Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus
Abstract
1. Introduction
2. Results
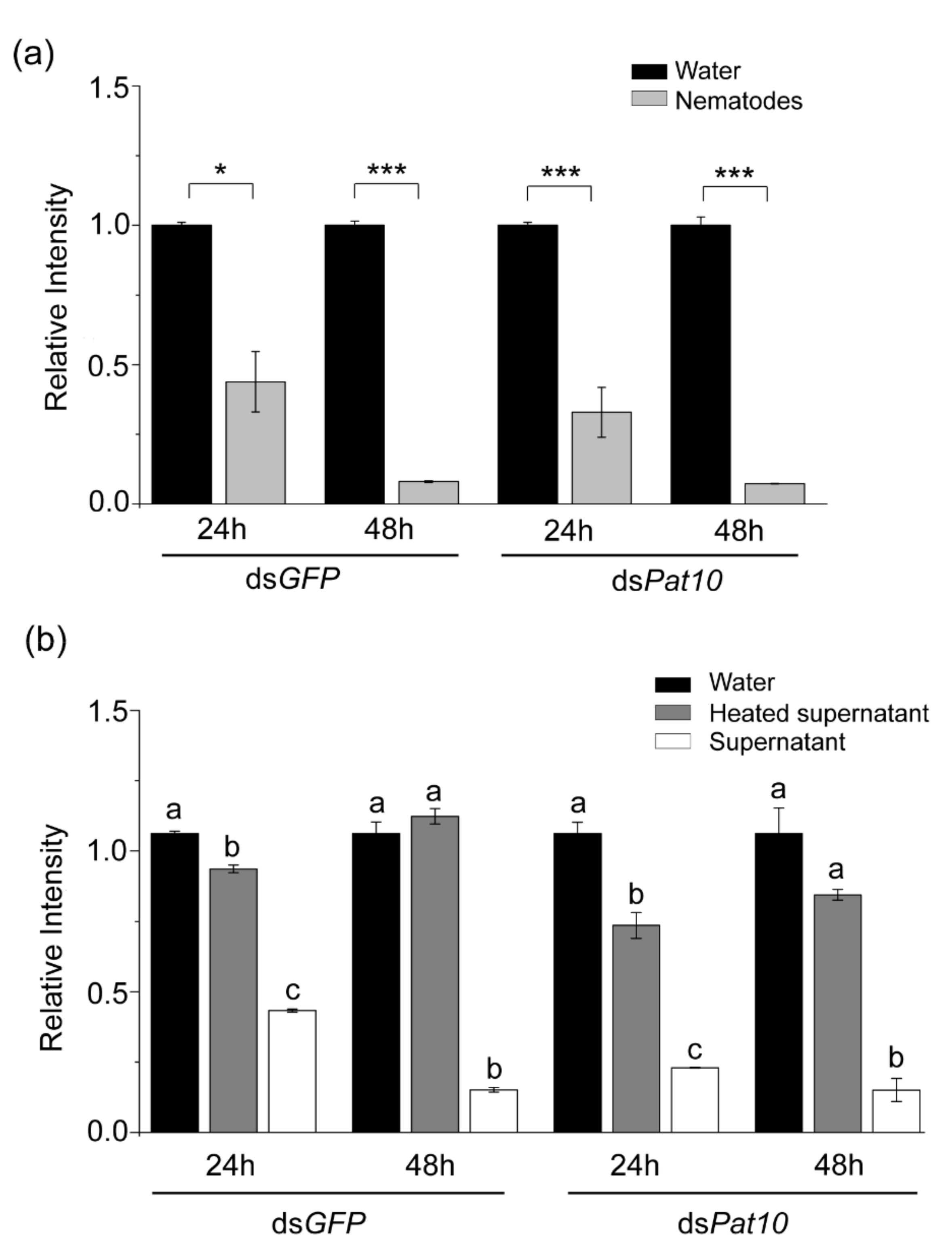
2.1. In Vitro Degradation of dsRNA by Nematodes
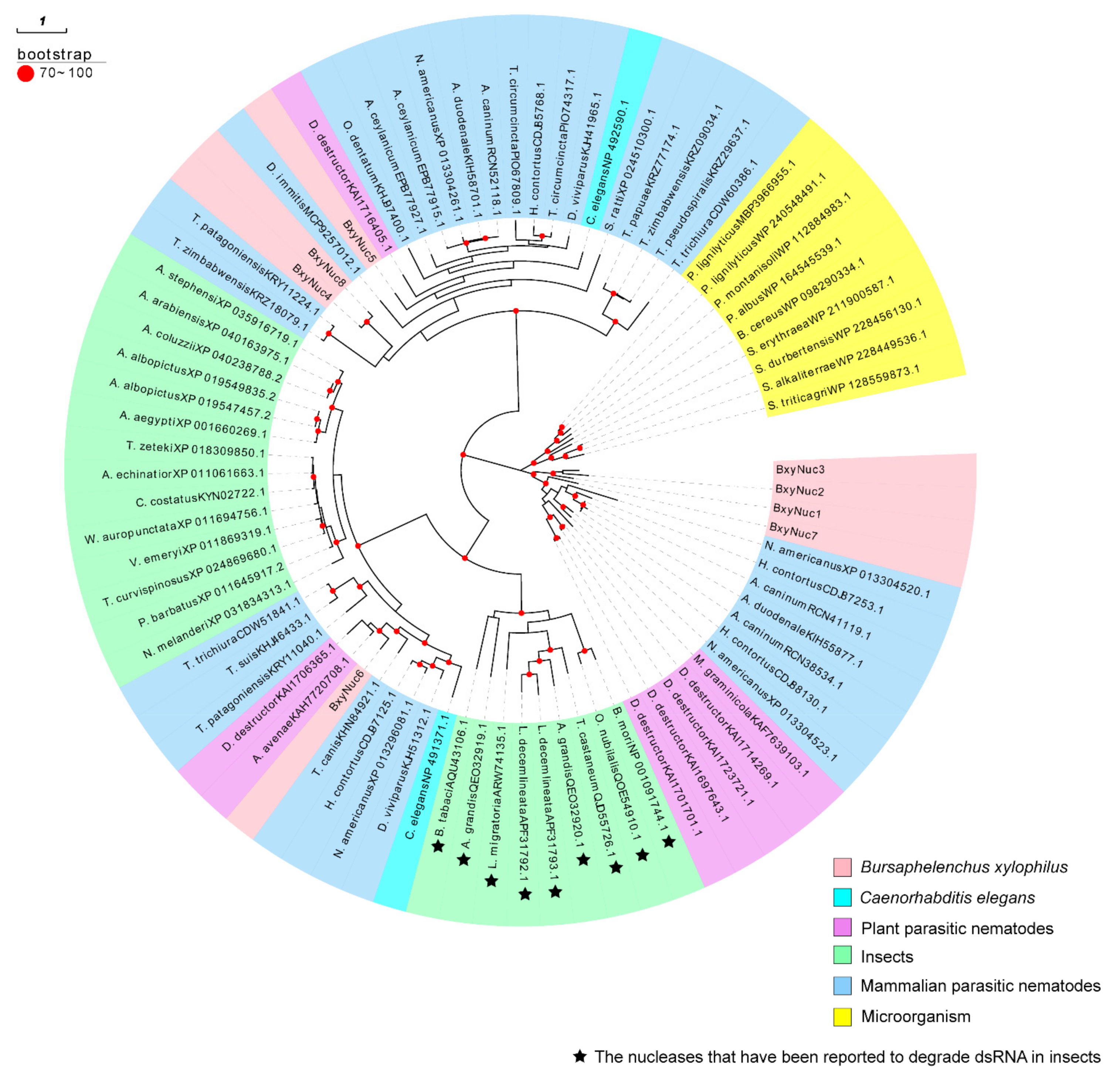
2.2. Screening of Extracellular Nuclease Genes
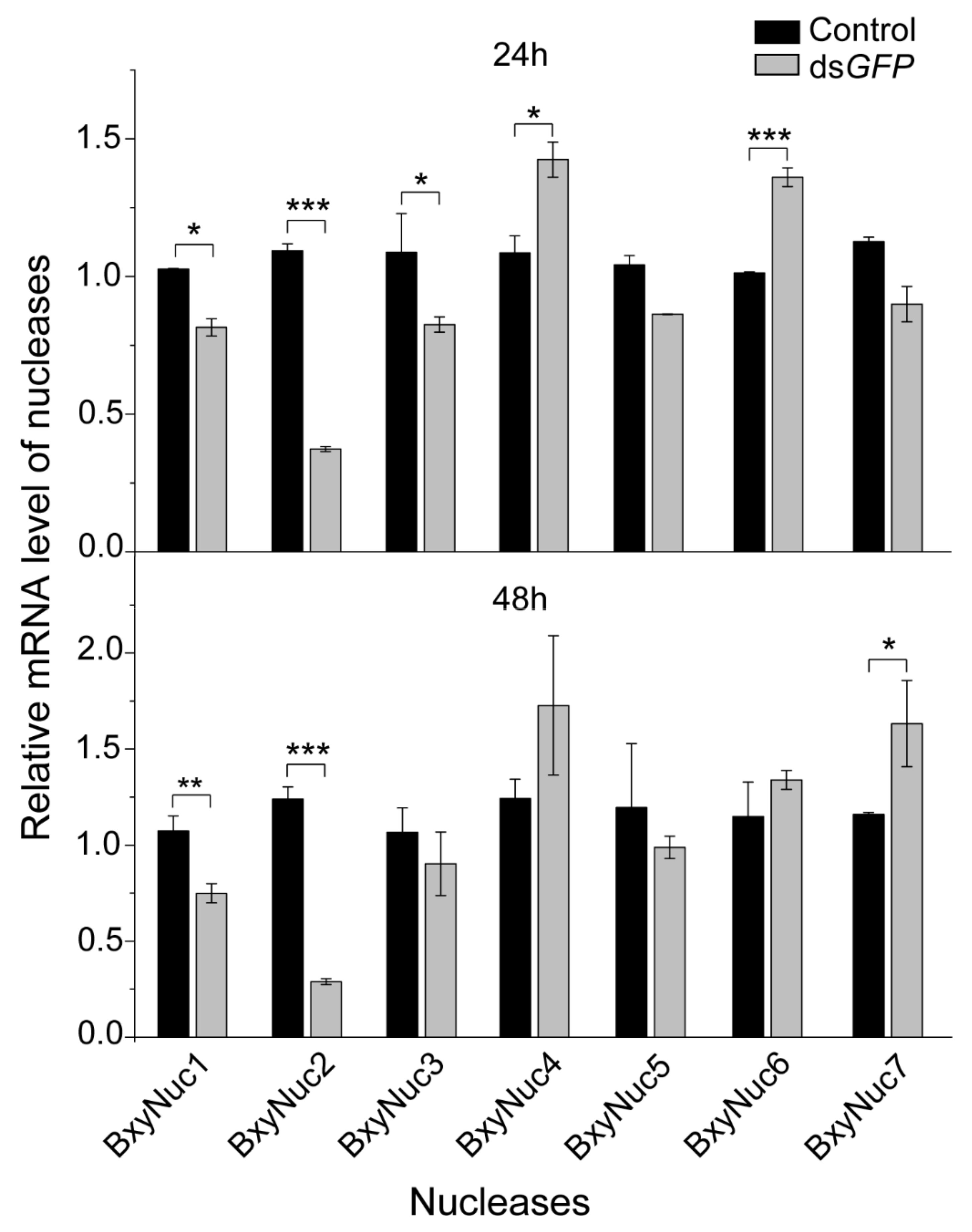
2.3. Extracellular Nuclease Gene Response to dsGFP
2.4. The Silencing of Extracellular Nuclease Genes Improves RNAi Efficiency
3. Discussion
4. Materials and Methods
4.1. Cultivation of Nematodes
4.2. Synthesis of dsRNA
4.3. Degradation of dsRNA Detected by Agarose Gel Electrophoresis
4.4. Screening and Cloning of B. xylophilus Extracellular Nuclease Genes
4.5. Interference of Target Gene and Extracellular Nuclease Genes
4.6. Quantitative Reverse Transcription PCR (qRT-PCR)
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, J.T.; Haegeman, A.; Danchin, E.G.J.; Gaur, H.S.; Helder, J.; Jones, M.G.K.; Kikuchi, T.; Manzanilla-Lopez, R.; Palomares-Rius, J.E.; Wesemael, W.M.; et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Inácio, M.L.; Nóbrega, F.; Vieira, P.; Bonifácio, L.; Naves, P.; Sousa, E.; Mota, M. First detection of Bursaphelenchus xylophilus associated with Pinus nigrain in Portugal and in Europe. For. Pathol. 2014, 45, 235–238. [Google Scholar] [CrossRef]
- Wang, H.H.; Wang, Y.B.; Yin, C.; Gao, J.; Tao, R.; Sun, Y.L.; Wang, C.Y.; Wang, Z.; Li, Y.X.; Sung, C.K. In vivo infection of Bursaphelenchus xylophilus by the fungus Esteya vermicola. Pest Manag. Sci. 2020, 76, 2854–2864. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, N.; Ekino, T.; Maehara, N.; Aikawa, T.; Giblin-Davis, R.M. Bursaphelenchus acaloleptae n. sp. sharing tree and beetle carrier hosts with B. luxuriosae Kanzaki & Futai, 2003 in Japan. Nematology 2019, 22, 515–527. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, J.; Yan, J.; Fang, G. Economic Loss of Pine Wood Nematode Disease in Mainland China from 1998 to 2017. Forests 2020, 11, 1042. