Single-Cell Analysis to Better Understand the Mechanisms Involved in MS
Abstract
1. Introduction
2. Methods: Search Strategies and Inclusion Criteria
3. Single-Cell Technologies: Innovative Methods for Analysis
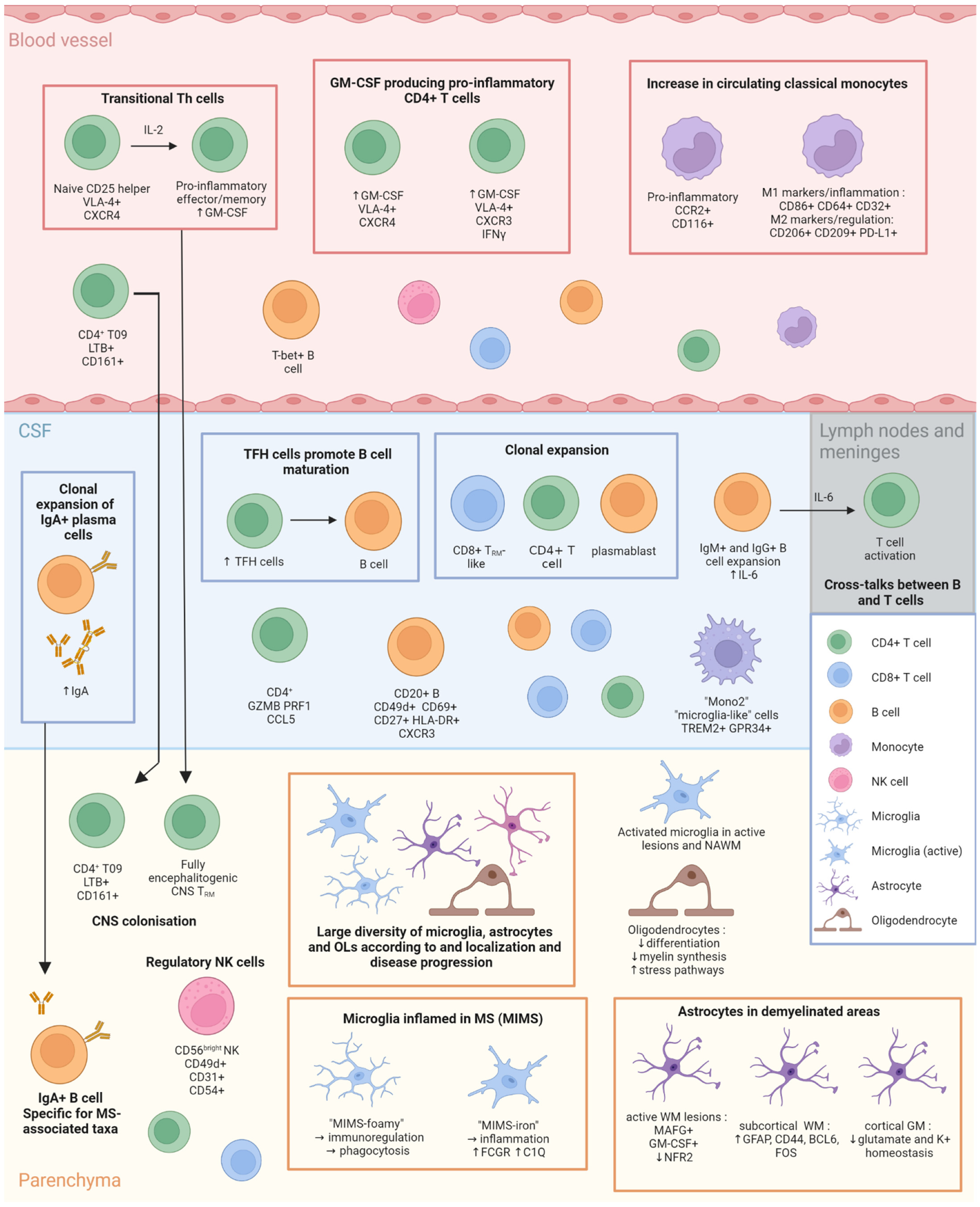
4. Latest Findings in MS Cellular Immunology
4.1. Lymphocytes
4.2. Monocytes
4.3. Microglia
4.4. Oligodendrocytes and OPC
4.5. Astrocytes
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Reich, D.S.; Lucchinetti, C.F.; Calabresi, P.A. Multiple Sclerosis. N. Engl. J. Med. 2018, 378, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Beltrán, E.; Gerdes, L.A.; Hansen, J.; Flierl-Hecht, A.; Krebs, S.; Blum, H.; Ertl-Wagner, B.; Barkhof, F.; Kümpfel, T.; Hohlfeld, R.; et al. Early Adaptive Immune Activation Detected in Monozygotic Twins with Prodromal Multiple Sclerosis. J. Clin. Investig. 2019, 129, 4758–4768. [Google Scholar] [CrossRef] [PubMed]
- Gerdes, L.A.; Janoschka, C.; Eveslage, M.; Mannig, B.; Wirth, T.; Schulte-Mecklenbeck, A.; Lauks, S.; Glau, L.; Gross, C.C.; Tolosa, E.; et al. Immune Signatures of Prodromal Multiple Sclerosis in Monozygotic Twins. Proc. Natl. Acad. Sci. USA 2020, 117, 21546–21556. [Google Scholar] [CrossRef] [PubMed]
- Alfredsson, L.; Olsson, T. Lifestyle and Environmental Factors in Multiple Sclerosis. Cold Spring Harb. Perspect. Med. 2019, 9, a028944. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Baranzini, S.E.; Geurts, J.; Hemmer, B.; Ciccarelli, O. Multiple Sclerosis. Lancet 2018, 391, 1622–1636. [Google Scholar] [CrossRef]
- Bjornevik, K.; Cortese, M.; Healy, B.C.; Kuhle, J.; Mina, M.J.; Leng, Y.; Elledge, S.J.; Niebuhr, D.W.; Scher, A.I.; Munger, K.L.; et al. Longitudinal Analysis Reveals High Prevalence of Epstein-Barr Virus Associated with Multiple Sclerosis. Science 2022, 375, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Hacohen, Y.; Ciccarelli, O. New Evidence for EBV Infection as a Cause of Multiple Sclerosis. Neurology 2022, 98, 605–606. [Google Scholar] [CrossRef]
- Lanz, T.V.; Brewer, R.C.; Ho, P.P.; Moon, J.-S.; Jude, K.M.; Fernandez, D.; Fernandes, R.A.; Gomez, A.M.; Nadj, G.-S.; Bartley, C.M.; et al. Clonally Expanded B Cells in Multiple Sclerosis Bind EBV EBNA1 and GlialCAM. Nature 2022, 603, 321–327. [Google Scholar] [CrossRef]
- Akaishi, T.; Takahashi, T.; Nakashima, I. Peripheral Blood Monocyte Count at Onset May Affect the Prognosis in Multiple Sclerosis. J. Neuroimmunol. 2018, 319, 37–40. [Google Scholar] [CrossRef]
- Vogel, D.Y.S.; Heijnen, P.D.A.M.; Breur, M.; de Vries, H.E.; Tool, A.T.J.; Amor, S.; Dijkstra, C.D. Macrophages Migrate in an Activation-Dependent Manner to Chemokines Involved in Neuroinflammation. J. Neuroinflammation 2014, 11, 23. [Google Scholar] [CrossRef]
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Pollard, J.W. Trophic Macrophages in Development and Disease. Nat. Rev. Immunol. 2009, 9, 259–270. [Google Scholar] [CrossRef]
- Booss, J.; Esiri, M.M.; Tourtellotte, W.W.; Mason, D.Y. Immunohistological Analysis of T Lymphocyte Subsets in the Central Nervous System in Chronic Progressive Multiple Sclerosis. J. Neurol. Sci. 1983, 62, 219–232. [Google Scholar] [CrossRef]
- Crawford, M.P.; Yan, S.X.; Ortega, S.B.; Mehta, R.S.; Hewitt, R.E.; Price, D.A.; Stastny, P.; Douek, D.C.; Koup, R.A.; Racke, M.K.; et al. High Prevalence of Autoreactive, Neuroantigen-Specific CD8+ T Cells in Multiple Sclerosis Revealed by Novel Flow Cytometric Assay. Blood 2004, 103, 4222–4231. [Google Scholar] [CrossRef]
- Salou, M.; Nicol, B.; Garcia, A.; Laplaud, D.-A. Involvement of CD8(+) T Cells in Multiple Sclerosis. Front. Immunol. 2015, 6, 604. [Google Scholar] [CrossRef]
- Hohlfeld, R.; Dornmair, K.; Meinl, E.; Wekerle, H. The Search for the Target Antigens of Multiple Sclerosis, Part 2: CD8+ T Cells, B Cells, and Antibodies in the Focus of Reverse-Translational Research. Lancet Neurol. 2016, 15, 317–331. [Google Scholar] [CrossRef]
