Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis
Abstract
1. Introduction
2. Polycomb Protein Complexes
3. Recruitment of Polycomb Complexes to Specific Sites in the Genome
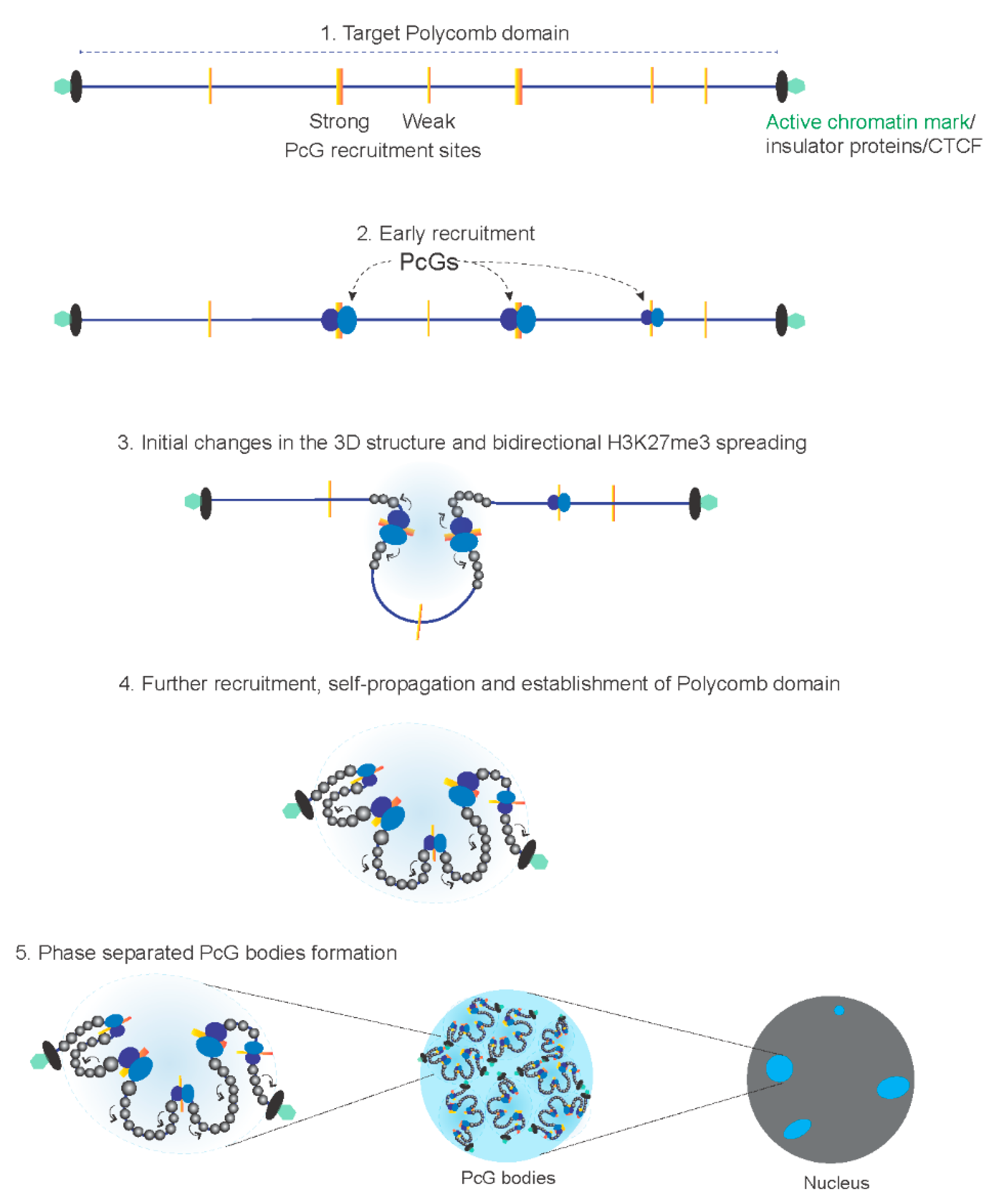
4. Mechanism of Polycomb Domain Establishment and Target Gene Repression
5. Role of Histones in Pathogenic Infection
6. Role of PcGs in Pathogenic Infection
6.1. HIV Latency and Polycomb
6.2. HPV and Polycomb
6.3. Herpes Virus and Polycomb
6.4. Other Pathogens and Polycomb Regulation
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Slifer, E.H. A mutant stock of Drosophila with extra sex combs. J. Exp. Zool. 1942, 90, 31–40. [Google Scholar] [CrossRef]
- Lewis, P.H.; Mislove, R.F. New mutants report. Drosoph. Inf. Serv. 1947, 21. [Google Scholar]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Shilatifard, A. The roles of Polycomb repressive complexes in mammalian development and cancer. Nat. Rev. Mol. Cell Biol. 2021, 22, 326–345. [Google Scholar] [CrossRef] [PubMed]
- Parreno, V.; Martinez, A.M.; Cavalli, G. Mechanisms of Polycomb group protein function in cancer. Cell Res. 2022, 32, 231–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, J.J.; Voruganti, S.; Nag, S.; Zhou, J.; Zhang, R. Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med. Res. Rev. 2015, 35, 1220–1267. [Google Scholar] [CrossRef]
- Chan, H.L.; Morey, L. Emerging Roles for Polycomb-Group Proteins in Stem Cells and Cancer. Trends Biochem. Sci. 2019, 44, 688–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ordonez-Rubiano, S.C.; Dhiman, A.; Jiao, G.; Strohmier, B.P.; Krusemark, C.J.; Dykhuizen, E.C. Polycomb group proteins in cancer: Multifaceted functions and strategies for modulation. NAR Cancer 2021, 3, zcab039. [Google Scholar] [CrossRef] [PubMed]
- de Napoles, M.; Mermoud, J.E.; Wakao, R.; Tang, Y.A.; Endoh, M.; Appanah, R.; Nesterova, T.B.; Silva, J.; Otte, A.P.; Vidal, M.; et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 2004, 7, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.; Verrijzer, C.P. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev. 2008, 22, 2799–2810. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196. [Google Scholar] [CrossRef]
- Muller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef]
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002, 16, 2893–2905. [Google Scholar] [CrossRef]
- Messmer, S.; Franke, A.; Paro, R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992, 6, 1241–1254. [Google Scholar] [CrossRef][Green Version]
- Gorfinkiel, N.; Fanti, L.; Melgar, T.; Garcia, E.; Pimpinelli, S.; Guerrero, I.; Vidal, M. The Drosophila Polycomb group gene Sex combs extra encodes the ortholog of mammalian Ring1 proteins. Mech. Dev. 2004, 121, 449–462. [Google Scholar] [CrossRef]
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol. Cell. Biol. 2002, 22, 6070–6078. [Google Scholar] [CrossRef]
- Robinson, A.K.; Leal, B.Z.; Chadwell, L.V.; Wang, R.; Ilangovan, U.; Kaur, Y.; Junco, S.E.; Schirf, V.; Osmulski, P.A.; Gaczynska, M.; et al. The growth-suppressive function of the polycomb group protein polyhomeotic is mediated by polymerization of its sterile alpha motif (SAM) domain. J. Biol. Chem. 2012, 287, 8702–8713. [Google Scholar] [CrossRef]
- van Wijnen, A.J.; Bagheri, L.; Badreldin, A.A.; Larson, A.N.; Dudakovic, A.; Thaler, R.; Paradise, C.R.; Wu, Z. Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone 2021, 143, 115659. [Google Scholar] [CrossRef]
- O’Loghlen, A.; Munoz-Cabello, A.M.; Gaspar-Maia, A.; Wu, H.A.; Banito, A.; Kunowska, N.; Racek, T.; Pemberton, H.N.; Beolchi, P.; Lavial, F.; et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 2012, 10, 33–46. [Google Scholar] [CrossRef]
- Morey, L.; Pascual, G.; Cozzuto, L.; Roma, G.; Wutz, A.; Benitah, S.A.; Di Croce, L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012, 10, 47–62. [Google Scholar] [CrossRef]
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975. [Google Scholar] [CrossRef]
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol. Cell 2019, 74, 1020–1036.e1028. [Google Scholar] [CrossRef]
- Tie, F.; Stratton, C.A.; Kurzhals, R.L.; Harte, P.J. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol. Cell Biol. 2007, 27, 2014–2026. [Google Scholar] [CrossRef]
