Evolution of Gene Arrangements in the Mitogenomes of Ensifera and Characterization of the Complete Mitogenome of Schizodactylus jimo
Abstract
:1. Introduction
2. Results
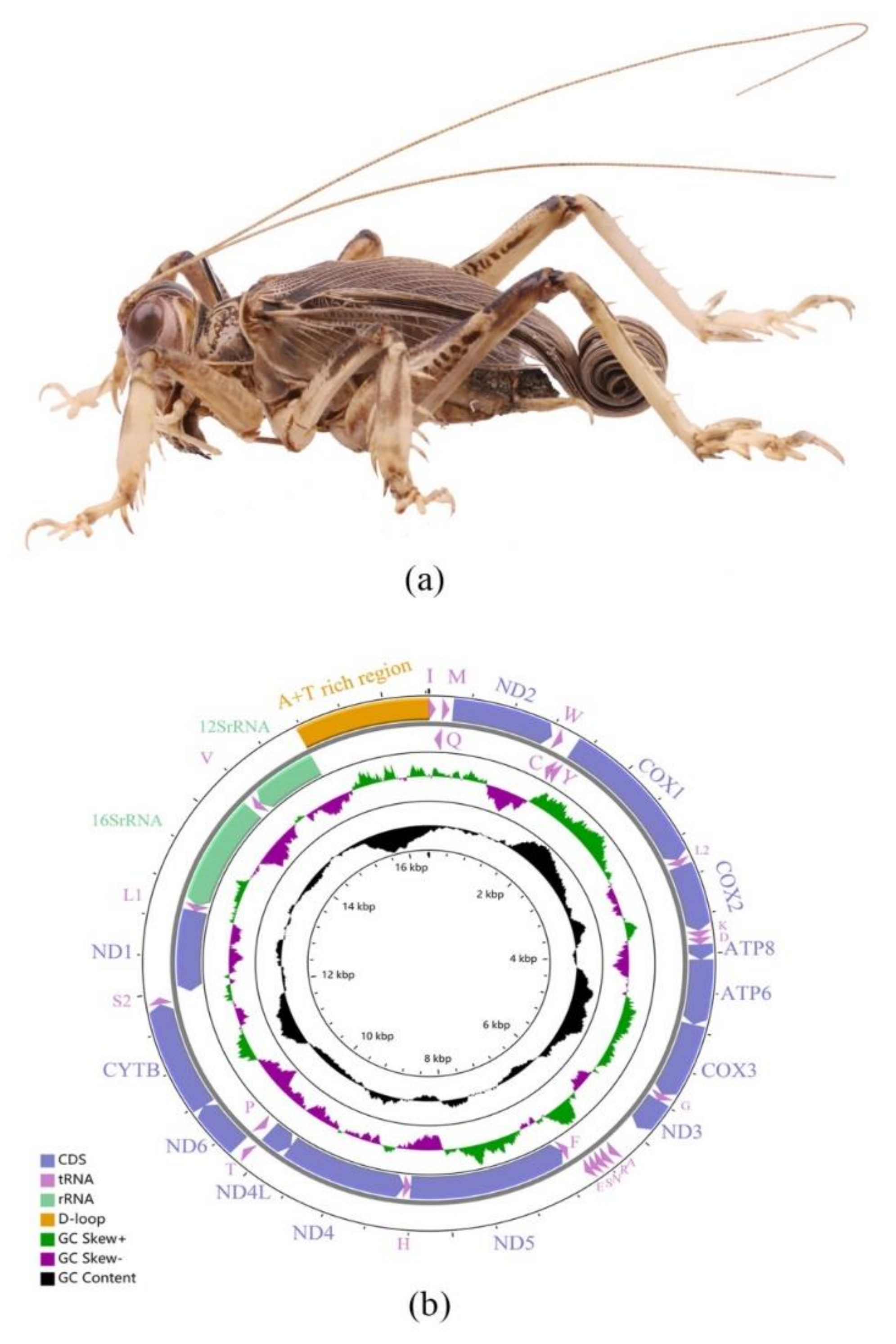
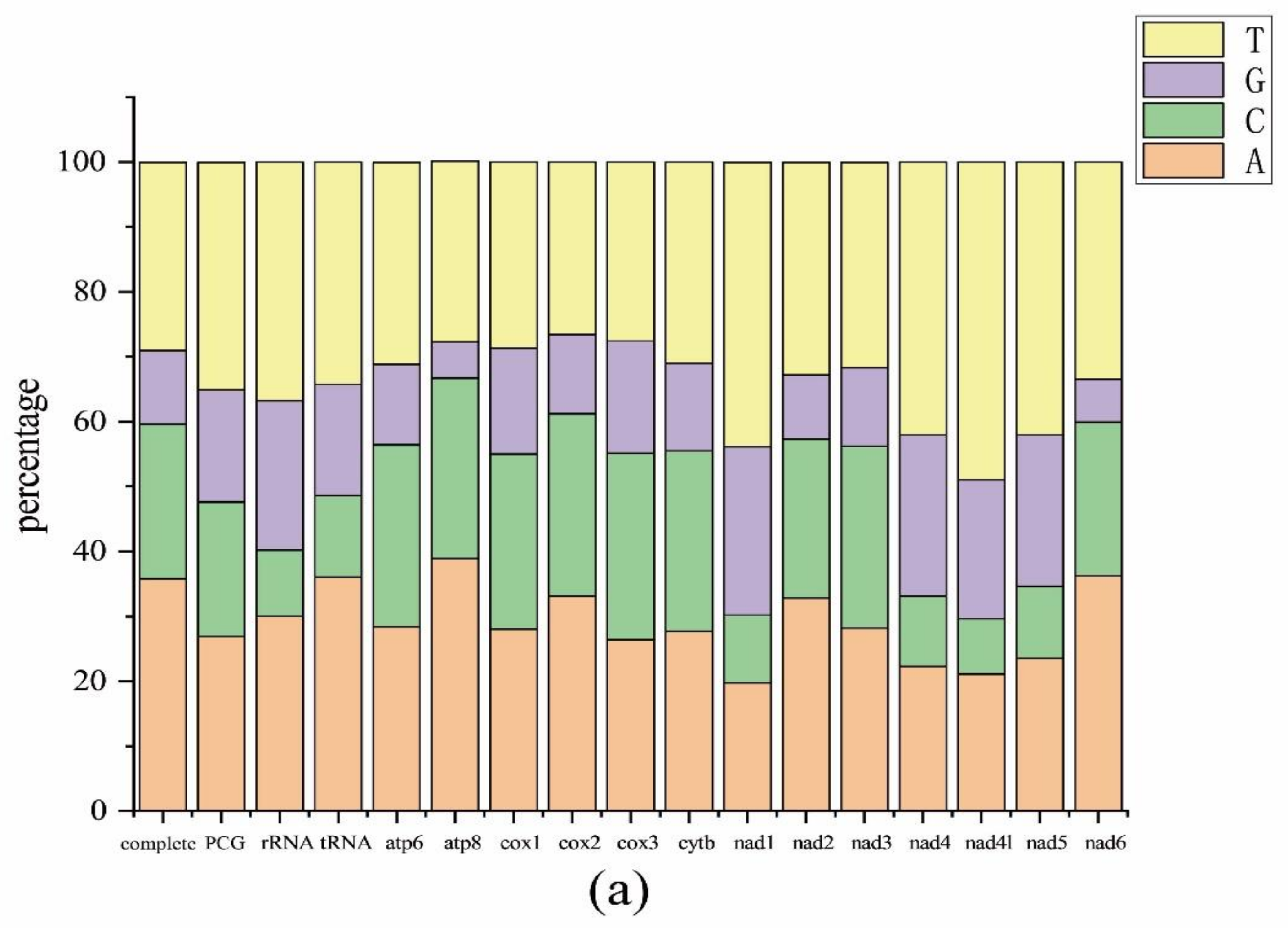
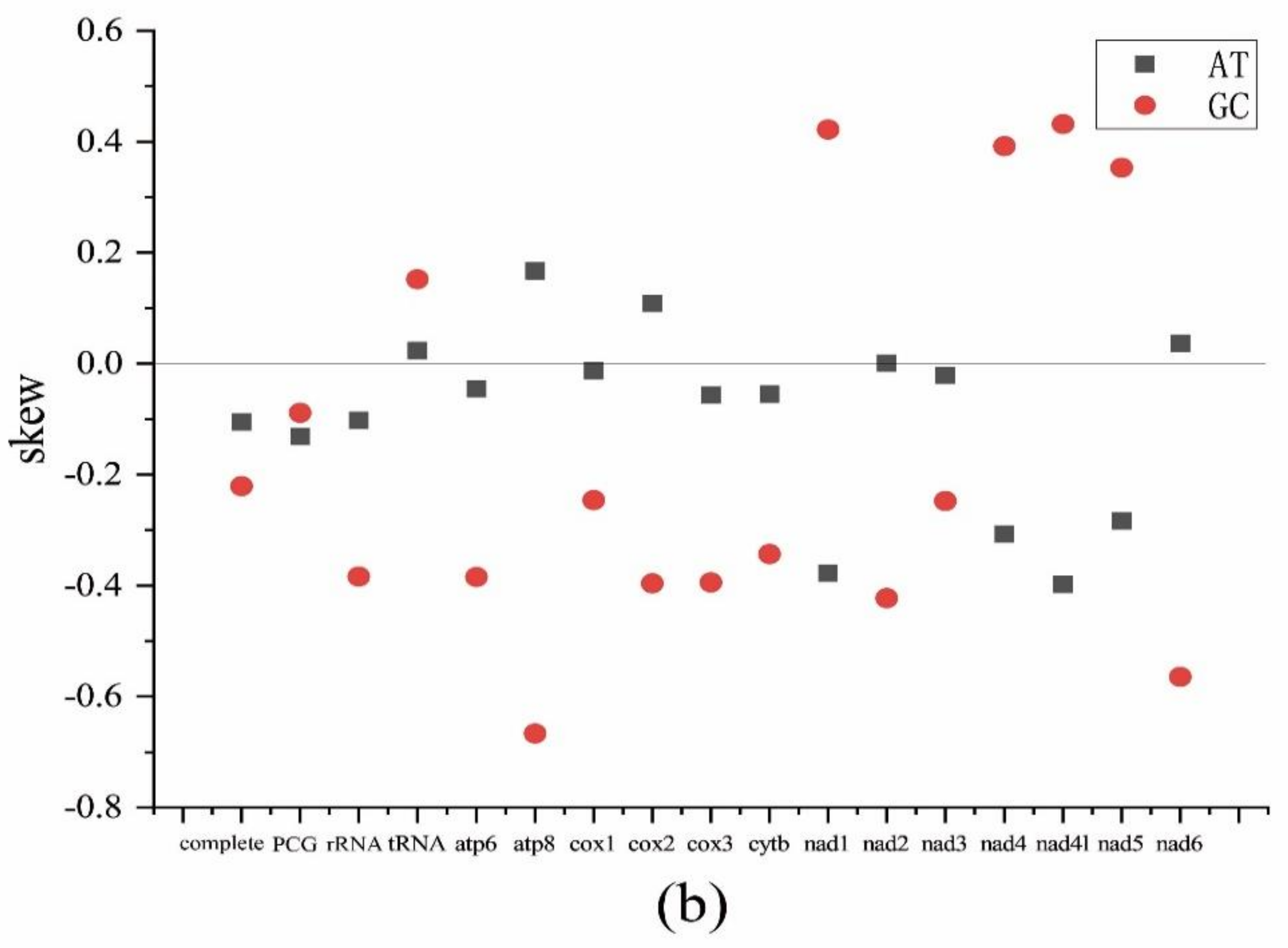
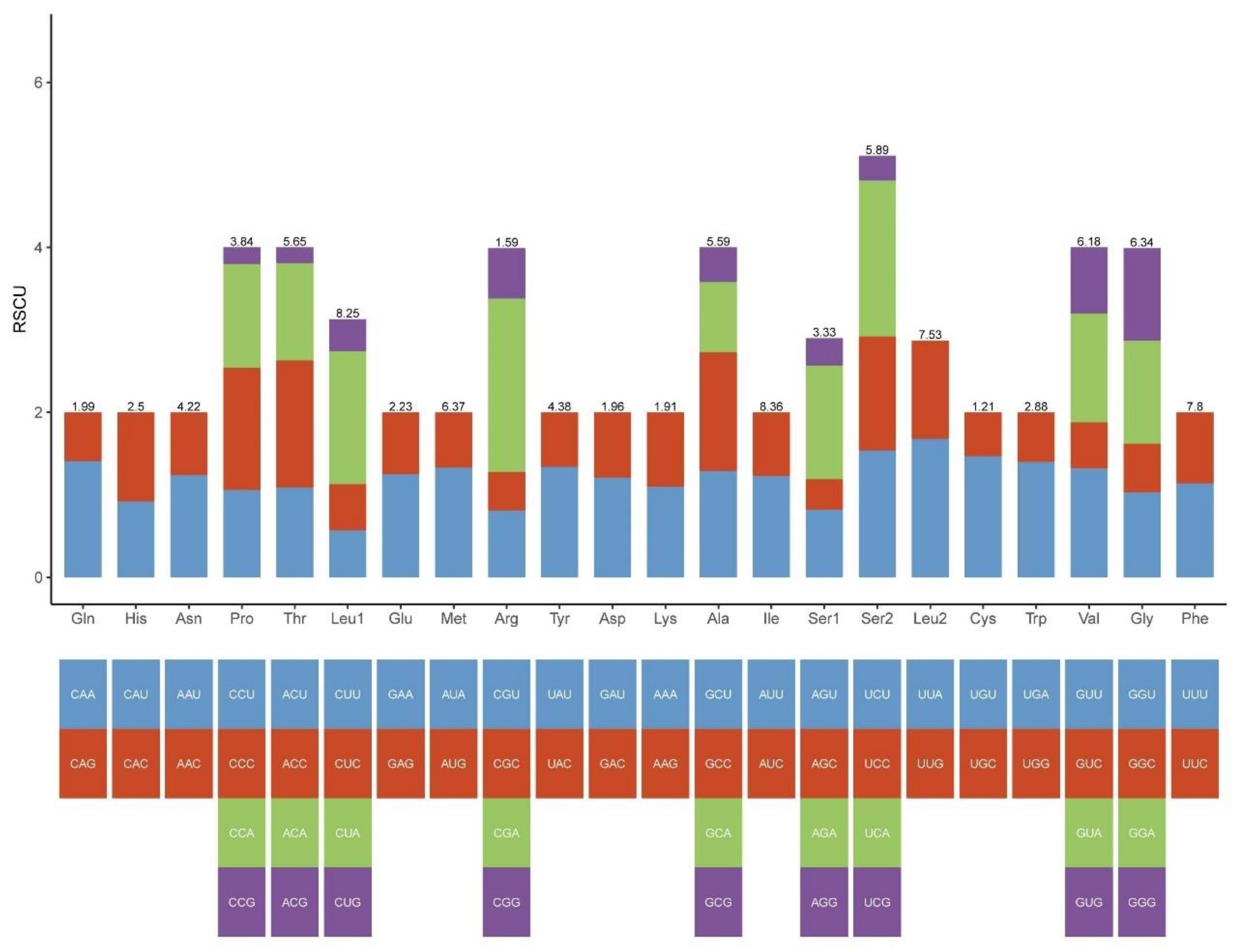
2.1. Mitogenome Features of Schizodactylus jimo
2.2. Phylogenetic and Divergence Time Analyses
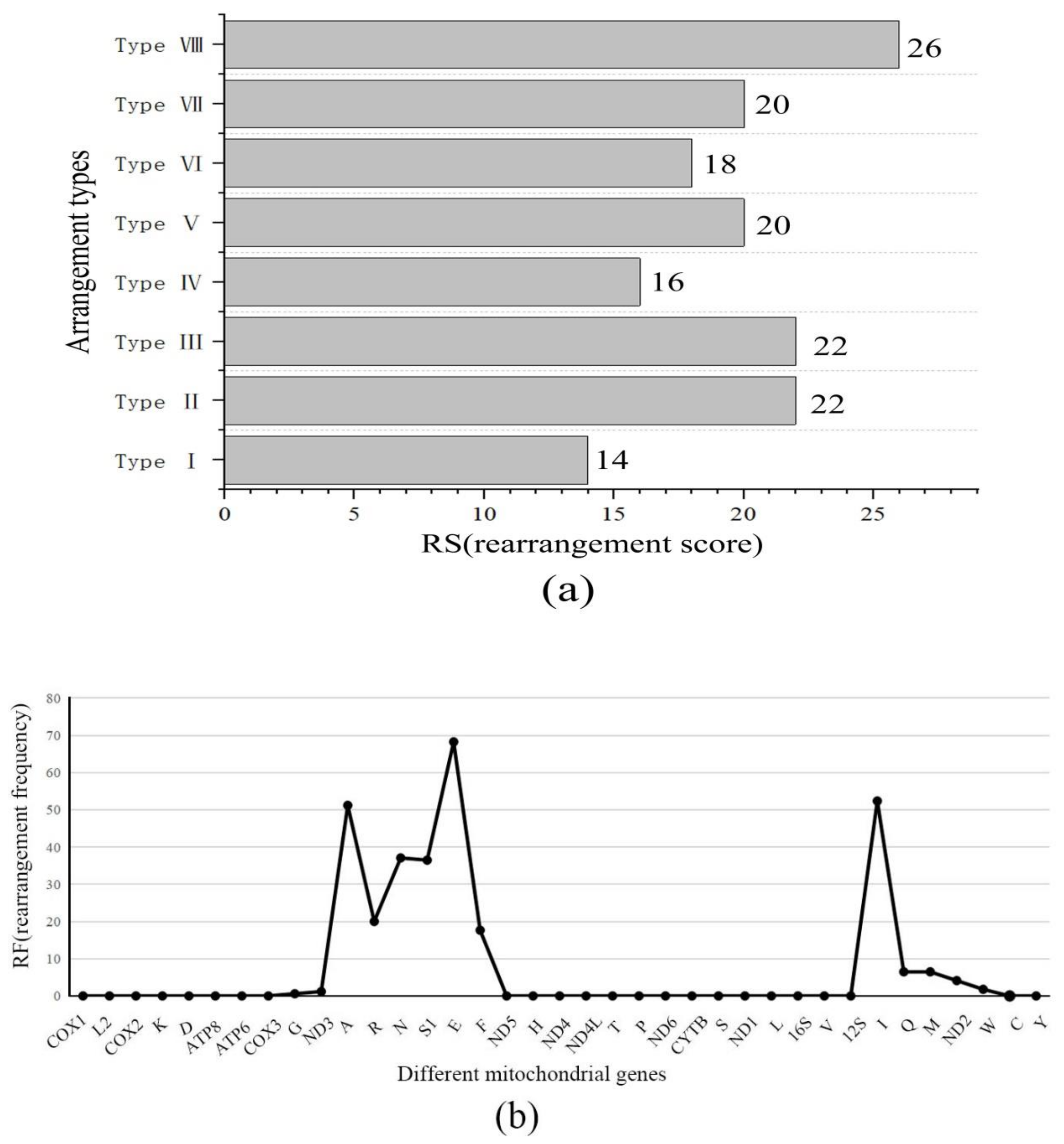
2.3. Evolution of Gene Rearrangements
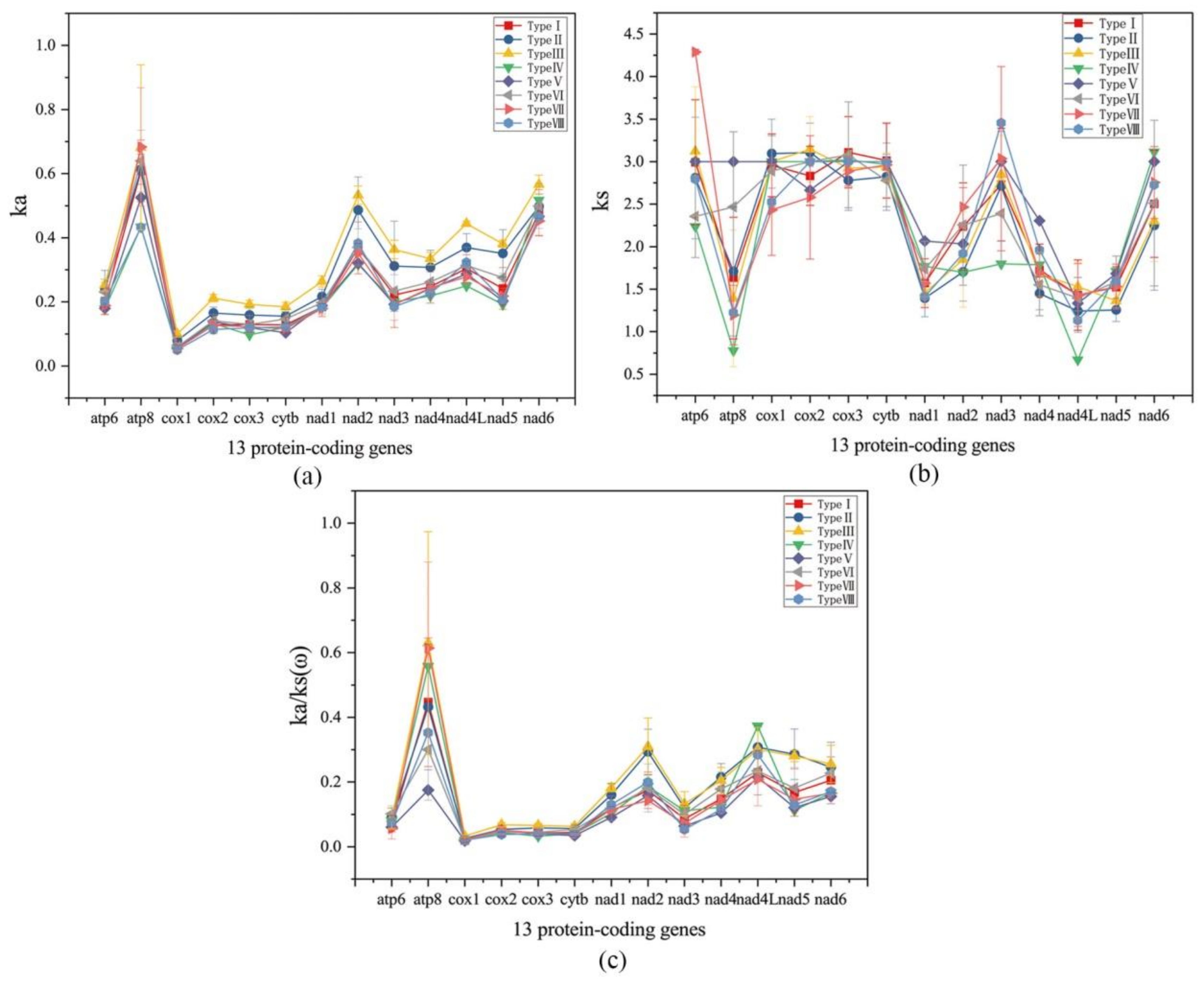
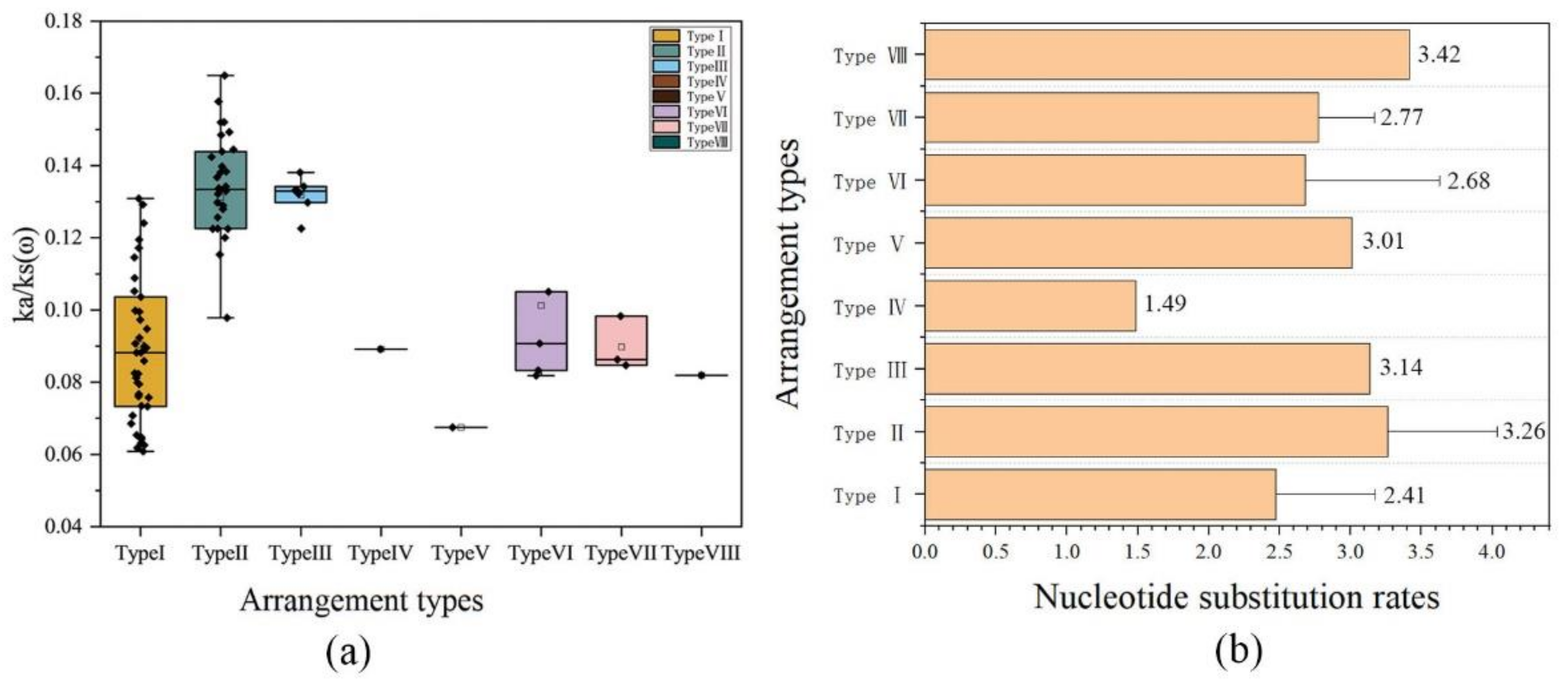
2.4. Evolutionary Analysis of Ensifera Mitogenomes
3. Discussion
3.1. Evolution of Mitogenomic Gene Rearrangements in Ensifera
3.2. Selection Constraints on the Mitogenome of Ensifera
4. Material and Methods
4.1. Sample Collection and DNA Extraction
4.2. Mitogenome Assembly, Annotation, and Analysis
4.3. Phylogenetic Analyses and Estimation of Divergence Time
4.4. Gene Arrangement Analysis in the Mitogenome
4.5. Analysis of Natural Selection Strength and Nucleotide Substitution Rates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boore, J.L. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol. Evol. 2006, 21, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.F.; Zhang, L.P.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. The complete mitochondrial genomes of four cockroaches (Insecta: Blattodea) and phylogenetic analyses within cockroaches. Gene 2016, 586, 115–122. [Google Scholar] [CrossRef]
- Ma, Y.; He, K.; Yu, P.; Yu, D.; Cheng, X.; Zhang, J. The complete mitochondrial genomes of three bristletails (Insecta: Archaeognatha): The paraphyly of Machilidae and insights into archaeognathan phylogeny. PLoS ONE 2015, 10, e0117669. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Johnson, K.P.; Sweet, A.D.; Yao, I.; Ferreira, R.L.; Cameron, S.L. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol. Phylogenetics Evol. 2018, 119, 118–127. [Google Scholar] [CrossRef] [Green Version]
