Emerging Advances of Detection Strategies for Tumor-Derived Exosomes
Abstract
:1. Introduction
2. Exosomes
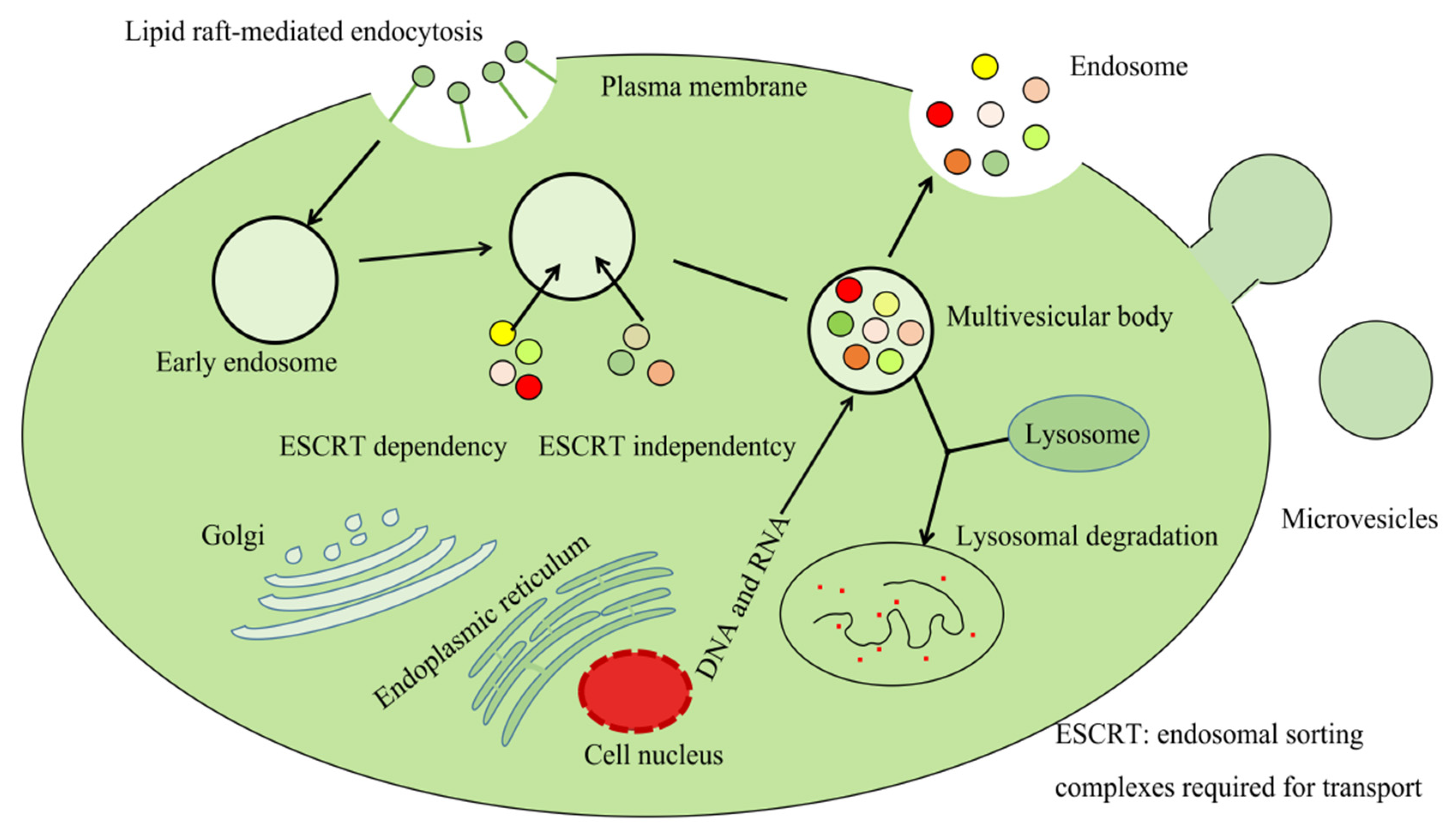
2.1. Synthesis and Release of Exosomes
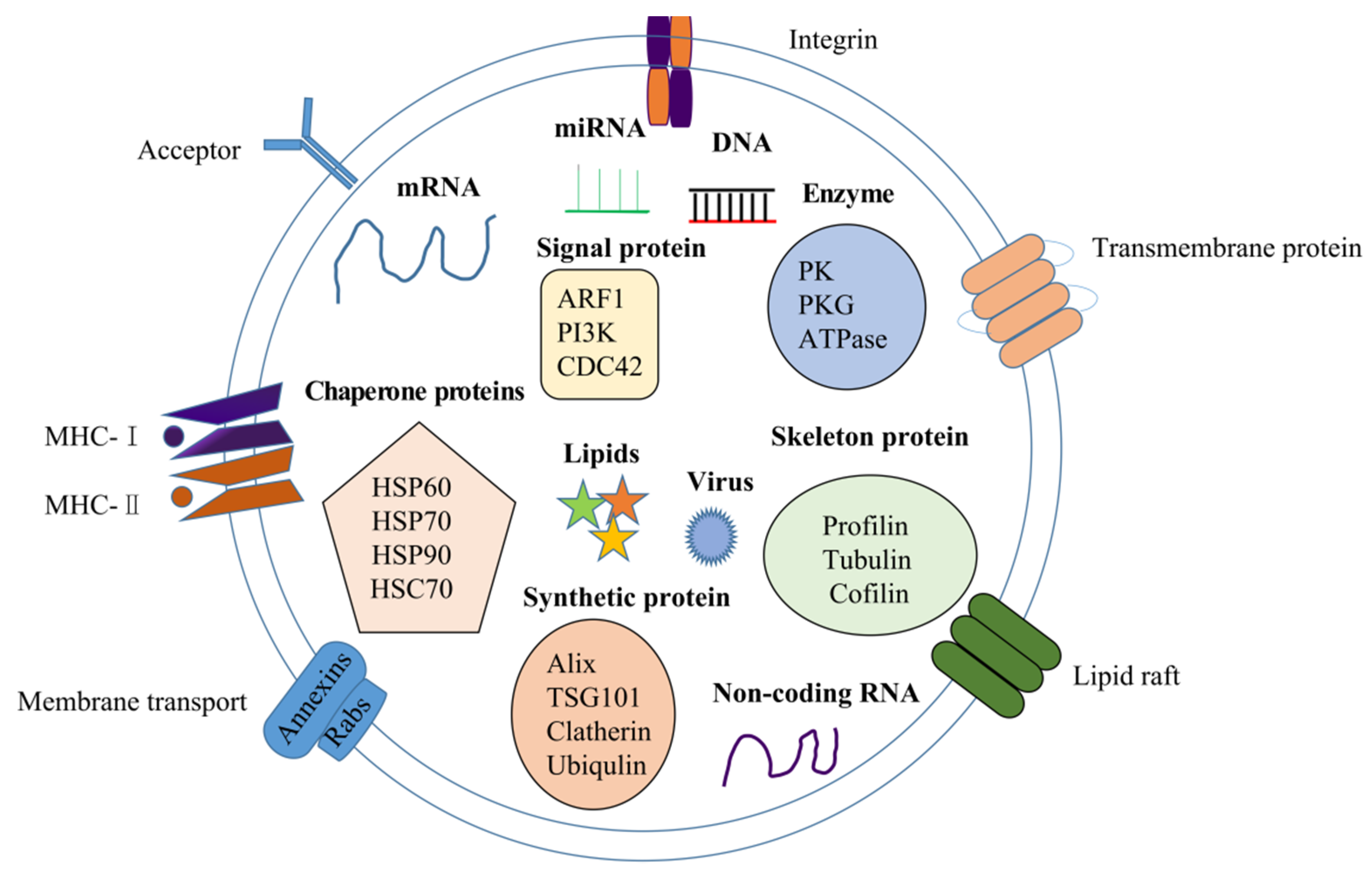
2.2. Components in Exosomes
2.2.1. Exosomal Nucleic Acid
2.2.2. Exosomal Proteins
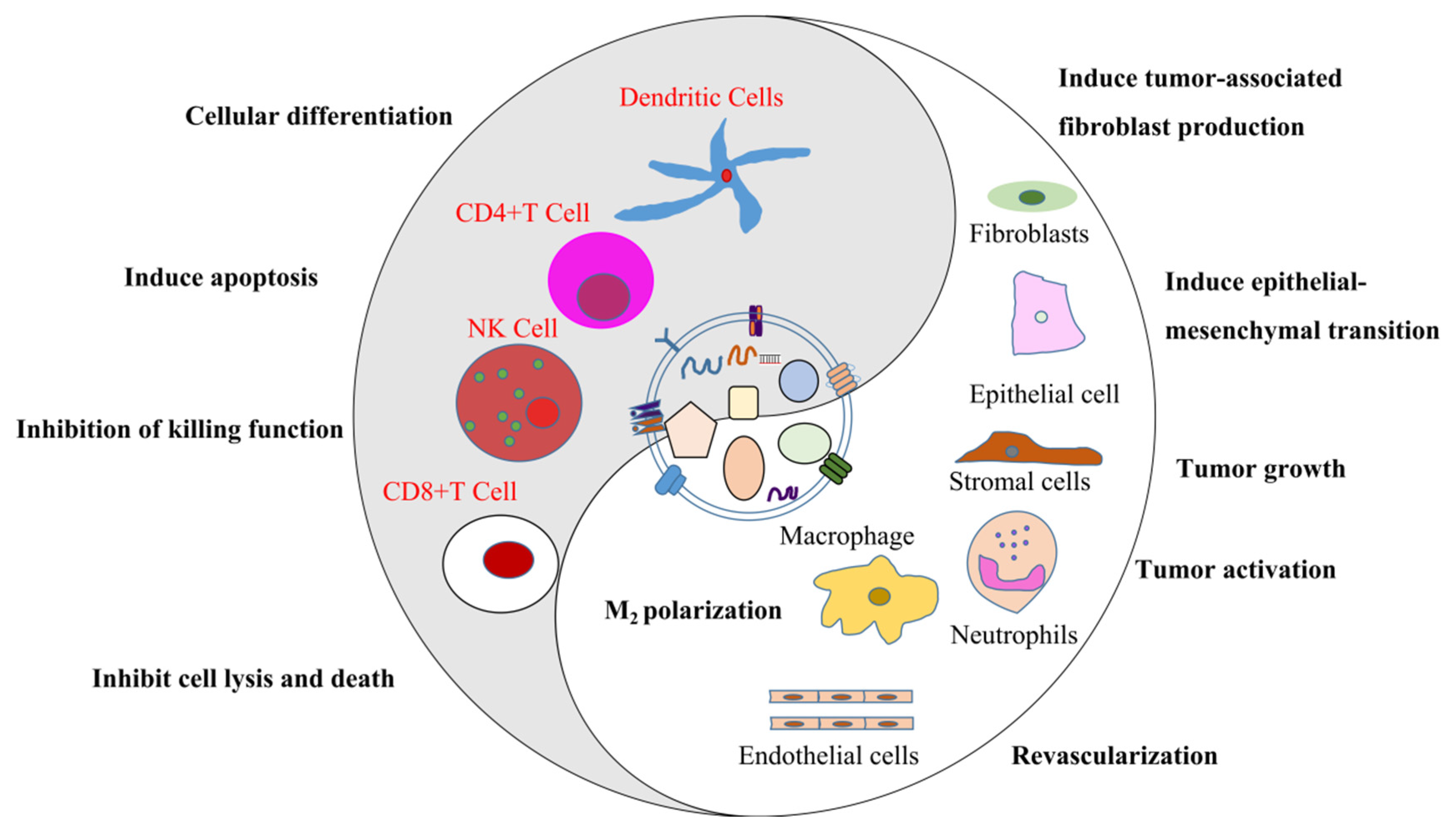
3. Exosomes and Tumor Development
4. Novel Detection Methods of Tumor-Derived Exosomes
4.1. Separation and Purification
4.2. Digital Detection of Proteins and Nucleic Acids
4.3. Microfluidic Technology
4.3.1. Acoustics-Based Microfluidics
4.3.2. Immunoaffinity-Based Microfluidics
4.4. Surface-Enhanced Raman Scattering Technology
4.5. Aptamer-Based Separation Method
4.6. Quantum Dot-Based Exosome Quantification
4.7. Emerging Nanomaterial Detection Technology
5. Single Exosome Phenotyping Technique
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MVB | multivesicular bodies |
| microRNAs | miRNAs |
| ESCRT | endosomal sorting complexes required for the transport |
| hnRNPH1 | heterogeneous nuclear ribonucleoproteins |
| SUFU | suppressor of fused protein |
| GBM | glioblastoma multiforme |
| PDAC | pancreatic ductal adenocarcinoma |
| MHC | major histocompatibility complexes |
| ARF1 | ADP-ribosylation factors 1 |
| PI3K | phosphatidylinositol 3-kinase |
| CDC42 | cell division cycle 42 |
| PK | pyruvate kinase |
| PKG | protein kinase G |
| GPC-1 | glypican-1 |
| EpCAM | epithelial cell adhesion molecule |
| PD-L1 | programmed death ligand 1 |
| EGFR | epidermal growth factor receptor |
| NK | natural killer |
| ELISA | enzyme-linked immunosorbent assay |
| PCR | polymerase chain reaction |
| ddELISA | digital droplet ELISA |
| ddPCR | droplet digital PCR |
| SERS | surface-enhanced Raman scattering |