- Hojfeldt, J.W.; Laugesen, A.; Willumsen, B.M.; Damhofer, H.; Hedehus, L.; Tvardovskiy, A.; Mohammad, F.; Jensen, O.N.; Helin, K. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Biol. 2018, 25, 225–232. [Google Scholar] [CrossRef]
- Verreault, A.; Kaufman, P.D.; Kobayashi, R.; Stillman, B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998, 8, 96–108. [Google Scholar] [CrossRef]
- Kuroda, M.I.; Kang, H.; De, S.; Kassis, J.A. Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming. Annu. Rev. Biochem. 2020, 89, 235–253. [Google Scholar] [CrossRef]
- Toth, Z.; Brulois, K.; Lee, H.R.; Izumiya, Y.; Tepper, C.; Kung, H.J.; Jung, J.U. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog. 2013, 9, e1003813. [Google Scholar] [CrossRef]
- Toth, Z.; Papp, B.; Brulois, K.; Choi, Y.J.; Gao, S.J.; Jung, J.U. LANA-Mediated Recruitment of Host Polycomb Repressive Complexes onto the KSHV Genome during De Novo Infection. PLoS Pathog. 2016, 12, e1005878. [Google Scholar] [CrossRef]
- Hyland, P.L.; McDade, S.S.; McCloskey, R.; Dickson, G.J.; Arthur, K.; McCance, D.J.; Patel, D. Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J. Virol. 2011, 85, 10999–11006. [Google Scholar] [CrossRef] [PubMed]
- Hopcraft, S.E.; Pattenden, S.G.; James, L.I.; Frye, S.; Dittmer, D.P.; Damania, B. Chromatin remodeling controls Kaposi’s sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog. 2018, 14, e1007267. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Dunphy, P.S.; Das, S.; Zhu, B.; Luo, T.; McBride, J.W. Ehrlichia chaffeensis TRP120 Effector Targets and Recruits Host Polycomb Group Proteins for Degradation To Promote Intracellular Infection. Infect. Immun. 2018, 86, e00845-17. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Kuriakose, J.A.; Zhu, B.; Wakeel, A.; McBride, J.W. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect. Immun. 2011, 79, 4382–4391. [Google Scholar] [CrossRef]
- Vargas-Ayala, R.C.; Jay, A.; Manara, F.; Maroui, M.A.; Hernandez-Vargas, H.; Diederichs, A.; Robitaille, A.; Sirand, C.; Ceraolo, M.G.; Romero-Medina, M.C.; et al. Interplay between the Epigenetic Enzyme Lysine (K)-Specific Demethylase 2B and Epstein-Barr Virus Infection. J. Virol. 2019, 93, e00273-19. [Google Scholar] [CrossRef]
- Naik, N.G.; Nguyen, T.H.; Roberts, L.; Fischer, L.T.; Glickman, K.; Golas, G.; Papp, B.; Toth, Z. Epigenetic factor siRNA screen during primary KSHV infection identifies novel host restriction factors for the lytic cycle of KSHV. PLoS Pathog. 2020, 16, e1008268. [Google Scholar] [CrossRef]
- Violot, S.; Hong, S.S.; Rakotobe, D.; Petit, C.; Gay, B.; Moreau, K.; Billaud, G.; Priet, S.; Sire, J.; Schwartz, O.; et al. The human polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J. Virol. 2003, 77, 12507–12522. [Google Scholar] [CrossRef] [PubMed]
- Rakotobe, D.; Tardy, J.C.; Andre, P.; Hong, S.S.; Darlix, J.L.; Boulanger, P. Human Polycomb group EED protein negatively affects HIV-1 assembly and release. Retrovirology 2007, 4, 37. [Google Scholar] [CrossRef]
- Friedman, J.; Cho, W.K.; Chu, C.K.; Keedy, K.S.; Archin, N.M.; Margolis, D.M.; Karn, J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 2011, 85, 9078–9089. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, K.; Das, B.; Dobrowolski, C.; Karn, J. Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency. mBio 2017, 8, e00133-17. [Google Scholar] [CrossRef]
- Holland, D.; Hoppe-Seyler, K.; Schuller, B.; Lohrey, C.; Maroldt, J.; Durst, M.; Hoppe-Seyler, F. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 2008, 68, 9964–9972. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, T.; Bao, X.; He, M.; Li, L.; Yang, X. Increased EZH2 expression is associated with proliferation and progression of cervical cancer and indicates a poor prognosis. Int. J. Gynecol. Pathol. 2014, 33, 218–224. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Tarrant-Elorza, M.; Verma, S.; Purushothaman, P.; Pari, G.S. Regulation of viral and cellular gene expression by Kaposi′s sarcoma-associated herpesvirus polyadenylated nuclear RNA. J. Virol. 2013, 87, 5540–5553. [Google Scholar] [CrossRef] [PubMed]
- Toth, Z.; Maglinte, D.T.; Lee, S.H.; Lee, H.R.; Wong, L.Y.; Brulois, K.F.; Lee, S.; Buckley, J.D.; Laird, P.W.; Marquez, V.E.; et al. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 2010, 6, e1001013. [Google Scholar] [CrossRef] [PubMed]