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, W.; Ding, P.; Wu, J.; Li, Y. The research progress of pine wilt disease control. Xiang Cun Ke Ji 2022, 13, 116–118. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Li, Y.; Liu, Z.; Li, D.; Wen, X.; Feng, Y.; Zhang, X. Pinewood Nematode Alters the Endophytic and Rhizospheric Microbial Communities of Pinus massoniana. Microb. Ecol. 2020, 81, 807–817. [Google Scholar] [CrossRef]
- Kim, N.; Jeon, H.W.; Mannaa, M.; Jeong, S.-I.; Kim, J.; Kim, J.; Lee, C.; Park, A.R.; Kim, J.-C.; Seo, Y.-S. Induction of resistance against pine wilt disease caused by Bursaphelenchus xylophilus using selected pine endophytic bacteria. Plant Pathol. 2019, 68, 434–444. [Google Scholar] [CrossRef]
- Rosso, M.N.; Jones, J.T.; Abad, P. RNAi and functional genomics in plant parasitic nematodes. Annu. Rev. Phytopathol. 2009, 47, 207–232. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Liu, S.; Jaouannet, M.; Dempsey, D.A.; Imani, J.; Coustau, C.; Kogel, K.H. RNA-based technologies for insect control in plant production. Biotechnol. Adv. 2019, 39, 107463. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.C.H.; Jones, M.G.K.; Fosu-Nyarko, J. Gene silencing in root lesion nematodes (Pratylenchus spp.) significantly reduces reproduction in a plant host. Exp. Parasitol. 2013, 133, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, S.; Fosu-Nyarko, J.; Jones, M.G.K. Attempt to Silence Genes of the RNAi Pathways of the Root-Knot Nematode, Meloidogyne incognita Results in Diverse Responses Including Increase and No Change in Expression of Some Genes. Front. Plant Sci. 2020, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Banerjee, A.; Gill, S.S.; Gupta, O.P.; Dahuja, A.; Jain, P.K.; Sirohi, A. RNA Interference: A Novel Source of Resistance to Combat Plant Parasitic Nematodes. Front. Plant Sci. 2017, 8, 834. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, K.Y.; Lee, S.; Oh, W.; Jeong, P.; Woo, T.; Kim, C.; Paik, Y.; Koo, H. The efficiency of RNA interference in Bursaphelenchus xylophilus. Mol. Cells 2008, 26, 81–86. [Google Scholar]