- Ifergan, I.; Kebir, H.; Alvarez, J.I.; Marceau, G.; Bernard, M.; Bourbonnière, L.; Poirier, J.; Duquette, P.; Talbot, P.J.; Arbour, N.; et al. Central Nervous System Recruitment of Effector Memory CD8+ T Lymphocytes during Neuroinflammation Is Dependent on A4 Integrin. Brain 2011, 134, 3560–3577. [Google Scholar] [CrossRef]
- Nicol, B.; Salou, M.; Vogel, I.; Garcia, A.; Dugast, E.; Morille, J.; Kilens, S.; Charpentier, E.; Donnart, A.; Nedellec, S.; et al. An Intermediate Level of CD161 Expression Defines a Novel Activated, Inflammatory, and Pathogenic Subset of CD8+ T Cells Involved in Multiple Sclerosis. J. Autoimmun. 2018, 88, 61–74. [Google Scholar] [CrossRef]
- Van Nierop, G.P.; van Luijn, M.M.; Michels, S.S.; Melief, M.-J.; Janssen, M.; Langerak, A.W.; Ouwendijk, W.J.D.; Hintzen, R.Q.; Verjans, G.M.G.M. Phenotypic and Functional Characterization of T Cells in White Matter Lesions of Multiple Sclerosis Patients. Acta Neuropathol. 2017, 134, 383–401. [Google Scholar] [CrossRef]
- Salou, M.; Nicol, B.; Garcia, A.; Baron, D.; Michel, L.; Elong-Ngono, A.; Hulin, P.; Nedellec, S.; Jacq-Foucher, M.; Le Frère, F.; et al. Neuropathologic, Phenotypic and Functional Analyses of Mucosal Associated Invariant T Cells in Multiple Sclerosis. Clin. Immunol. 2016, 166–167, 1–11. [Google Scholar] [CrossRef]
- Karandikar, N.J.; Crawford, M.P.; Yan, X.; Ratts, R.B.; Brenchley, J.M.; Ambrozak, D.R.; Lovett-Racke, A.E.; Frohman, E.M.; Stastny, P.; Douek, D.C.; et al. Glatiramer Acetate (Copaxone) Therapy Induces CD8(+) T Cell Responses in Patients with Multiple Sclerosis. J. Clin. Investig. 2002, 109, 641–649. [Google Scholar] [CrossRef]
- Li, J.; Zaslavsky, M.; Su, Y.; Guo, J.; Sikora, M.J.; van Unen, V.; Christophersen, A.; Chiou, S.-H.; Chen, L.; Li, J.; et al. KIR+CD8+ T Cells Suppress Pathogenic T Cells and Are Active in Autoimmune Diseases and COVID-19. Science 2022, 376, eabi9591. [Google Scholar] [CrossRef]
- Saligrama, N.; Zhao, F.; Sikora, M.J.; Serratelli, W.S.; Fernandes, R.A.; Louis, D.M.; Yao, W.; Ji, X.; Idoyaga, J.; Mahajan, V.B.; et al. Opposing T Cell Responses in Experimental Autoimmune Encephalomyelitis. Nature 2019, 572, 481–487. [Google Scholar] [CrossRef]
- Tennakoon, D.K.; Mehta, R.S.; Ortega, S.B.; Bhoj, V.; Racke, M.K.; Karandikar, N.J. Therapeutic Induction of Regulatory, Cytotoxic CD8+ T Cells in Multiple Sclerosis. J. Immunol. 2006, 176, 7119–7129. [Google Scholar] [CrossRef]
- Petereit, H.F.; Richter, N.; Pukrop, R.; Bamborschke, S. Interferon Gamma Production in Blood Lymphocytes Correlates with Disability Score in Multiple Sclerosis Patients. Mult. Scler. 2000, 6, 19–23. [Google Scholar] [CrossRef]
- Tzartos, J.S.; Friese, M.A.; Craner, M.J.; Palace, J.; Newcombe, J.; Esiri, M.M.; Fugger, L. Interleukin-17 Production in Central Nervous System-Infiltrating T Cells and Glial Cells Is Associated with Active Disease in Multiple Sclerosis. Am. J. Pathol. 2008, 172, 146–155. [Google Scholar] [CrossRef]
- Tahmasebinia, F.; Pourgholaminejad, A. The Role of Th17 Cells in Auto-Inflammatory Neurological Disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 2017, 79, 408–416. [Google Scholar] [CrossRef]
- Murphy, A.C.; Lalor, S.J.; Lynch, M.A.; Mills, K.H.G. Infiltration of Th1 and Th17 Cells and Activation of Microglia in the CNS during the Course of Experimental Autoimmune Encephalomyelitis. Brain Behav. Immun. 2010, 24, 641–651. [Google Scholar] [CrossRef]
- Haque, R.; Kim, Y.; Park, K.; Jang, H.; Kim, S.Y.; Lee, H.; Kim, H.J. Altered Distributions in Circulating Follicular Helper and Follicular Regulatory T Cells Accountable for Imbalanced Cytokine Production in Multiple Sclerosis. Clin. Exp. Immunol. 2021, 205, 75–88. [Google Scholar] [CrossRef]