- Asaka, M.N.; Kawaguchi, A.; Sakai, Y.; Mori, K.; Nagata, K. Polycomb repressive complex 2 facilitates the nuclear export of the influenza viral genome through the interaction with M1. Sci. Rep. 2016, 6, 33608. [Google Scholar] [CrossRef] [PubMed]
- Jha, H.C.; Mehta, D.; Lu, J.; El-Naccache, D.; Shukla, S.K.; Kovacsics, C.; Kolson, D.; Robertson, E.S. Gammaherpesvirus Infection of Human Neuronal Cells. mBio 2015, 6, e01844-15. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Tan, X.; Wang, X.; Wang, X.; Yang, L.; Robertson, E.S.; Lan, K. Epigenetic Landscape of Kaposi’s Sarcoma-Associated Herpesvirus Genome in Classic Kaposi’s Sarcoma Tissues. PLoS Pathog. 2017, 13, e1006167. [Google Scholar] [CrossRef] [PubMed]
- Gunther, T.; Grundhoff, A. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 2010, 6, e1000935. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Cheng, Y.; Sun, M.A.; Gehred, N.D.; Kassis, J.A. Structure and function of an ectopic Polycomb chromatin domain. Sci. Adv. 2019, 5, eaau9739. [Google Scholar] [CrossRef]
- Dorafshan, E.; Kahn, T.G.; Schwartz, Y.B. Hierarchical recruitment of Polycomb complexes revisited. Nucleus 2017, 8, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Sing, A.; Pannell, D.; Karaiskakis, A.; Sturgeon, K.; Djabali, M.; Ellis, J.; Lipshitz, H.D.; Cordes, S.P. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 2009, 138, 885–897. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 2010, 140, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. Variable requirements for DNA-binding proteins at polycomb-dependent repressive regions in human HOX clusters. Mol. Cell Biol. 2013, 33, 3274–3285. [Google Scholar] [CrossRef] [PubMed]
- Mendenhall, E.M.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.K.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. eLife 2012, 1, e00205. [Google Scholar] [CrossRef]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.P.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Stielow, B.; Finkernagel, F.; Stiewe, T.; Nist, A.; Suske, G. MGA, L3MBTL2 and E2F6 determine genomic binding of the non-canonical Polycomb repressive complex PRC1.6. PLoS Genet. 2018, 14, e1007193. [Google Scholar] [CrossRef] [PubMed]
- Endoh, M.; Endo, T.A.; Shinga, J.; Hayashi, K.; Farcas, A.; Ma, K.W.; Ito, S.; Sharif, J.; Endoh, T.; Onaga, N.; et al. Correction: PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife 2017, 6, e27970. [Google Scholar] [CrossRef] [PubMed]
- Scelfo, A.; Fernandez-Perez, D.; Tamburri, S.; Zanotti, M.; Lavarone, E.; Soldi, M.; Bonaldi, T.; Ferrari, K.J.; Pasini, D. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol. Cell 2019, 74, 1037–1052.e1037. [Google Scholar] [CrossRef] [PubMed]
- Kaustov, L.; Ouyang, H.; Amaya, M.; Lemak, A.; Nady, N.; Duan, S.; Wasney, G.A.; Li, Z.; Vedadi, M.; Schapira, M.; et al. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 2011, 286, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 2017, 549, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Hunkapiller, J.; Shen, Y.; Diaz, A.; Cagney, G.; McCleary, D.; Ramalho-Santos, M.; Krogan, N.; Ren, B.; Song, J.S.; Reiter, J.F. Polycomb-like 3 promotes polycomb repressive complex 2 binding to CpG islands and embryonic stem cell self-renewal. PLoS Genet. 2012, 8, e1002576. [Google Scholar] [CrossRef] [PubMed]
- Perino, M.; van Mierlo, G.; Karemaker, I.D.; van Genesen, S.; Vermeulen, M.; Marks, H.; van Heeringen, S.J.; Veenstra, G.J.C. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018, 50, 1002–1010. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Margueron, R.; Ku, M.; Chambon, P.; Bernstein, B.E.; Reinberg, D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010, 24, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Hojfeldt, J.W.; Hedehus, L.; Laugesen, A.; Tatar, T.; Wiehle, L.; Helin, K. Non-core Subunits of the PRC2 Complex Are Collectively Required for Its Target-Site Specificity. Mol. Cell 2019, 76, 423–436.e423. [Google Scholar] [CrossRef]