- Wang, M.; Wang, D.; Zhang, X.; Wang, X.; Liu, W.; Hou, X.; Huang, X.; Xie, B.; Cheng, X. Double-stranded RNA-mediated interference of dumpy genes in Bursaphelenchus xylophilus by feeding on filamentous fungal transformants. Int. J. Parasitol. 2016, 46, 351–360. [Google Scholar] [CrossRef]
- Xue, Q.; Wu, X.Q.; Zhang, W.J.; Deng, L.N.; Wu, M.M. Cathepsin L-like Cysteine Proteinase Genes Are Associated with the Development and Pathogenicity of Pine Wood Nematode, Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2019, 20, 215. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, X.; Zhou, L.; Hu, J.; Guo, K. Application of RNA Interference in the Pinewood Nematode, Bursaphelenchus xylophilus. J. Vis. Exp. 2022, 181, e63645. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, X.; Li, Y.; Zhang, J.; Zhang, Z.; Wu, H. Cloning arginine kinase gene and its RNAi in Bursaphelenchus xylophilus causing pine wilt disease. Eur. J. Plant. Pathol. 2012, 134, 521–532. [Google Scholar] [CrossRef]
- Garcia, R.A.; Macedo, L.L.P.; Nascimento, D.C.; Gillet, F.X.; Moreira-Pinto, C.E.; Faheem, M.; Basso, A.M.M.; Silva, M.C.M.; Grossi-de-Sa, M.F. Nucleases as a barrier to gene silencing in the cotton boll weevil, Anthonomus grandis. PLoS ONE 2017, 12, e0189600. [Google Scholar] [CrossRef]
- Fan, Y.H.; Song, H.F.; Abbas, M.; Wang, Y.L.; Li, T.; Ma, E.B.; Cooper, A.M.W.; Silver, K.; Zhu, K.Y.; Zhang, J.Z. A dsRNA-degrading nuclease (dsRNase2) limits RNAi efficiency in the Asian corn borer (Ostrinia furnacalis). Insect Sci. 2021, 28, 1677–1689. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.S.; Ahn, S.J.; Flinn, C.M.; Choi, M.Y. Identification and functional analysis of dsRNases in spotted-wing drosophila, Drosophila suzukii. Arch. Insect Biochem. Physiol. 2021, 107, e21822. [Google Scholar] [CrossRef] [PubMed]