| HDL | high-density lipoprotein |
| LDL | low-density lipoprotein |
| VLDL | very-low-density lipoprotein |
| Exo-PD-L1 | exosomal PD-L1 |
| PDMS | polydimethylsiloxane |
| FITC | fluorescein isothiocyanate |
| AUC | area under the curve |
| MPC | methacryloyloxyethyl phosphorylcholine |
| QDs | quantum dots |
| HRP | horseradish peroxidase |
| MFBPs | magnetic and fluorescent biological probes |
| HER2 | human epidermalgrowth factor receptor-2 |
| COF | covalent organic framework |
| AuNPs | gold nanoparticles |
| pSC4 | para-sulfocalix[4]arene hydrate |
| PBS | phosphate buffer |
References
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Johnstone, R.M. Maturation of reticulocytes: Formation of exosomes as a mechanism for shedding membrane proteins. Biochem. Cell Biol. Biochim. Biol. Cell 1992, 70, 179. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ji, S.; Shao, G.; Zhang, J.; Zhao, K.; Wang, Z.; Wu, A. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clin. Transl. Oncol. 2018, 20, 906–911. [Google Scholar] [CrossRef]
- Nilsson, J.; Skog, J.; Nordstrand, A.; Baranov, V.; Mincheva-Nilsson, L.; Breakefield, X.O.; Widmark, A. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 2009, 100, 1603–1607. [Google Scholar] [CrossRef]
- Petersen, K.E.; Manangon, E.; Hood, J.L.; Wickline, S.A.; Fernandez, D.P.; Johnson, W.P.; Gale, B.K. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal. Bioanal. Chem. 2014, 406, 7855–7866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimers Dement. 2015, 11, 600–607.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraser, K.B.; Moehle, M.S.; Daher, J.P.L.; Webber, P.J.; Williams, J.Y.; Stewart, C.A.; Yacoubian, T.A.; Cowell, R.M.; Dokland, T.; Ye, T.; et al. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum. Mol. Genet. 2013, 22, 4988–5000. [Google Scholar] [CrossRef] [Green Version]