- Huber, J.E.; Chang, Y.; Meinl, I.; Kümpfel, T.; Meinl, E.; Baumjohann, D. Fingolimod Profoundly Reduces Frequencies and Alters Subset Composition of Circulating T Follicular Helper Cells in Multiple Sclerosis Patients. J. Immunol. 2020, 204, 1101–1110. [Google Scholar] [CrossRef] [PubMed]
- Schafflick, D.; Xu, C.A.; Hartlehnert, M.; Cole, M.; Schulte-Mecklenbeck, A.; Lautwein, T.; Wolbert, J.; Heming, M.; Meuth, S.G.; Kuhlmann, T.; et al. Integrated Single Cell Analysis of Blood and Cerebrospinal Fluid Leukocytes in Multiple Sclerosis. Nat. Commun. 2020, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Morille, J.; Mandon, M.; Rodriguez, S.; Roulois, D.; Leonard, S.; Garcia, A.; Wiertlewski, S.; Le Page, E.; Berthelot, L.; Nicot, A.; et al. Multiple sclerosis cerebrospinal fluid is enriched with follicular T cells displaying a Th1/Eomes signature. Neurol. Neuroimmunol. Neuroinflamm. 2022, in press. [Google Scholar]
- Haas, J.; Hug, A.; Viehöver, A.; Fritzsching, B.; Falk, C.S.; Filser, A.; Vetter, T.; Milkova, L.; Korporal, M.; Fritz, B.; et al. Reduced Suppressive Effect of CD4+CD25high Regulatory T Cells on the T Cell Immune Response against Myelin Oligodendrocyte Glycoprotein in Patients with Multiple Sclerosis. Eur. J. Immunol. 2005, 35, 3343–3352. [Google Scholar] [CrossRef]
- Venken, K.; Hellings, N.; Broekmans, T.; Hensen, K.; Rummens, J.-L.; Stinissen, P. Natural Naive CD4+CD25+CD127low Regulatory T Cell (Treg) Development and Function Are Disturbed in Multiple Sclerosis Patients: Recovery of Memory Treg Homeostasis during Disease Progression. J. Immunol. 2008, 180, 6411–6420. [Google Scholar] [CrossRef]
- Absinta, M.; Maric, D.; Gharagozloo, M.; Garton, T.; Smith, M.D.; Jin, J.; Fitzgerald, K.C.; Song, A.; Liu, P.; Lin, J.-P.; et al. A Lymphocyte-Microglia-Astrocyte Axis in Chronic Active Multiple Sclerosis. Nature 2021, 597, 709–714. [Google Scholar] [CrossRef]
- Böttcher, C.; van der Poel, M.; Fernández-Zapata, C.; Schlickeiser, S.; Leman, J.K.H.; Hsiao, C.-C.; Mizee, M.R.; Adelia; Vincenten, M.C.J.; Kunkel, D.; et al. Single-Cell Mass Cytometry Reveals Complex Myeloid Cell Composition in Active Lesions of Progressive Multiple Sclerosis. Acta Neuropathol. Commun. 2020, 8, 136. [Google Scholar] [CrossRef]
- Couloume, L.; Ferrant, J.; Le Gallou, S.; Mandon, M.; Jean, R.; Bescher, N.; Zephir, H.; Edan, G.; Thouvenot, E.; Ruet, A.; et al. Mass Cytometry Identifies Expansion of T-Bet+ B Cells and CD206+ Monocytes in Early Multiple Sclerosis. Front. Immunol. 2021, 12, 653577. [Google Scholar] [CrossRef]
- Diebold, M.; Galli, E.; Kopf, A.; Sanderson, N.S.R.; Callegari, I.; Benkert, P.; Gonzalo Núñez, N.; Ingelfinger, F.; Herms, S.; Cichon, S.; et al. High-Dimensional Immune Profiling Identifies a Biomarker to Monitor Dimethyl Fumarate Response in Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 2022, 119, e2205042119. [Google Scholar] [CrossRef]
- Galli, E.; Hartmann, F.J.; Schreiner, B.; Ingelfinger, F.; Arvaniti, E.; Diebold, M.; Mrdjen, D.; van der Meer, F.; Krieg, C.; Nimer, F.A.; et al. GM-CSF and CXCR4 Define a T Helper Cell Signature in Multiple Sclerosis. Nat. Med. 2019, 25, 1290–1300. [Google Scholar] [CrossRef]
- Ingelfinger, F.; Gerdes, L.A.; Kavaka, V.; Krishnarajah, S.; Friebel, E.; Galli, E.; Zwicky, P.; Furrer, R.; Peukert, C.; Dutertre, C.-A.; et al. Twin Study Reveals Non-Heritable Immune Perturbations in Multiple Sclerosis. Nature 2022, 603, 152–158. [Google Scholar] [CrossRef]
- Jäkel, S.; Agirre, E.; Mendanha Falcão, A.; van Bruggen, D.; Lee, K.W.; Knuesel, I.; Malhotra, D.; Ffrench-Constant, C.; Williams, A.; Castelo-Branco, G. Altered Human Oligodendrocyte Heterogeneity in Multiple Sclerosis. Nature 2019, 566, 543–547. [Google Scholar] [CrossRef]