- Zheng, B.Y.; Cao, L.J.; Tang, P.; van Achterberg, K.; Hoffmann, A.A.; Chen, H.Y.; Chen, X.X.; Wei, S.J. Gene arrangement and sequence of mitochondrial genomes yield insights into the phylogeny and evolution of bees and sphecid wasps (Hymenoptera: Apoidea). Mol. Phylogenetics Evol. 2018, 124, 1–9. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, J.; Rouse, G.W.; Wiklund, H.; Pleijel, F.; Watanabe, H.K.; Chen, C.; Qian, P.Y.; Qiu, J.W. Phylogeny, evolution and mitochondrial gene order rearrangement in scale worms (Aphroditiformia, Annelida). Mol. Phylogenetics Evol. 2018, 125, 220–231. [Google Scholar] [CrossRef]
- Li, H.; Hui, L.; Shi, A.; Štys, P.; Zhou, X.; Cai, W. The Complete Mitochondrial Genome and Novel Gene Arrangement of the Unique-Headed Bug Stenopirates sp. (Hemiptera: Enicocephalidae). PLoS ONE 2012, 7, e29419. [Google Scholar]
- Riyaz, M.; Shah, R.A.; Savarimuthu, I.; Kuppusamy, S. Comparative mitochondrial genome analysis of Eudocima salaminia (Cramer, 1777) (Lepidoptera: Noctuoidea), novel gene rearrangement and phylogenetic relationship within the superfamily Noctuoidea. Mol. Biol. Rep. 2021, 48, 4449–4463. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, H.; Feng, Z.; Li, B.; Cai, W. Novel gene rearrangement in the mitochondrial genome of Pachyneuron aphidis (Hymenoptera: Pteromalidae). Int. J. Biol. Macromol. 2020, 149, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.P.; Suleman; Zhang, Y.; Nie, Y.; Fu, Y.T.; Liu, G.H. The complete mitochondrial genome of capillariid nematodes (Eucoleus annulatus): A novel gene arrangement and phylogenetic implications. Vet. Parasitol. 2021, 296, 109476. [Google Scholar] [CrossRef]