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Muller, C.W.; Vermeulen, M.; Muller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Kanhere, A.; Viiri, K.; Araujo, C.C.; Rasaiyaah, J.; Bouwman, R.D.; Whyte, W.A.; Pereira, C.F.; Brookes, E.; Walker, K.; Bell, G.W.; et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell 2010, 38, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Sun, B.K.; Erwin, J.A.; Song, J.J.; Lee, J.T. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 2008, 322, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Pintacuda, G.; Masui, O.; Koseki, Y.; Gdula, M.; Cerase, A.; Brown, D.; Mould, A.; Innocent, C.; Nakayama, M.; et al. PCGF3/5-PRC1 initiates Polycomb recruitment in X chromosome inactivation. Science 2017, 356, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Bousard, A.; Raposo, A.C.; Zylicz, J.J.; Picard, C.; Pires, V.B.; Qi, Y.; Gil, C.; Syx, L.; Chang, H.Y.; Heard, E.; et al. The role of Xist-mediated Polycomb recruitment in the initiation of X-chromosome inactivation. EMBO Rep. 2019, 20, e48019. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Cooper, S.; Farcas, A.M.; Blackledge, N.P.; Brockdorff, N. Chromatin sampling—An emerging perspective on targeting polycomb repressor proteins. PLoS Genet. 2013, 9, e1003717. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.L.; Son, J.; Drury, W.J., 3rd; Voigt, P.; Martin, S.R.; Taylor, W.R.; De Marco, V.; et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009, 461, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Poepsel, S.; Kasinath, V.; Nogales, E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol. 2018, 25, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Harrison, M.M.; Villalta, J.E.; Kaplan, T.; Eisen, M.B. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 2014, 3, e03737. [Google Scholar] [CrossRef]
- Zenk, F.; Loeser, E.; Schiavo, R.; Kilpert, F.; Bogdanovic, O.; Iovino, N. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 2017, 357, 212–216. [Google Scholar] [CrossRef]
- Alabert, C.; Barth, T.K.; Reveron-Gomez, N.; Sidoli, S.; Schmidt, A.; Jensen, O.N.; Imhof, A.; Groth, A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015, 29, 585–590. [Google Scholar] [CrossRef]
- Coleman, R.T.; Struhl, G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 2017, 356, eaai8236. [Google Scholar] [CrossRef]
- Laprell, F.; Finkl, K.; Muller, J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 2017, 352, 85–88. [Google Scholar] [CrossRef]
- Pengelly, A.R.; Copur, O.; Jackle, H.; Herzig, A.; Muller, J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 2013, 339, 698–699. [Google Scholar] [CrossRef]
- Illingworth, R.S.; Moffat, M.; Mann, A.R.; Read, D.; Hunter, C.J.; Pradeepa, M.M.; Adams, I.R.; Bickmore, W.A. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev. 2015, 29, 1897–1902. [Google Scholar] [CrossRef] [PubMed]
- Pengelly, A.R.; Kalb, R.; Finkl, K.; Muller, J. Transcriptional repression by PRC1 in the absence of H2A monoubiquitylation. Genes Dev. 2015, 29, 1487–1492. [Google Scholar] [CrossRef] [PubMed]
- Francis, N.J.; Kingston, R.E.; Woodcock, C.L. Chromatin compaction by a polycomb group protein complex. Science 2004, 306, 1574–1577. [Google Scholar] [CrossRef]
- Lau, M.S.; Schwartz, M.G.; Kundu, S.; Savol, A.J.; Wang, P.I.; Marr, S.K.; Grau, D.J.; Schorderet, P.; Sadreyev, R.I.; Tabin, C.J.; et al. Mutation of a nucleosome compaction region disrupts Polycomb-mediated axial patterning. Science 2017, 355, 1081–1084. [Google Scholar] [CrossRef]
- Eagen, K.P.; Aiden, E.L.; Kornberg, R.D. Polycomb-mediated chromatin loops revealed by a subkilobase-resolution chromatin interaction map. Proc. Natl. Acad. Sci. USA 2017, 114, 8764–8769. [Google Scholar] [CrossRef]
- Peterson, A.J.; Kyba, M.; Bornemann, D.; Morgan, K.; Brock, H.W.; Simon, J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol. Cell Biol. 1997, 17, 6683–6692. [Google Scholar] [CrossRef]
- Kim, C.A.; Gingery, M.; Pilpa, R.M.; Bowie, J.U. The SAM domain of polyhomeotic forms a helical polymer. Nat. Struct. Biol. 2002, 9, 453–457. [Google Scholar] [CrossRef]
- Isono, K.; Endo, T.A.; Ku, M.; Yamada, D.; Suzuki, R.; Sharif, J.; Ishikura, T.; Toyoda, T.; Bernstein, B.E.; Koseki, H. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell 2013, 26, 565–577. [Google Scholar] [CrossRef]
- Kundu, S.; Ji, F.; Sunwoo, H.; Jain, G.; Lee, J.T.; Sadreyev, R.I.; Dekker, J.; Kingston, R.E. Polycomb Repressive Complex 1 Generates Discrete Compacted Domains that Change during Differentiation. Mol. Cell 2018, 71, 191. [Google Scholar] [CrossRef] [PubMed]