- Johansson, D.; Rauld, C.; Roux, J.; Regairaz, C.; Galli, E.; Callegari, I.; Raad, L.; Waldt, A.; Cuttat, R.; Roma, G.; et al. Mass Cytometry of CSF Identifies an MS-Associated B-Cell Population. Neurol. Neuroimmunol. Neuroinflamm. 2021, 8, e943. [Google Scholar] [CrossRef]
- Kaufmann, M.; Evans, H.; Schaupp, A.-L.; Engler, J.B.; Kaur, G.; Willing, A.; Kursawe, N.; Schubert, C.; Attfield, K.E.; Fugger, L.; et al. Identifying CNS-Colonizing T Cells as Potential Therapeutic Targets to Prevent Progression of Multiple Sclerosis. Med 2021, 2, 296–312.e8. [Google Scholar] [CrossRef]
- Kihara, Y.; Zhu, Y.; Jonnalagadda, D.; Romanow, W.; Palmer, C.; Siddoway, B.; Rivera, R.; Dutta, R.; Trapp, B.D.; Chun, J. Single-Nucleus RNA-Seq of Normal-Appearing Brain Regions in Relapsing-Remitting vs. Secondary Progressive Multiple Sclerosis: Implications for the Efficacy of Fingolimod. Front. Cell Neurosci. 2022, 16, 918041. [Google Scholar] [CrossRef]
- Masuda, T.; Sankowski, R.; Staszewski, O.; Böttcher, C.; Amann, L.; Sagar; Scheiwe, C.; Nessler, S.; Kunz, P.; van Loo, G.; et al. Spatial and Temporal Heterogeneity of Mouse and Human Microglia at Single-Cell Resolution. Nature 2019, 566, 388–392. [Google Scholar] [CrossRef]
- Pröbstel, A.-K.; Zhou, X.; Baumann, R.; Wischnewski, S.; Kutza, M.; Rojas, O.L.; Sellrie, K.; Bischof, A.; Kim, K.; Ramesh, A.; et al. Gut Microbiota–Specific IgA + B Cells Traffic to the CNS in Active Multiple Sclerosis. Sci. Immunol. 2020, 5, eabc7191. [Google Scholar] [CrossRef]
- Ramaglia, V.; Sheikh-Mohamed, S.; Legg, K.; Park, C.; Rojas, O.L.; Zandee, S.; Fu, F.; Ornatsky, O.; Swanson, E.C.; Pitt, D.; et al. Multiplexed Imaging of Immune Cells in Staged Multiple Sclerosis Lesions by Mass Cytometry. eLife 2019, 8, e48051. [Google Scholar] [CrossRef]
- Ramesh, A.; Schubert, R.D.; Greenfield, A.L.; Dandekar, R.; Loudermilk, R.; Sabatino, J.J.; Koelzer, M.T.; Tran, E.B.; Koshal, K.; Kim, K.; et al. A Pathogenic and Clonally Expanded B Cell Transcriptome in Active Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 2020, 117, 22932–22943. [Google Scholar] [CrossRef]
- Rodríguez-Lorenzo, S.; van Olst, L.; Rodriguez-Mogeda, C.; Kamermans, A.; van der Pol, S.M.; Rodríguez, E.; Kooij, G.; de Vries, H.E. Single-Cell Profiling Reveals Periventricular CD56bright NK Cell Accumulation in Multiple Sclerosis. eLife 2022, 11, e73849. [Google Scholar] [CrossRef]
- Schirmer, L.; Velmeshev, D.; Holmqvist, S.; Kaufmann, M.; Werneburg, S.; Jung, D.; Vistnes, S.; Stockley, J.H.; Young, A.; Steindel, M.; et al. Neuronal Vulnerability and Multilineage Diversity in Multiple Sclerosis. Nature 2019, 573, 75–82. [Google Scholar] [CrossRef]
- Wheeler, M.A.; Clark, I.C.; Tjon, E.C.; Li, Z.; Zandee, S.E.J.; Couturier, C.P.; Watson, B.R.; Scalisi, G.; Alkwai, S.; Rothhammer, V.; et al. MAFG-Driven Astrocytes Promote CNS Inflammation. Nature 2020, 578, 593–599. [Google Scholar] [CrossRef]
- Kharchenko, P.V.; Silberstein, L.; Scadden, D.T. Bayesian Approach to Single-Cell Differential Expression Analysis. Nat. Methods 2014, 11, 740–742. [Google Scholar] [CrossRef]
- Luecken, M.D.; Büttner, M.; Chaichoompu, K.; Danese, A.; Interlandi, M.; Mueller, M.F.; Strobl, D.C.; Zappia, L.; Dugas, M.; Colomé-Tatché, M.; et al. Benchmarking Atlas-Level Data Integration in Single-Cell Genomics. Nat. Methods 2022, 19, 41–50. [Google Scholar] [CrossRef]
- Spitzer, M.H.; Nolan, G.P. Mass Cytometry: Single Cells, Many Features. Cell 2016, 165, 780–791. [Google Scholar] [CrossRef]
- Leipold, M.D.; Newell, E.W.; Maecker, H.T. Multiparameter Phenotyping of Human PBMCs Using Mass Cytometry. In Immunosenescence; Shaw, A.C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1343, pp. 81–95. ISBN 978-1-4939-2962-7. [Google Scholar]