- Cooper, A.M.; Silver, K.; Zhang, J.; Park, Y.; Zhu, K.Y. Molecular mechanisms influencing efficiency of RNA interference in insects. Pest Manag. Sci. 2019, 75, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Fu, W.; Sheng, C.; Han, Z. Biochemical Comparison of dsRNA Degrading Nucleases in Four Different Insects. Front. Physiol. 2018, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Vieira, P.; Shao, J.; Vijayapalani, P.; Maier, T.R.; Pellegrin, C.; Akker, S.E.; Baum, T.J.; Nemchinov, L.G. A new esophageal gland transcriptome reveals signatures of large scale de novo effector birth in the root lesion nematode Pratylenchus penetrans. BMC Genom. 2020, 21, 738. [Google Scholar] [CrossRef]
- Shinya, R.; Morisaka, H.; Kikuchi, T.; Takeuchi, Y.; Ueda, M.; Futai, K. Secretome Analysis of the Pine Wood Nematode Bursaphelenchus xylophilus Reveals the Tangled Roots of Parasitism and Its Potential for Molecular Mimicry. PLoS ONE 2013, 8, e67377. [Google Scholar] [CrossRef]
- Maule, A.G.; McVeigh, P.; Dalzell, J.J.; Atkinson, L.; Mousley, A.; Marks, N.J. An eye on RNAi in nematode parasites. Trends Parasitol. 2011, 27, 505–513. [Google Scholar] [CrossRef]
- Britton, C.; Samarasinghe, B.; Knox, D.P. Ups and downs of RNA interference in parasitic nematodes. Exp. Parasitol. 2011, 132, 56–61. [Google Scholar] [CrossRef]
- Cooper, A.M.W.; Song, H.; Shi, X.; Yu, Z.; Lorenzen, M.; Silver, K.; Zhang, J.; Zhu, K.Y. Molecular Characterizations of Double-Stranded RNA Degrading Nuclease Genes from Ostrinia nubilalis. Insects 2020, 11, E652. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Eck, J.V.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef]
- Spit, J.; Philips, A.; Wynant, N.; Santos, D.; Plaetinck, G.; Broeck, J.V. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017, 81, 103–116. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, K.; Chen, J.; Wang, J.; Zhang, H.; Ze, L.; Zhu, G.; Zhao, C.; Xiao, H.; Han, Z. Identification of a double-stranded RNA-degrading nuclease influencing both ingestion and injection RNA interference efficiency in the red flour beetle Tribolium castaneum. Insect Biochem. Mol. Biol. 2020, 125, 103440. [Google Scholar] [CrossRef] [PubMed]