- Oksuz, O.; Narendra, V.; Lee, C.H.; Descostes, N.; LeRoy, G.; Raviram, R.; Blumenberg, L.; Karch, K.; Rocha, P.P.; Garcia, B.A.; et al. Capturing the Onset of PRC2-Mediated Repressive Domain Formation. Mol. Cell 2018, 70, 1149–1162. [Google Scholar] [CrossRef]
- De, S.; Mitra, A.; Cheng, Y.; Pfeifer, K.; Kassis, J.A. Formation of a Polycomb-Domain in the Absence of Strong Polycomb Response Elements. PLoS Genet. 2016, 12, e1006200. [Google Scholar] [CrossRef]
- Ogiyama, Y.; Schuettengruber, B.; Papadopoulos, G.L.; Chang, J.M.; Cavalli, G. Polycomb-Dependent Chromatin Looping Contributes to Gene Silencing during Drosophila Development. Mol. Cell 2018, 71, 73–88. [Google Scholar] [CrossRef]
- Trojer, P.; Cao, A.R.; Gao, Z.; Li, Y.; Zhang, J.; Xu, X.; Li, G.; Losson, R.; Erdjument-Bromage, H.; Tempst, P.; et al. L3MBTL2 protein acts in concert with PcG protein-mediated monoubiquitination of H2A to establish a repressive chromatin structure. Mol. Cell 2011, 42, 438–450. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Fu, C.; Stratton, C.A.; Fang, M.; Harte, P.J. Polycomb inhibits histone acetylation by CBP by binding directly to its catalytic domain. Proc. Natl. Acad. Sci. USA 2016, 113, E744–E753. [Google Scholar] [CrossRef] [PubMed]
- Dellino, G.I.; Schwartz, Y.B.; Farkas, G.; McCabe, D.; Elgin, S.C.; Pirrotta, V. Polycomb silencing blocks transcription initiation. Mol. Cell 2004, 13, 887–893. [Google Scholar] [CrossRef]
- Zhou, W.; Zhu, P.; Wang, J.; Pascual, G.; Ohgi, K.A.; Lozach, J.; Glass, C.K.; Rosenfeld, M.G. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell 2008, 29, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Kajitani, T.; Togo, S.; Masuko, N.; Ohdan, H.; Hishikawa, Y.; Koji, T.; Matsuyama, T.; Ikura, T.; Muramatsu, M.; et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008, 22, 37–49. [Google Scholar] [CrossRef]
- Loubiere, V.; Martinez, A.M.; Cavalli, G. Cell Fate and Developmental Regulation Dynamics by Polycomb Proteins and 3D Genome Architecture. Bioessays 2019, 41, e1800222. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Bourbon, H.M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef] [PubMed]
- Cheutin, T.; Cavalli, G. The multiscale effects of polycomb mechanisms on 3D chromatin folding. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.A.; Batsche, E.; Regnault, B.; Tham, T.N.; Seveau, S.; Muchardt, C.; Cossart, P. Histone modifications induced by a family of bacterial toxins. Proc. Natl. Acad. Sci. USA 2007, 104, 13467–13472. [Google Scholar] [CrossRef]
- Dong, W.; Rasid, O.; Chevalier, C.; Connor, M.; Eldridge, M.J.G.; Hamon, M.A. Streptococcus pneumoniae Infection Promotes Histone H3 Dephosphorylation by Modulating Host PP1 Phosphatase. Cell Rep. 2020, 30, 4016–4026. [Google Scholar] [CrossRef] [PubMed]
- Watson, Z.; Dhummakupt, A.; Messer, H.; Phelan, D.; Bloom, D. Role of polycomb proteins in regulating HSV-1 latency. Viruses 2013, 5, 1740–1757. [Google Scholar] [CrossRef] [PubMed]
- Harth-Hertle, M.L.; Scholz, B.A.; Erhard, F.; Glaser, L.V.; Dolken, L.; Zimmer, R.; Kempkes, B. Inactivation of intergenic enhancers by EBNA3A initiates and maintains polycomb signatures across a chromatin domain encoding CXCL10 and CXCL9. PLoS Pathog. 2013, 9, e1003638. [Google Scholar] [CrossRef] [PubMed]
- Baxter, J.; Sauer, S.; Peters, A.; John, R.; Williams, R.; Caparros, M.L.; Arney, K.; Otte, A.; Jenuwein, T.; Merkenschlager, M.; et al. Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. EMBO J. 2004, 23, 4462–4472. [Google Scholar] [CrossRef] [PubMed]
- Dufour, C.; Gantner, P.; Fromentin, R.; Chomont, N. The multifaceted nature of HIV latency. J. Clin. Investig. 2020, 130, 3381–3390. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Iqbal, M.; Tariq, M.; Baig, S.M.; Abbas, W. Epigenetic regulation of HIV-1 latency: Focus on polycomb group (PcG) proteins. Clin. Epigenetics 2018, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Yoon, C.H.; Jang, D.H.; Kim, K.C.; Park, S.Y.; Kim, H.Y.; Kim, S.S.; Chi, S.G.; Choi, B.S. Disruption of polycomb repressor complex-mediated gene silencing reactivates HIV-1 provirus in latently infected cells. Intervirology 2014, 57, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, J.; Yang, Z.; Lu, X.; Jin, C.; Cheng, L.; Wu, N. RbAp48, a novel inhibitory factor that regulates the transcription of human immunodeficiency virus type 1. Int. J. Mol. Med. 2016, 38, 267–274. [Google Scholar] [CrossRef]