- Pappalardo, J.L.; Zhang, L.; Pecsok, M.K.; Perlman, K.; Zografou, C.; Raddassi, K.; Abulaban, A.; Krishnaswamy, S.; Antel, J.; van Dijk, D.; et al. Transcriptomic and Clonal Characterization of T Cells in the Human Central Nervous System. Sci. Immunol. 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Noster, R.; Riedel, R.; Mashreghi, M.-F.; Radbruch, H.; Harms, L.; Haftmann, C.; Chang, H.-D.; Radbruch, A.; Zielinski, C.E. IL-17 and GM-CSF Expression Are Antagonistically Regulated by Human T Helper Cells. Sci. Transl. Med. 2014, 6, 241ra80. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, F.J.; Khademi, M.; Aram, J.; Ammann, S.; Kockum, I.; Constantinescu, C.; Gran, B.; Piehl, F.; Olsson, T.; Codarri, L.; et al. Multiple Sclerosis-Associated IL2RA Polymorphism Controls GM-CSF Production in Human TH Cells. Nat. Commun. 2014, 5, 5056. [Google Scholar] [CrossRef] [PubMed]
- Nurieva, R.I.; Chung, Y.; Martinez, G.J.; Yang, X.O.; Tanaka, S.; Matskevitch, T.D.; Wang, Y.-H.; Dong, C. Bcl6 Mediates the Development of T Follicular Helper Cells. Science 2009, 325, 1001–1005. [Google Scholar] [CrossRef] [PubMed]
- Van Langelaar, J.; Rijvers, L.; Janssen, M.; Wierenga-Wolf, A.F.; Melief, M.; Siepman, T.A.; de Vries, H.E.; Unger, P.A.; van Ham, S.M.; Hintzen, R.Q.; et al. Induction of Brain-infiltrating T-bet–Expressing B Cells in Multiple Sclerosis. Ann. Neurol. 2019, 86, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Mimpen, M.; Smolders, J.; Hupperts, R.; Damoiseaux, J. Natural Killer Cells in Multiple Sclerosis: A Review. Immunol. Lett. 2020, 222, 1–11. [Google Scholar] [CrossRef]
- Esaulova, E.; Cantoni, C.; Shchukina, I.; Zaitsev, K.; Bucelli, R.C.; Wu, G.F.; Artyomov, M.N.; Cross, A.H.; Edelson, B.T. Single-Cell RNA-Seq Analysis of Human CSF Microglia and Myeloid Cells in Neuroinflammation. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7, e732. [Google Scholar] [CrossRef]
- Farhadian, S.F.; Mehta, S.S.; Zografou, C.; Robertson, K.; Price, R.W.; Pappalardo, J.; Chiarella, J.; Hafler, D.A.; Spudich, S.S. Single-Cell RNA Sequencing Reveals Microglia-like Cells in Cerebrospinal Fluid during Virologically Suppressed HIV. JCI Insight 2018, 3, 121718. [Google Scholar] [CrossRef]
- Haas, J.; Schwarz, A.; Korporal-Kuhnke, M.; Jarius, S.; Wildemann, B. Myeloid Dendritic Cells Exhibit Defects in Activation and Function in Patients with Multiple Sclerosis. J. Neuroimmunol. 2016, 301, 53–60. [Google Scholar] [CrossRef]
- Li, Q.; Barres, B.A. Microglia and Macrophages in Brain Homeostasis and Disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef]
- Lawson, L.J.; Perry, V.H.; Dri, P.; Gordon, S. Heterogeneity in the Distribution and Morphology of Microglia in the Normal Adult Mouse Brain. Neuroscience 1990, 39, 151–170. [Google Scholar] [CrossRef]
- Schmid, C.D.; Sautkulis, L.N.; Danielson, P.E.; Cooper, J.; Hasel, K.W.; Hilbush, B.S.; Sutcliffe, J.G.; Carson, M.J. Heterogeneous Expression of the Triggering Receptor Expressed on Myeloid Cells-2 on Adult Murine Microglia. J. Neurochem. 2002, 83, 1309–1320. [Google Scholar] [CrossRef]
- Plemel, J.R.; Stratton, J.A.; Michaels, N.J.; Rawji, K.S.; Zhang, E.; Sinha, S.; Baaklini, C.S.; Dong, Y.; Ho, M.; Thorburn, K.; et al. Microglia Response Following Acute Demyelination Is Heterogeneous and Limits Infiltrating Macrophage Dispersion. Sci. Adv. 2020, 6, eaay6324. [Google Scholar] [CrossRef]
- Olah, M.; Patrick, E.; Villani, A.-C.; Xu, J.; White, C.C.; Ryan, K.J.; Piehowski, P.; Kapasi, A.; Nejad, P.; Cimpean, M.; et al. A Transcriptomic Atlas of Aged Human Microglia. Nat. Commun. 2018, 9, 539. [Google Scholar] [CrossRef]
- Böttcher, C.; Schlickeiser, S.; Sneeboer, M.A.M.; Kunkel, D.; Knop, A.; Paza, E.; Fidzinski, P.; Kraus, L.; Snijders, G.J.L.; Kahn, R.S.; et al. Human Microglia Regional Heterogeneity and Phenotypes Determined by Multiplexed Single-Cell Mass Cytometry. Nat. Neurosci. 2019, 22, 78–90. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Fransen, N.L.; Touil, H.; Michailidou, I.; Huitinga, I.; Gommerman, J.L.; Bar-Or, A.; Ramaglia, V. Accumulation of Meningeal Lymphocytes Correlates with White Matter Lesion Activity in Progressive Multiple Sclerosis. JCI Insight 2022, 7, e151683. [Google Scholar] [CrossRef]