- Fraser, K.B.; Rawlins, A.B.; Clark, R.G.; Alcalay, R.N.; Standaert, D.G.; Liu, N.J.; West, A.B.; Parkinson’s Dis Biomarker Program. Ser(P)-1292 LRRK2 in urinary exosomes is elevated in idiopathic Parkinson’s disease. Mov. Disord. 2016, 31, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Tomasetti, M.; Lee, W.; Santarelli, L.; Neuzil, J. Exosome-derived microRNAs in cancer metabolism: Possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017, 49, e285. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Boriachek, K.; Islam, M.N.; Moller, A.; Salomon, C.; Nguyen, N.T.; Hossain, M.S.A.; Yamauchi, Y.; Shiddiky, M.J.A. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small 2018, 14, 1702153. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.Y.; Chen, C.C. Toward characterizing extracellular vesicles at a single-particle level. J. Biomed. Sci. 2019, 26, 9. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, H.; Im, H.; Castro, C.M.; Breakefield, X.; Weissleder, R.; Lee, H. New Technologies for Analysis of Extracellular Vesicles. Chem. Rev. 2018, 118, 1917–1950. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef] [Green Version]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Babst, M.; Katzmann, D.J.; Estepa-Sabal, E.J.; Meerloo, T.; Emr, S.D. Escrt-III: An endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 2002, 3, 271–282. [Google Scholar] [CrossRef] [Green Version]
- Wollert, T.; Hurley, J.H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 2010, 464, 864–869. [Google Scholar] [CrossRef] [Green Version]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef] [PubMed]