- Kim, H.G.; Kim, K.C.; Roh, T.Y.; Park, J.; Jung, K.M.; Lee, J.S.; Choi, S.Y.; Kim, S.S.; Choi, B.S. Gene silencing in HIV-1 latency by polycomb repressive group. Virol. J. 2011, 8, 179. [Google Scholar] [CrossRef]
- Sharma, A.L.; Hokello, J.; Sonti, S.; Zicari, S.; Sun, L.; Alqatawni, A.; Bukrinsky, M.; Simon, G.; Chauhan, A.; Daniel, R.; et al. CBF-1 Promotes the Establishment and Maintenance of HIV Latency by Recruiting Polycomb Repressive Complexes, PRC1 and PRC2, at HIV LTR. Viruses 2020, 12, 1040. [Google Scholar] [CrossRef] [PubMed]
- Coull, J.J.; He, G.; Melander, C.; Rucker, V.C.; Dervan, P.B.; Margolis, D.M. Targeted derepression of the human immunodeficiency virus type 1 long terminal repeat by pyrrole-imidazole polyamides. J. Virol. 2002, 76, 12349–12354. [Google Scholar] [CrossRef]
- Coull, J.J.; Romerio, F.; Sun, J.M.; Volker, J.L.; Galvin, K.M.; Davie, J.R.; Shi, Y.; Hansen, U.; Margolis, D.M. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 2000, 74, 6790–6799. [Google Scholar] [CrossRef] [PubMed]
- Miranda, T.B.; Cortez, C.C.; Yoo, C.B.; Liang, G.; Abe, M.; Kelly, T.K.; Marquez, V.E.; Jones, P.A. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol. Cancer Ther. 2009, 8, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, M.K.; McManamy, M.E.; Burch, B.D.; Archin, N.M.; Margolis, D.M. H3K27 Demethylation at the Proviral Promoter Sensitizes Latent HIV to the Effects of Vorinostat in Ex Vivo Cultures of Resting CD4+ T Cells. J. Virol. 2015, 89, 8392–8405. [Google Scholar] [CrossRef]
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human papillomavirus and cervical cancer. Lancet 2007, 370, 890–907. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Huh, K.W.; Munger, K. Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J. Virol. 2008, 82, 8695–8705. [Google Scholar] [CrossRef]
- Trimarchi, J.M.; Fairchild, B.; Wen, J.; Lees, J.A. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1519–1524. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Crum, C.P.; Munger, K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. USA 2011, 108, 2130–2135. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Munger, K. Biochemical and functional interactions of human papillomavirus proteins with polycomb group proteins. Viruses 2013, 5, 1231–1249. [Google Scholar] [CrossRef]
- Menges, C.W.; Kadariya, Y.; Altomare, D.; Talarchek, J.; Neumann-Domer, E.; Wu, Y.; Xiao, G.H.; Shapiro, I.M.; Kolev, V.N.; Pachter, J.A.; et al. Tumor suppressor alterations cooperate to drive aggressive mesotheliomas with enriched cancer stem cells via a p53-miR-34a-c-Met axis. Cancer Res. 2014, 74, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.L.; Zhou, B.P.; Xia, W.; Wu, Y.; Yang, C.C.; Chen, C.T.; Ping, B.; Otte, A.P.; Hung, M.C. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005, 310, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Dochnal, S.A.; Francois, A.K.; Cliffe, A.R. De Novo Polycomb Recruitment: Lessons from Latent Herpesviruses. Viruses 2021, 13, 1470. [Google Scholar] [CrossRef] [PubMed]
- Knipe, D.M.; Cliffe, A. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 2008, 6, 211–221. [Google Scholar] [CrossRef]
- Reeves, M.; Sinclair, J. Regulation of human cytomegalovirus transcription in latency: Beyond the major immediate-early promoter. Viruses 2013, 5, 1395–1413. [Google Scholar] [CrossRef]
- Reeves, M.B.; Lehner, P.J.; Sissons, J.G.P.; Sinclair, J.H. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 2005, 86, 2949–2954. [Google Scholar] [CrossRef]
- Murata, T.; Kondo, Y.; Sugimoto, A.; Kawashima, D.; Saito, S.; Isomura, H.; Kanda, T.; Tsurumi, T. Epigenetic histone modification of Epstein-Barr virus BZLF1 promoter during latency and reactivation in Raji cells. J. Virol. 2012, 86, 4752–4761. [Google Scholar] [CrossRef]
- Ramasubramanyan, S.; Osborn, K.; Flower, K.; Sinclair, A.J. Dynamic chromatin environment of key lytic cycle regulatory regions of the Epstein-Barr virus genome. J. Virol. 2012, 86, 1809–1819. [Google Scholar] [CrossRef][Green Version]