- Fransen, N.L.; Hsiao, C.-C.; van der Poel, M.; Engelenburg, H.J.; Verdaasdonk, K.; Vincenten, M.C.J.; Remmerswaal, E.B.M.; Kuhlmann, T.; Mason, M.R.J.; Hamann, J.; et al. Tissue-Resident Memory T Cells Invade the Brain Parenchyma in Multiple Sclerosis White Matter Lesions. Brain 2020, 143, 1714–1730. [Google Scholar] [CrossRef]
- Krajnc, N.; Berger, T.; Bsteh, G. Measuring Treatment Response in Progressive Multiple Sclerosis-Considerations for Adapting to an Era of Multiple Treatment Options. Biomolecules 2021, 11, 1342. [Google Scholar] [CrossRef]
- Crawford, A.H.; Chambers, C.; Franklin, R.J.M. Remyelination: The True Regeneration of the Central Nervous System. J. Comp. Pathol. 2013, 149, 242–254. [Google Scholar] [CrossRef]
- Marques, S.; Zeisel, A.; Codeluppi, S.; van Bruggen, D.; Mendanha Falcão, A.; Xiao, L.; Li, H.; Häring, M.; Hochgerner, H.; Romanov, R.A.; et al. Oligodendrocyte Heterogeneity in the Mouse Juvenile and Adult Central Nervous System. Science 2016, 352, 1326–1329. [Google Scholar] [CrossRef]
- Falcão, A.M.; van Bruggen, D.; Marques, S.; Meijer, M.; Jäkel, S.; Agirre, E.; Samudyata; Floriddia, E.M.; Vanichkina, D.P.; Ffrench-Constant, C.; et al. Disease-Specific Oligodendrocyte Lineage Cells Arise in Multiple Sclerosis. Nat. Med. 2018, 24, 1837–1844. [Google Scholar] [CrossRef] [PubMed]
- Garland, E.F.; Hartnell, I.J.; Boche, D. Microglia and Astrocyte Function and Communication: What Do We Know in Humans? Front. Neurosci. 2022, 16, 824888. [Google Scholar] [CrossRef] [PubMed]
- Liddelow, S.A.; Barres, B.A. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity 2017, 46, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Batiuk, M.Y.; Martirosyan, A.; Wahis, J.; de Vin, F.; Marneffe, C.; Kusserow, C.; Koeppen, J.; Viana, J.F.; Oliveira, J.F.; Voet, T.; et al. Identification of Region-Specific Astrocyte Subtypes at Single Cell Resolution. Nat. Commun. 2020, 11, 1220. [Google Scholar] [CrossRef]
- Lake, B.B.; Chen, S.; Sos, B.C.; Fan, J.; Kaeser, G.E.; Yung, Y.C.; Duong, T.E.; Gao, D.; Chun, J.; Kharchenko, P.V.; et al. Integrative Single-Cell Analysis of Transcriptional and Epigenetic States in the Human Adult Brain. Nat. Biotechnol. 2018, 36, 70–80. [Google Scholar] [CrossRef]
| Authors | Single-Cell Technology | Samples | Studied Cells | Key Findings |
|---|---|---|---|---|
| Absinta et al. [36] | snRNAseq | Frozen brain tissue from progressive MS patients (n = 5) and age- and sex-matched non-affected, non-demented controls (n = 3) | Microglia, immune cells, lymphocytes, oligodendrocytes, OPC, astrocytes, neurons and vascular cells | Large diversity of cells. Microglia inflamed in MS (MIMS) are divided into 2 subsets: “MIMS-foamy” and “MIMS-iron”. Strong inter-communication between cell-subsets and especially with astrocytes. Importance of the C1q component, central in microglia activation and upregulated in MIMS cells on the edge of chronic active lesions. |
| Beltràn et al. [2] | scRNAseq | CSF samples and PBMCs from clinically discordant monozygotic twin pairs (n = 4), co-twins with SCNI from whom CSF of the corresponding MS twin was not available (n = 4), subjects with encephalitis (n = 2), NIC controls with intracranial idiopathic hypertension (n = 2) | Immune cells in CSF, mainly lymphocytes | Clonal expansion of CD8+ T cells, plasmablasts and CD4+ T cells. Clonally expanded T cells showed tissue-resident memory phenotype. |
| Böttcher et al. [37] | CyTOF | WM from control brains (n = 8) and NAWM (n = 10) and WM (n = 10) lesions from progressive MS patient brains | Myeloid cells | Fewer homeostatic microglia at the lesion site but an increase in activated cells. Cells found in lesions are activated with a strong phagocytic profile. A cluster of cells that expresses a high level of TNF is decreased in active lesions. |