- Giesbrecht, D.; Heschuk, D.; Wiens, I.; Boguski, D.; LaChance, P.; Whyard, S. RNA Interference Is Enhanced by Knockdown of double-stranded RNases in the Yellow Fever Mosquito Aedes aegypti. Insects 2020, 11, E327. [Google Scholar] [CrossRef] [PubMed]
- The C. Elegans Sequencing Consortium. Genome Sequence of the Nematode C. elegan: A Platform for Investigating Biology. Science 1998, 282, 2012–2018. [Google Scholar] [CrossRef]
- Xu, L.; Xu, S.; Sun, L.; Zhang, Y.; Luo, J.; Bock, R.; Zhang, J. Synergistic action of the gut microbiota in environmental RNA interference in a leaf beetle. Microbiome 2021, 9, 98. [Google Scholar] [CrossRef]
- Wynant, N.; Santos, D.; Verdonck, R.; Spit, J.; Wielendaele, P.V.; Broeck, J.V. Identification, functional characterization and phylogenetic analysis of double stranded RNA degrading enzymes present in the gut of the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2014, 46, 1–8. [Google Scholar] [CrossRef]
- Véleza, A.M.; Fishilevich, E. The mysteries of insect RNAi: A focus on dsRNA uptake and transport. Pestic. Biochem. Phys. 2018, 151, 25–31. [Google Scholar] [CrossRef]
- Cardoso, J.M.S.; Fonseca, L.; Gomes, P.; Egas, C.; Abrantes, I. Molecular characterization and functional analysis of a calponin gene from the pinewood nematode. For. Pathol. 2015, 45, 467–473. [Google Scholar] [CrossRef]
- Wang, M.; Song, S.; Wang, X.; Tian, Y.; Zhang, Z.; Hu, S.; Cheng, X. Double Stranded RNA Expression of Bx-apa-2 and Bx-apm-2 Genes in Fusarium oxysporum Mediated RNAi for Bursaphelenchus xylophilus. J. Southwest For. Univ. 2022, 42, 72–79. [Google Scholar] [CrossRef]