- Xu, W.; Ding, J.; Lin, S.; Xu, R.; Liu, H. Comparative mitogenomes of three species in Moenkhausia: Rare irregular gene rearrangement within Characidae. Int. J. Biol. Macromol. 2021, 183, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.Q.; Ma, C.; Chen, J.Y.; Yang, D.R. The complete mitochondrial genomes of two ghost moths, Thitarodes renzhiensis and Thitarodes yunnanensis: The ancestral gene arrangement in Lepidoptera. BMC Genomics 2012, 13, 276. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.D.; Jia, Y.Y.; Cao, S.S.; Zhang, Z.Y.; Storey, K.B.; Yu, D.N.; Zhang, J.Y. Six complete mitochondrial genomes of mayflies from three genera of Ephemerellidae (Insecta: Ephemeroptera) with inversion and translocation of trnI rearrangement and their phylogenetic relationships. PeerJ 2020, 8, e9740. [Google Scholar] [CrossRef]
- Gao, X.Y.; Cai, Y.Y.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. Characteristics of the complete mitochondrial genome of Suhpalacsa longialata (Neuroptera, Ascalaphidae) and its phylogenetic implications. PeerJ 2018, 6, e5914. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.Y.; Zhang, S.S.; Zhang, L.P.; Yu, D.N.; Zhang, J.Y.; Cheng, H.Y. The complete mitochondrial genome of Epeorus herklotsi (Ephemeroptera: Heptageniidae) and its phylogeny. Mitochondrial DNA B Resour. 2018, 3, 303–304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.P.; Cai, Y.Y.; Yu, D.N.; Storey, K.B.; Zhang, J.Y. Gene characteristics of the complete mitochondrial genomes of Paratoxodera polyacantha and Toxodera hauseri (Mantodea: Toxoderidae). PeerJ 2018, 6, e4595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.P.; Yu, D.N.; Storey, K.B.; Cheng, H.Y.; Zhang, J.Y. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int. J. Biol. Macromol. 2018, 111, 787–795. [Google Scholar] [CrossRef]