| Couloume et al. [38] | CyTOF | Frozen PBMCs from MS patients at first relapse (n = 11) and healthy controls (n = 8) | Myeloid and lymphoid cells | Increase in T-bet-expressing B cells and CD206+ classical monocytes in early MS. T-bet+ B cells enriched in aggressive MS patients. |
| Diebold et al. [39] | CyTOF | Frozen PBMCs from RRMS patients (n = 31) before and at 12 weeks and 48 weeks of DMF treatment | Lymphocytes | Identification of GM-CSF+ IFNγ+ CXCR3+ memory helper T cells linked to axonal damage in MS and reduced with DMF treatment. |
| Galli et al. [40] | CyTOF | Frozen PBMCs and CSF cells from RRMS patients (n = 39), healthy controls (n = 29) and non-inflammatory disease controls (n = 31) | Lymphoid and myeloid cells | Identification of GM-CSF+ VLA-4+ CXCR4+ memory helper T cells expanded in blood and enriched in CNS of MS patients. This population is reduced by DMF treatment. |
| Ingelfinger et al. [41] | CyTOF scRNAseq | Frozen PBMCs from monozygotic twin pairs clinically discordant for MS (n = 61) | Lymphoid and myeloid cells | MS patients show an increase in inflammatory classical monocytes and transitional helper T cells hyper-responsive to IL-2 with expressing migration and proliferation markers. These immune perturbations are non-heritable. |
| Jäkel et al. [42] | snRNAseq | WM from human controls (n = 5) and progressive MS patients. For MS block, different WM areas were used: NAWM (n = 3), active (A) (n = 2), chronic active (CA) (n = 4), chronic inactive (CI) (n = 3) and remyelinated (RM) (n = 2) lesions | Neurons, OLs, OPCs, committed OL precursors, astrocytes, vascular smooth muscle cells, pericytes, endothelial cells and immune cells | Loss of the Olig1 mature OL in MS patients. Modification of the transcriptional profile of the other OL. Depletion of Olig6 and OPC in MS and increased expression of myelin genes in mature OL. |
| Johansson et al. [43] | CyTOF | Frozen PBMCs and CSF cells from MS patients (n = 14) and controls (n = 25) | Lymphoid and myeloid cells | Identification of a CD49d+ CD69+ CD27+ CXCR3+ HLA-DR+ B cell population associated with MS. |
| Kaufmann et al. [44] | scRNAseq spatial RNAseq | Frozen PBMCs from RRMS patients during (n = 10) and after (n = 9) natalizumab treatment, RRMS (n = 11) and PPMS (n = 10) patients without immunomodulatory treatment and matched healthy controls (n = 31). Fresh-frozen brain tissue from progressive MS patients and controls. | Lymphoid and myeloid cells | Identification of population of pathogenic CD161+ LTB+ CD4+ T cells (T09) in peripheral blood of RRMS and PPMS patients and in CNS lesions of progressive MS patients. These cells are likely present at disease initiation and become CNS-resident in cortical brain regions. |
| Kihara et al. [45] | snRNAseq | Non-lesioned samples from frozen brains of RRMS (n = 5) and SPMS patients (n = 5) | Astrocytes, endothelial cells, excitatory and inhibitory neurons, OLs, OPCs, lymphocytes, myeloid cells (microglia/macrophages) and pericytes | When comparing RRMS and SPMS: lower expression of excitatory neuronal markers in RRMS. Decreased expression of OL markers in SPMS. OPC maturation and myelination gene signature is greater in RRMS. RRMS astrocytes showed upregulation of marker genes for pan-reactive astrocytes. Immediate–early genes were increased in RRMS astrocytes. |
| Masuda et al. [46] | scRNA-seq | Healthy brains from epileptic patients (n = 5); brains from RR-MS patients or patients with first manifestation of MS (n = 5) | Isolated microglia | Microglia associated with MS brain expressed lower level of core genes but higher level of other genes including cytokines and chemokines |