- Christ, L.; Raiborg, C.; Wenzel, E.M.; Campsteijn, C.; Stenmark, H. Cellular Functions and Molecular Mechanisms of the ESCRT Membrane-Scission Machinery. Trends Biochem. Sci. 2017, 42, 42–56. [Google Scholar] [CrossRef]
- Raiborg, C.; Stenmark, H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009, 458, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Schoneberg, J.; Lee, I.H.; Iwasa, J.H.; Hurley, J.H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 2017, 18, 5–17. [Google Scholar] [CrossRef]
- Xu, H.; Dong, X.Y.; Chen, Y.M.; Wang, X.J. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 2018, 56, 479–484. [Google Scholar] [CrossRef]
- Wani, S.; Kaul, D.; Mavuduru, R.S.; Kakkar, N.; Bhatia, A. Urinary-exosomal miR-2909, A novel pathognomonic trait of prostate cancer severity. J. Biotechnol. 2017, 259, 135–139. [Google Scholar] [CrossRef]
- Yang, H.; Fu, H.L.; Wang, B.; Zhang, X.; Mao, J.H.; Li, X.; Wang, M.; Sun, Z.X.; Qian, H.; Xu, W.R. Exosomal miR-423-5p targets SUFU to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol. Carcinogen. 2018, 57, 1223–1236. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Iinuma, H.; Umemoto, Y.; Yanagisawa, T.; Matsumoto, A.; Jinno, H. Exosome-encapsulated microRNA-223-3p as a minimally invasive biomarker for the early detection of invasive breast cancer. Oncol. Lett. 2018, 15, 9584–9592. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.B.; Seth, S.; Koch, M.; Zhang, J.H.; Weitz, J.; Chin, L.; Futreal, A.; et al. Identification of Double-stranded Genomic DNA Spanning All Chromosomes with Mutated KRAS and p53 DNA in the Serum Exosomes of Patients with Pancreatic Cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.; Rodriguez-Barrueco, R.; Silva, J.M.; Zhang, W.J.; Hearn, S.; Elemento, O.; Paknejad, N.; Manova-Todorova, K.; Welte, K.; Bromberg, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar]