- Shao, R.; Barker, S.C. The highly rearranged mitochondrial genome of the plague thrips, Thrips imaginis (Insecta:thysanoptera): Convergence of two novel gene boundaries and an extraordinary arrangement of rRNA genes. Mol. Biol. Evol. 2003, 3, 362–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, R.; Zhu, X.Q.; Barker, S.C.; Kate, H. Evolution of Extensively Fragmented Mitochondrial Genomes in the Lice of Humans. Genome Biol. Evol. 2012, 4, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Gibson, T.; Dowton, M. Evolutionary Dynamics of the Mitochondrial Genome in the Evaniomorpha (Hymenoptera)—A Group with an Intermediate Rate of Gene Rearrangement. Genome Biol. Evol. 2014, 6, 1862–1874. [Google Scholar] [CrossRef]
- Mao, M.; Dowton, M. Complete mitochondrial genomes of Ceratobaeus sp. and Idris sp. (Hymenoptera: Scelionidae): Shared gene rearrangements as potential phylogenetic markers at the tribal level. Mol. Biol. Rep. 2014, 41, 6419–6427. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.L.; Li, Q.; Gu, Y.; Shi, B.C.; Achterberg, C.V.; Wei, S.J.; Chen, X.X. The complete mitochondrial genome of Taeniogonalos taihorina (Bischoff) (Hymenoptera: Trigonalyidae) reveals a novel gene rearrangement pattern in the Hymenoptera. Gene 2014, 543, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shao, R.; Song, F.; Zhou, X.; Yang, Q.; Li, Z.; Cai, W. Mitochondrial Genomes of Two Barklice, Psococerastis albimaculata and Longivalvus hyalospilus (Psocoptera: Psocomorpha): Contrasting Rates in Mitochondrial Gene Rearrangement between Major Lineages of Psocodea. PLoS ONE 2013, 8, e61685. [Google Scholar]
- Zhang, H.; Lu, C.; Liu, Q.; Zou, T.; Qiao, G.; Huang, X. Insights into the evolution of aphid mitogenome features from new data and comparative analysis. Animals 2022, 12, 1970. [Google Scholar] [CrossRef]
- Dowton, M.; Cameron, S.L.; Dowavic, J.I.; Austin, A.D.; Whiting, M.F. Characterization of 67 mitochondrial tRNA gene rearrangements in the Hymenoptera suggests that mitochondrial tRNA gene position is selectively neutral. Mol. Biol. Evol. 2009, 26, 1607–1617. [Google Scholar] [CrossRef] [Green Version]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef]