| Pröbstel et al. [47] | scRNAseq | Fecal samples, PBMCs and CSF cells from CIS (n = 4) and RRMS patients (n = 39) and healthy controls (n = 31). Snap-frozen brain tissue from MS patients (n = 12) (cortical and subcortical areas with acute and chronic active lesions) and controls (n = 5). | B cells | IgA enrichment in inflamed CNS. IgA-producing B cells specific to MS-associated gut microbiota traffic to the CNS of active MS patients. |
| Ramaglia et al. [48] | Multiplexing imaging by mass cytometry | Different kinds of lesions in one RR-MS patient (n = 1) and a non-neurological control case (n = 1) that died of cardiac arrest. Study performed on frozen samples | Macrophages, microglia, T cells and B cells | Modification in cell morphology regarding the lesion type; activated microglial cells are not only present in active lesions but also in the NAWM |
| Ramesh et al. [49] | scRNAseq | Paired PBMCs and CSF from RRMS patients (n = 18), non-MS neurological controls (n = 3) and healthy controls (n = 3) | Lymphoid and myeloid cells | Polyclonal IgM+ and IgG+ B cells are expanded in CSF of MS patients and polarized towards an inflammatory and memory phenotype. |
| Rodríguez-Lorenzo et al. [50] | CyTOF | Immune cells from fresh brain tissue of progressive MS patients (n = 12), patients with dementia (n = 8) and controls (n = 10). FFPE brain tissue from MS patients (choroid plexus, n = 10; periventricular areas, n = 7) and non-neurological controls (choroid plexus, n = 8; periventricular areas, n = 5). Blood and CSF were also collected. | Lymphoid and myeloid cells | CD56bright NK cells with a migratory profile and NK cell activation markers are increased in MS septum. These cells may be immunoregulatory. |
| Schafflick et al. [32] | scRNAseq | Fresh PBMCs and CSF cells from RRMS patients (n = 6) and control patients (n = 6) | Lymphoid and myeloid cells | “Mono2” myeloid cells that express intermediate monocyte, perivascular macrophage, border-associated macrophage and microglia markers are specific to CSF of MS patients. Cytotoxic CD4+ helper T cells and Tregs expanded in CSF of MS patients. |
| Schirmer et al. [51] | snRNAseq; multiplex in situ hybridization | Frozen human brain samples with cortical and subcortical lesions or lesion-free zones, from MS cases (n = 17) and controls (n = 16) | Excitatory and inhibitory cortical neurons, astrocytes, OL lineage cells, microglia and smaller cell populations | Activation transcriptional profile of microglia from MS; expression of phagocytic markers. Modification of the astrocyte transcriptional profile depending on the demyelinating areas. |
| Wheeler et al. [52] | scRNA-seq | Fresh control (n = 4) and MS brains from surgery (n = 6) | Astrocytes | Large increase in astrocytes in MS vs. CTRL associated with a decrease in NRF2 activation but increases in MAFG activation, DNA methylation, GM-CSF signaling and pro-inflammatory pathway activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dugast, E.; Shah, S.; Laplaud, D.-A. Single-Cell Analysis to Better Understand the Mechanisms Involved in MS. Int. J. Mol. Sci. 2022, 23, 12142. https://doi.org/10.3390/ijms232012142
Dugast E, Shah S, Laplaud D-A. Single-Cell Analysis to Better Understand the Mechanisms Involved in MS. International Journal of Molecular Sciences. 2022; 23(20):12142. https://doi.org/10.3390/ijms232012142
Chicago/Turabian StyleDugast, Emilie, Sita Shah, and David-Axel Laplaud. 2022. "Single-Cell Analysis to Better Understand the Mechanisms Involved in MS" International Journal of Molecular Sciences 23, no. 20: 12142. https://doi.org/10.3390/ijms232012142
APA StyleDugast, E., Shah, S., & Laplaud, D.-A. (2022). Single-Cell Analysis to Better Understand the Mechanisms Involved in MS. International Journal of Molecular Sciences, 23(20), 12142. https://doi.org/10.3390/ijms232012142





