152 | 365 | 143 | Biallelic | Identification of regulators for drug resistance, morphological variation, and iron acquisition | |
| Noble et al., 2010 [7] | gene knockout (HIS1 and LEU2 marker) | SN152 | ~3000 | 115 | Biallelic | Morphogenetic switching and pathogenicity | |
| Shapiro et al., 2018 [8] | CRISPR–Cas9 | GZY803, GZY892 | 250 | 250 | Biallelic | Targeting antifungal efflux and biofilm adhesion factors | |
| Transposon insertion | Davis et al., 2002 [12] | Tn7-UAU1 | CAI4 | 353 | 3 | Biallelic | Identification of pH-response regulators |
| Uhl et al., 2003 [19] | Tn7-URA3 | CAI4 | 18000 | 146 | Monoallelic | Analysis of genes for filamentous growth | |
| Nobile and Mitchell et al., 2005 [16] | Tn7-UAU1 | DAY185 | 83 | 2 | Biallelic | Regulation of cell-surface genes and biofilm formation | |
| Norice et al., 2007 [17] | Tn7-UAU1 | CAI4 | 25 | 21 | Biallelic | Validation of essential genes for biofilm formation and virulence | |
| Oh et al., 2010 [18] | Tn5-UAU1 | BWP17 | 3633 | 269 | Monoallelic | Identification of gene annotation and drug target | |
| Blankenship et al., 2010 [11] | Tn7-UAU1 | BWP17 | 108 | 80 | Biallelic | Analysis of protein kinase signaling for cell wall regulation | |
| Epp E et al., 2010 [13] | Tn7-UAU1 | SN95 | 4700 | 20 | Biallelic | Identification of genes for morphological transformation | |
| Bharucha N et al., 2011 [10] | Tn7-URA3 | cbk1Δ/CBK1 | 6528 | 441 | Monoallelic | Analysis of the RAM Network during Morphogenesis | |
| Gao J. et al., (2018) [14] | PB-URA3 | GZY892 | 4791 | 4791 | Monoallelic | Identification of regulators of the white-to-opaque switching; identification of mutants resistant to caspofungin | |
| Mielich K et al., 2018 [15] | AS/Ds-NAT1 | GZY896 | 1610 | 1195 | Monoallelic | Analysis of C. albicans essential genes | |
| Conditional expression | Roemer et al., 2003 [22] | GRACE | CaSS1 | 1152 | 567 | Biallelic | Identification and prioritization of essential genes as antifungal drug targets |
| Sahni et al., 2010 [23] | Ptet induced overexpression | P37005 | 107 | 107 | Monoallelic | Analysis of the evolution of new signal transduction pathways | |
| Chauvel et al., 2012 [20]; Znaidi et al., 2018 [25] | Ptet and PPCK1 induced overexpression | BWP17 | 956 | 572 | Monoallelic | Identification of regulators of morphogenesis and fitness; colonization of the mammalian GI tract; cell-wall function | |
| Schillig, R; Morschhäuser, J; et al., 2013 [24] | PGAL4 induced overexpression | SC5314 | 82 | 82 | Monoallelic | Identification of regulators of invasive filamentous growth and mediators of fluconazole resistance | |
| O’Meara et al., 2015 [21] | GRACE | unreported | 2356 | 974 | Monoallelic | Global analysis of morphogenesis; macrophage pyroptosis | |
| Chen Y et al., 2018 [26] | GRACE | CaSS1 | 887 | unreported | Biallelic | Screening GRACE collection of strains against classic antifungal drugs. | |
| Fu C et al., 2021 [27] | GRACE | CaLC6106 | 866 | 149 | Monoallelic | Identification of fungal-specific essential genes and an antifungal compound that targets C. albicans glutaminyl-tRNA synthetase. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Liu, J.; Liu, Y.; Lv, Q.; Yan, L. Application of the Mutant Libraries for Candida albicans Functional Genomics. Int. J. Mol. Sci. 2022, 23, 12307. https://doi.org/10.3390/ijms232012307
Wang R, Liu J, Liu Y, Lv Q, Yan L. Application of the Mutant Libraries for Candida albicans Functional Genomics. International Journal of Molecular Sciences. 2022; 23(20):12307. https://doi.org/10.3390/ijms232012307
Chicago/Turabian StyleWang, Ruina, Jiacun Liu, Yu Liu, Quanzhen Lv, and Lan Yan. 2022. "Application of the Mutant Libraries for Candida albicans Functional Genomics" International Journal of Molecular Sciences 23, no. 20: 12307. https://doi.org/10.3390/ijms232012307
APA StyleWang, R., Liu, J., Liu, Y., Lv, Q., & Yan, L. (2022). Application of the Mutant Libraries for Candida albicans Functional Genomics. International Journal of Molecular Sciences, 23(20), 12307. https://doi.org/10.3390/ijms232012307