- Terami, H.; Williams, B.D.; Kitamura, S.; Sakube, Y.; Matsumoto, S.; Doi, S.; Obinata, T.; Kagawa, H. Genomic Organization, Expression, and Analysis of the Troponin C Gene pat-10 of Caenorhabditis elegans. J. Cell Biol. 1999, 146, 193–202. [Google Scholar] [CrossRef]




| Name | Accession Number | ORF Length | Amino Acid Sequence Length | Signal Peptide | Theoretical pI/Mw |
|---|---|---|---|---|---|
| BxyNuc1 | OP482151 | 993 | 330 | 20–21.VAS-KS | 5.38/37,069.64 |
| BxyNuc2 | OP482152 | 1068 | 355 | 21–22.AEA-AG | 6.37/40,630.98 |
| BxyNuc3 | OP482153 | 1035 | 344 | 20–21.VAA-SH | 7.26/39,173.47 |
| BxyNuc4 | OP482154 | 933 | 310 | 17–18.GNA-AI | 8.38/34,944.08 |
| BxyNuc5 | OP482155 | 890 | 296 | 15–16.TSA-QI | 8.85/33,692.55 |
| BxyNuc6 | OP482156 | 927 | 308 | 18–19.ICG-TQ | 9.12/35,178.94 |
| BxyNuc7 | OP482157 | 963 | 320 | 19–20.AQG-IG | 5.72/36,009.80 |
| BxyNuc8 | OP482158 | 933 | 310 | 17–18.GNA-VI | 8.58/34,493.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Li, Y.; Li, D.; Zhang, W.; Wang, X.; Wen, X.; Liu, Z.; Feng, Y.; Zhang, X. Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus. Int. J. Mol. Sci. 2022, 23, 12278. https://doi.org/10.3390/ijms232012278
Wang R, Li Y, Li D, Zhang W, Wang X, Wen X, Liu Z, Feng Y, Zhang X. Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus. International Journal of Molecular Sciences. 2022; 23(20):12278. https://doi.org/10.3390/ijms232012278
Chicago/Turabian StyleWang, Ruijiong, Yongxia Li, Dongzhen Li, Wei Zhang, Xuan Wang, Xiaojian Wen, Zhenkai Liu, Yuqian Feng, and Xingyao Zhang. 2022. "Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus" International Journal of Molecular Sciences 23, no. 20: 12278. https://doi.org/10.3390/ijms232012278
APA StyleWang, R., Li, Y., Li, D., Zhang, W., Wang, X., Wen, X., Liu, Z., Feng, Y., & Zhang, X. (2022). Identification of the Extracellular Nuclease Influencing Soaking RNA Interference Efficiency in Bursaphelenchus xylophilus. International Journal of Molecular Sciences, 23(20), 12278. https://doi.org/10.3390/ijms232012278