- Cliffe, A.R.; Garber, D.A.; Knipe, D.M. Transcription of the herpes simplex virus latency-associated transcript promotes the formation of facultative heterochromatin on lytic promoters. J. Virol. 2009, 83, 8182–8190. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Zhou, C.; Johnson, K.E.; Colgrove, R.C.; Coen, D.M.; Knipe, D.M. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 2005, 102, 16055–16059. [Google Scholar] [CrossRef]
- Kwiatkowski, D.L.; Thompson, H.W.; Bloom, D.C. The polycomb group protein Bmi1 binds to the herpes simplex virus 1 latent genome and maintains repressive histone marks during latency. J. Virol. 2009, 83, 8173–8181. [Google Scholar] [CrossRef] [PubMed]
- Kubat, N.J.; Amelio, A.L.; Giordani, N.V.; Bloom, D.C. The Herpes Simplex Virus Type 1 Latency-Associated Transcript (LAT) Enhancer/rcr Is Hyperacetylated during Latency Independently of LAT Transcription. J. Virol. 2004, 78, 12508–12518. [Google Scholar] [CrossRef] [PubMed]
- Kubat, N.J.; Tran, R.K.; McAnany, P.; Bloom, D.C. Specific Histone Tail Modification and Not DNA Methylation Is a Determinant of Herpes Simplex Virus Type 1 Latent Gene Expression. J. Virol. 2004, 78, 1139–1149. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, A.R.; Arbuckle, J.H.; Vogel, J.L.; Geden, M.J.; Rothbart, S.B.; Cusack, C.L.; Strahl, B.D.; Kristie, T.M.; Deshmukh, M. Neuronal Stress Pathway Mediating a Histone Methyl/Phospho Switch Is Required for Herpes Simplex Virus Reactivation. Cell Host Microbe 2015, 18, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Cliffe, A.R.; Coen, D.M.; Knipe, D.M. Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection. mBio 2013, 4, e00590-12. [Google Scholar] [CrossRef]
- Nicoll, M.P.; Hann, W.; Shivkumar, M.; Harman, L.E.; Connor, V.; Coleman, H.M.; Proenca, J.T.; Efstathiou, S. The HSV-1 Latency-Associated Transcript Functions to Repress Latent Phase Lytic Gene Expression and Suppress Virus Reactivation from Latently Infected Neurons. PLoS Pathog. 2016, 12, e1005539. [Google Scholar] [CrossRef]
- Messer, H.G.; Jacobs, D.; Dhummakupt, A.; Bloom, D.C. Inhibition of H3K27me3-specific histone demethylases JMJD3 and UTX blocks reactivation of herpes simplex virus 1 in trigeminal ganglion neurons. J. Virol. 2015, 89, 3417–3420. [Google Scholar] [CrossRef]
- Bertke, A.S.; Patel, A.; Krause, P.R. Herpes simplex virus latency-associated transcript sequence downstream of the promoter influences type-specific reactivation and viral neurotropism. J. Virol. 2007, 81, 6605–6613. [Google Scholar] [CrossRef]
- Yanez, A.A.; Harrell, T.; Sriranganathan, H.J.; Ives, A.M.; Bertke, A.S. Neurotrophic Factors NGF, GDNF and NTN Selectively Modulate HSV1 and HSV2 Lytic Infection and Reactivation in Primary Adult Sensory and Autonomic Neurons. Pathogens 2017, 6, 5. [Google Scholar] [CrossRef]
- Dukhovny, A.; Sloutskin, A.; Markus, A.; Yee, M.B.; Kinchington, P.R.; Goldstein, R.S. Varicella-zoster virus infects human embryonic stem cell-derived neurons and neurospheres but not pluripotent embryonic stem cells or early progenitors. J. Virol. 2012, 86, 3211–3218. [Google Scholar] [CrossRef]
- Laemmle, L.; Goldstein, R.S.; Kinchington, P.R. Modeling Varicella Zoster Virus Persistence and Reactivation—Closer to Resolving a Perplexing Persistent State. Front. Microbiol. 2019, 10, 1634. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, T.; Okuno, Y.; Sato, Y.; Goshima, F.; Yoshiyama, H.; Kanda, T.; Kimura, H.; Murata, T. Regulation of Epstein-Barr Virus Life Cycle and Cell Proliferation by Histone H3K27 Methyltransferase EZH2 in Akata Cells. mSphere 2018, 3, e00478-18. [Google Scholar] [CrossRef] [PubMed]
- Abraham, C.G.; Kulesza, C.A. Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J. Virol. 2013, 87, 13193–13205. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Wang, H.; Yu, Y.; Yi, W.; Zhu, S.; Li, E.; Liang, Y. Epigenetically repressing human cytomegalovirus lytic infection and reactivation from latency in THP-1 model by targeting H3K9 and H3K27 histone demethylases. PLoS ONE 2017, 12, e0175390. [Google Scholar] [CrossRef] [PubMed]