- Schierup, M.H.; Hein, J. Consequences of recombination on traditional phylogenetic analysis. Genetics 2000, 156, 879–891. [Google Scholar] [CrossRef]
- Wei, Y.E.; Dang, J.P.; Xie, L.D.; Huang, Y. Complete Mitochondrial Genome of Teleogryllus emma (Orthoptera:Gryllidae) with a New Gene Order in Orthoptera. Zool. Res. 2008, 29, 236–244. [Google Scholar]
- Liu, C.; Chang, J.; Ma, C.; Li, L.; Zhou, S. Mitochondrial genomes of two Sinochlora species (Orthoptera): Novel genome rearrangements and recognition sequence of replication origin. BMC Genomics 2013, 14, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gwynne, D.T. Phylogeny of the Ensifera (Orthoptera): A hypothesis supporting multiple origins of acoustical signalling, complex spermatophores and maternal care in crickets, katydids, and weta. J. Orthoptera Res. 1995, 4, 203–218. [Google Scholar] [CrossRef]
- Kevan, D.K.M. Orthoptera. In Synopsis and Classification of Living Organisms; Parker, S.P., Ed.; McGraw-Hill Inc.: New York, NY, USA, 1982; pp. 352–379. [Google Scholar]
- Leubner, F.; Bradler, S.; Wipfler, B. The thoracic morphology of the wingless dune cricket Comicus calcaris (Orthoptera: Schizodactylidae): Novel apomorphic characters for the group and adaptations to sand desert environments. Arthropod. Struct. Dev. 2017, 46, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Irish, J. The species of Comicus Brunner v. Wattenwyl (Orthoptera: Schizodactylidae) with a discussion of their origin: Introduction. Navors. Nas. Mus. Res. Natl. Mus. 1986, 5, 254–263. [Google Scholar]
- Heads, S.; Leuzinger, L. On the placement of the Cretaceous orthopteran Brauckmannia groeningae from Brazil, with notes on the relationships of Schizodactylidae (Orthoptera, Ensifera). Zookeys 2011, 77, 17–30. [Google Scholar] [CrossRef]