- Strenger, V.; Caselli, E.; Lautenschlager, I.; Schwinger, W.; Aberle, S.W.; Loginov, R.; Gentili, V.; Nacheva, E.; DiLuca, D.; Urban, C. Detection of HHV-6-specific mRNA and antigens in PBMCs of individuals with chromosomally integrated HHV-6 (ciHHV-6). Clin. Microbiol. Infect. 2014, 20, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Saviola, A.J.; Zimmermann, C.; Mariani, M.P.; Signorelli, S.A.; Gerrard, D.L.; Boyd, J.R.; Wight, D.J.; Morissette, G.; Gravel, A.; Dubuc, I.; et al. Chromatin Profiles of Chromosomally Integrated Human Herpesvirus-6A. Front. Microbiol. 2019, 10, 1408. [Google Scholar] [CrossRef]
- Frohlich, J.; Grundhoff, A. Epigenetic control in Kaposi sarcoma-associated herpesvirus infection and associated disease. Semin. Immunopathol. 2020, 42, 143–157. [Google Scholar] [CrossRef]
- Gunther, T.; Frohlich, J.; Herrde, C.; Ohno, S.; Burkhardt, L.; Adler, H.; Grundhoff, A. A comparative epigenome analysis of gammaherpesviruses suggests cis-acting sequence features as critical mediators of rapid polycomb recruitment. PLoS Pathog. 2019, 15, e1007838. [Google Scholar] [CrossRef]
- Rossetto, C.C.; Pari, G. KSHV PAN RNA associates with demethylases UTX and JMJD3 to activate lytic replication through a physical interaction with the virus genome. PLoS Pathog. 2012, 8, e1002680. [Google Scholar] [CrossRef]
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by Polycomb and COMPASS families. Science 2016, 352, aad9780. [Google Scholar] [CrossRef]
- Kassis, J.A.; Kennison, J.A.; Tamkun, J.W. Polycomb and Trithorax Group Genes in Drosophila. Genetics 2017, 206, 1699–1725. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, S.; Crea, F. Targeting SARS-CoV-2 using polycomb inhibitors as antiviral agents. Epigenomics 2020, 12, 811–812. [Google Scholar] [CrossRef] [PubMed]
- Menachery, V.D.; Eisfeld, A.J.; Schafer, A.; Josset, L.; Sims, A.C.; Proll, S.; Fan, S.; Li, C.; Neumann, G.; Tilton, S.C.; et al. Pathogenic influenza viruses and coronaviruses utilize similar and contrasting approaches to control interferon-stimulated gene responses. mBio 2014, 5, e01174-14. [Google Scholar] [CrossRef] [PubMed]
- Jennings, B.H. Drosophila—A versatile model in biology & medicine. Mater. Today 2011, 14, 6. [Google Scholar]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster—From microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Harnish, J.M.; Link, N.; Yamamoto, S. Drosophila as a Model for Infectious Diseases. Int. J. Mol. Sci. 2021, 22, 42. [Google Scholar] [CrossRef] [PubMed]
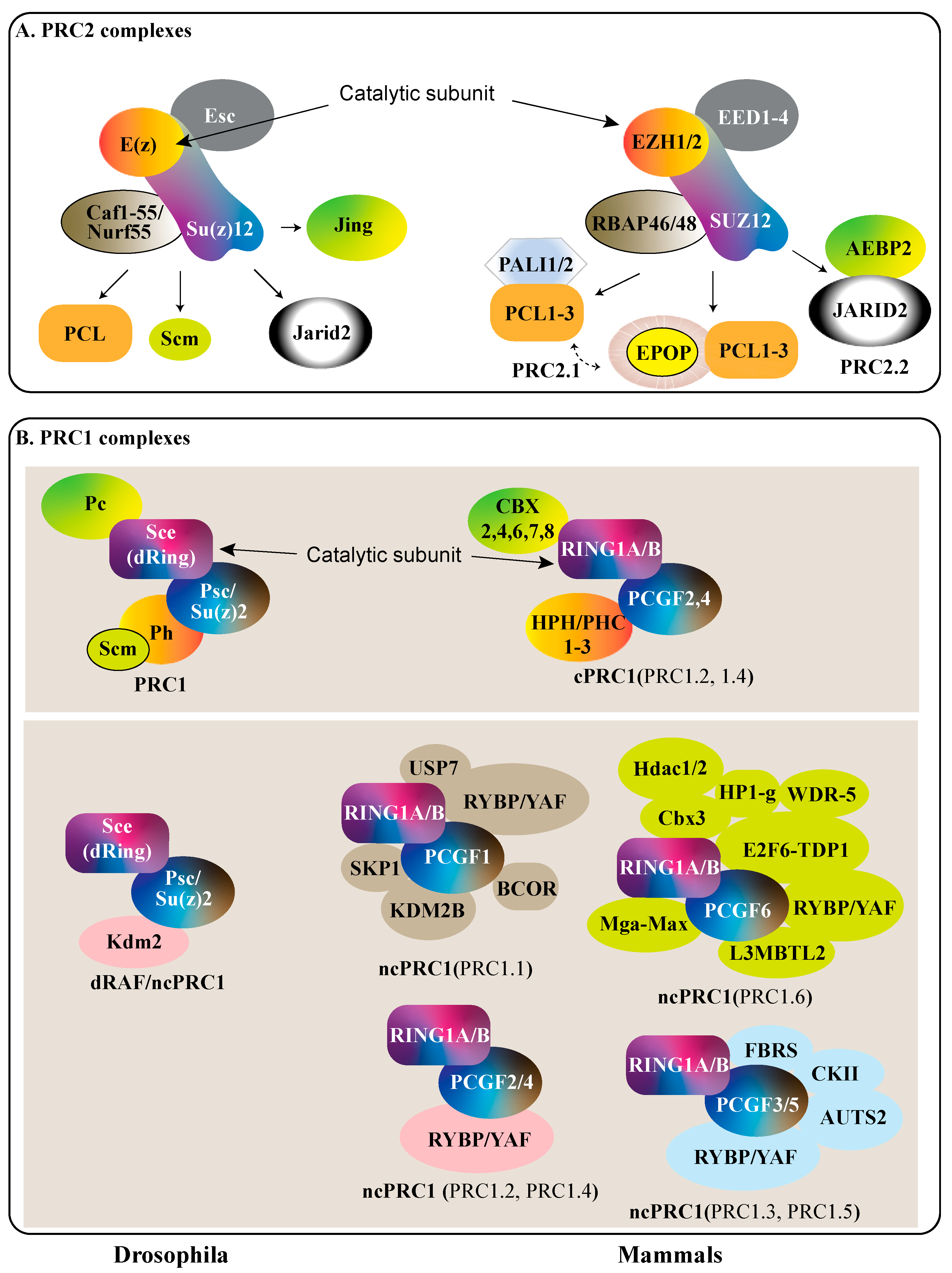
| D. melanogaster | H. sapiens | PcG Complex | Function | Pathogen Involvement |
|---|---|---|---|---|
| Pc | CBX-2,4,6,7,8 | cPRC1 | Interacts with histone H3K27me3 | TBD |
| Sce | RING1A/B | PRC1 | E3 ubiquitin ligase | KSHV [29,30] |
| Psc | BMI1/PCGF4 | PRC1 | Stabilize PRC1 complex | HPV [31], KSHV [32], E. chaffeensis [33,34] |
| Su(z)2 | MEL18/PCGF2 | PRC1 | Stabilize PRC1 complex | E. chaffeensis [33,34] |
| Ph-p and Ph-D | HPH/PHC-(1–3) | PRC1 | Establish High-order chromatin structure | TBD |
| Rybp | RYBP | ncPRC1 | Interacts with histone modification marks | KSHV [29] |
| Kdm2 | KDM2B/FBXL10 | ncPRC1 | DemethylatesLysine-36 of Histone H3 | EBV [35] KSHV [36] |
| Esc | EED | PRC2 | Binds to H3K27me3 | HIV-1 [37,38] |
| E(z) | EZH1/2 | PRC2 | Histone methyltransferase of H3K27 | HIV-1 [39,40], HPV [41,42], HHV5 [43], KSHV [29,44],Influenza [45] |
| Su(z)12 | SUZ12 | PRC2 | Stabilizes core complex | HHV5 [43], KSHV [46,47,48] |
| Caf1-55/Nurf55 | RBAP46/48 | PRC2 | Interact with Histone H4 tail | TBD |
| Jarid2 | JARID2 | PRC2 | Scaffold for recruitment of complexes | TBD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scholl, A.; De, S. Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis. Int. J. Mol. Sci. 2022, 23, 12285. https://doi.org/10.3390/ijms232012285
Scholl A, De S. Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis. International Journal of Molecular Sciences. 2022; 23(20):12285. https://doi.org/10.3390/ijms232012285
Chicago/Turabian StyleScholl, Aaron, and Sandip De. 2022. "Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis" International Journal of Molecular Sciences 23, no. 20: 12285. https://doi.org/10.3390/ijms232012285
APA StyleScholl, A., & De, S. (2022). Epigenetic Regulation by Polycomb Complexes from Drosophila to Human and Its Relation to Communicable Disease Pathogenesis. International Journal of Molecular Sciences, 23(20), 12285. https://doi.org/10.3390/ijms232012285