- Song, H.; Amédégnato, C.; Cigliano, M.M.; Desutter-Grandcolas, L.; Heads, S.W.; Huang, Y.; Otte, D.; Whiting, M.F. 300 million years of diversification: Elucidating the patterns of orthopteran evolution based on comprehensive taxon and gene sampling. Cladistics Int. J. Willi Hennig Soc. 2015, 31, 621–651. [Google Scholar] [CrossRef] [PubMed]
- He, Z.Q.; Liu, Y.J. A new species of Dune Cricket from China (Orthoptera: Ensifera: Schizodactylidae). Zootaxa 2021, 4999, 356–362. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Shi, F.M.; Zhao, L.; Fox, G.E. The First Mitochondrial Genome for the Superfamily Hagloidea and Implications for Its Systematic Status in Ensifera. PLoS ONE 2014, 9, e86027. [Google Scholar] [CrossRef]
- Shao, R.; Campbell, N.J.; Schmidt, E.R.; Barker, S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001, 18, 1828–1832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryuto, S.; Kosuke, K.; Shota, H.; Keigo, I.; Nguyen, C.; Nguyen, T.P.; Le, B.; Kim, O.; Katsuhiko, M.; Haruko, T. Comparative Analysis of Mitochondrial Genomes in Gryllidea (Insecta: Orthoptera): Implications for Adaptive Evolution in Ant-Loving Crickets. Genome Biol. Evol. 2021, 10, evab222. [Google Scholar]
- Yang, J.; Ye, F.; Huang, Y. Mitochondrial genomes of four katydids (Orthoptera: Phaneropteridae): New gene rearrangements and their phylogenetic implications. Gene 2016, 575, 702–711. [Google Scholar] [CrossRef]
- Yang, J.; Ren, Q.; Huang, Y. Complete mitochondrial genomes of three crickets (Orthoptera: Gryllidae) and comparative analyses within Ensifera mitogenomes. Zootaxa 2016, 4092, 529. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.J.; Zhao, L.; Liu, N.; Guo, H.; Guan, B.; Di, J.; Shi, F. Towards a higher-level Ensifera phylogeny inferred from mitogenome sequences. Mol. Phylogenetics Evol. 2017, 108, 22–33. [Google Scholar] [CrossRef]
- Arndt, A.; Smith, M.J. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria. Mol. Biol. Evol. 1998, 15, 1009–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Excoffier, L. Evolution of human mitochondrial DNA: Evidence for departure from a pure neutral model of populations at equilibrium. J. Mol. Evol. 1990, 30, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Li, Y.T.; Xin, Z.Z.; Tang, Y.Y.; Yang, T.T.; Tang, B.P.; Sun, Y.; Zhang, D.Z.; Zhou, C.L.; Liu, Q.N.; Yu, X.M. Comparative Mitochondrial Genome Analyses of Sesarmid and Other Brachyuran Crabs Reveal Gene Rearrangements and Phylogeny. Front. Genet. 2020, 11, 536640. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Miao, G.; Hu, S.; Sun, Q.; Ding, H.; Ji, Z.; Guo, P.; Yan, S.; Wang, C.; Kan, X.; et al. Quantification and evolution of mitochondrial genome rearrangement in Amphibians. BMC Ecol. Evol. 2021, 21, 19. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Boore, J.L.; Brown, W.M. Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: Duplication and nonrandom loss. Mol. Biol. Evol. 2002, 19, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.; Qiu, Z.; Yuan, H.; Wang, X.; Huang, Y. Evolutionary rates of and selective constraints on the mitochondrial genomes of Orthoptera insects with different wing types. Mol. Phylogenetics Evol. 2020, 145, 106734. [Google Scholar] [CrossRef] [PubMed]
- Yunyun, L.; Yanping, L.; Zhiqiang, R.; Chao, B.; Xinxin, Y.; Junxing, Y.; Wansheng, J.; Qiong, S. The Complete Mitochondrial Genome of Glyptothorax macromaculatus Provides a Well-Resolved Molecular Phylogeny of the Chinese Sisorid Catfishes. Genes 2018, 9, 282. [Google Scholar]
- Renfu, S.; Mark, D.; Anna, M.; Barker, S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003, 20, 1612–1619. [Google Scholar]
- Wang, C.Y.; Zhao, M.; Wang, S.J.; Xu, H.L.; Yang, Y.M.; Liu, L.N.; Feng, Y. The Complete Mitochondrial Genome of Lepidotrigona flavibasis (Hymenoptera: Meliponini) and High Gene Rearrangement in Lepidotrigona Mitogenomes. J. Insect Sci. 2021, 21, 10–11. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenetics Evol. 2013, 2, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772. [Google Scholar]
- Gary, B. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, 181–184. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partition Finder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [PubMed] [Green Version]
- Sanderson, M.J. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics 2003, 19, 301–302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Zhang, J.; Kan, X.; Miao, G.; Hu, S.; Sun, Q.; Tian, W. qMGR: A new approach for quantifying mitochondrial genome rearrangement. Mitochondrion 2020, 52, 20–23. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, Z.-C.; Guan, D.-L.; Jiang, T.; Wang, H.; Zhao, L.; Xu, S.-Q. Evolution of Gene Arrangements in the Mitogenomes of Ensifera and Characterization of the Complete Mitogenome of Schizodactylus jimo. Int. J. Mol. Sci. 2022, 23, 12094. https://doi.org/10.3390/ijms232012094
Dan Z-C, Guan D-L, Jiang T, Wang H, Zhao L, Xu S-Q. Evolution of Gene Arrangements in the Mitogenomes of Ensifera and Characterization of the Complete Mitogenome of Schizodactylus jimo. International Journal of Molecular Sciences. 2022; 23(20):12094. https://doi.org/10.3390/ijms232012094
Chicago/Turabian StyleDan, Zhi-Cuo, De-Long Guan, Tao Jiang, Hang Wang, Lu Zhao, and Sheng-Quan Xu. 2022. "Evolution of Gene Arrangements in the Mitogenomes of Ensifera and Characterization of the Complete Mitogenome of Schizodactylus jimo" International Journal of Molecular Sciences 23, no. 20: 12094. https://doi.org/10.3390/ijms232012094
APA StyleDan, Z.-C., Guan, D.-L., Jiang, T., Wang, H., Zhao, L., & Xu, S.-Q. (2022). Evolution of Gene Arrangements in the Mitogenomes of Ensifera and Characterization of the Complete Mitogenome of Schizodactylus jimo. International Journal of Molecular Sciences, 23(20), 12094. https://doi.org/10.3390/ijms232012094