- Garcia-Romero, N.; Carrion-Navarro, J.; Esteban-Rubio, S.; Lazaro-Ibanez, E.; Peris-Celda, M.; Alonso, M.M.; Guzman-De-Villoria, J.; Fernandez-Carballal, C.; de Mendivil, A.O.; Garcia-Duque, S.; et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget 2017, 8, 1416–1428. [Google Scholar] [CrossRef] [Green Version]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabro, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, Y.; Konishi, Y.; Kosaka, N.; Katsuda, T.; Kato, T.; Ochiya, T. Comparative marker analysis of extracellular vesicles in different human cancer types. J. Extracell. Vesicles 2013, 2, 20424. [Google Scholar] [CrossRef] [PubMed]
- Jo, W.; Jeong, D.; Kim, J.; Park, J. Self-Renewal of Bone Marrow Stem Cells by Nanovesicles Engineered from Embryonic Stem Cells. Adv. Healthc. Mater. 2016, 5, 3148–3156. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; Ritter, T. The Exosome—A Naturally Secreted Nanoparticle and its Application to Wound Healing. Adv. Mater. 2016, 28, 5542–5552. [Google Scholar] [CrossRef] [PubMed]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Andaloussi, S.E.; Mager, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef]
- Figueroa, J.M.; Skog, J.; Akers, J.; Li, H.Y.; Komotar, R.; Jensen, R.; Ringel, F.; Yang, I.; Kalkanis, S.; Thompson, R.; et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro-Oncology 2017, 19, 1494–1502. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Shao, N.Y.; Xue, L.; Wang, R.; Luo, K.M.; Zhi, F.; Lan, Q. miR-454-3p Is an Exosomal Biomarker and Functions as a Tumor Suppressor in Glioma. Mol. Cancer Ther. 2019, 18, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Manterola, L.; Guruceaga, E.; Perez-Larraya, J.G.; Gonzalez-Huarriz, M.; Jauregui, P.; Tejada, S.; Diez-Valle, R.; Segura, V.; Sampron, N.; Barrena, C.; et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-Oncology 2014, 16, 520–527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiao, H.; Zhou, H.; Santiago, S.; Lee, J.M.; Garon, E.B.; Yang, J.P.; Brinkmann, O.; Yan, X.M.; Akin, D.; et al. Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol. Life Sci. 2012, 69, 3341–3350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanaoka, R.; Iinuma, H.; Dejima, H.; Sakai, T.; Uehara, H.; Matsutani, N.; Kawamura, M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology 2018, 94, 311–323. [Google Scholar] [CrossRef]
- Yuwen, D.L.; Ma, Y.Z.; Wang, D.Q.; Gao, J.; Li, X.; Xue, W.W.; Fan, M.M.; Xu, Q.; Shen, Y.; Shu, Y.Q. Prognostic Role of Circulating Exosomal miR-425-3p for the Response of NSCLC to Platinum-Based Chemotherapy. Cancer Epidemiol. Prev. Biomark. 2019, 28, 163–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Qiu, X.; Li, X.; Fan, H.; Zhang, F.; Lv, T.; Song, Y. Expression profiles and clinical value of plasma exosomal Tim-3 and Galectin-9 in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 498, 409–415. [Google Scholar] [CrossRef]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, Y.; Qiu, F.; Qiu, Z. Proteomic identification of exosomal LRG1: A potential urinary biomarker for detecting NSCLC. Electrophoresis 2011, 32, 1976–1983. [Google Scholar] [CrossRef]
- Goldvaser, H.; Gutkin, A.; Beery, E.; Edel, Y.; Nordenberg, J.; Wolach, O.; Rabizadeh, E.; Uziel, O.; Lahav, M. Characterisation of blood-derived exosomal hTERT mRNA secretion in cancer patients: A potential pan-cancer marker. Br. J. Cancer 2017, 117, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, P.G.; Lee, J.E.; Cho, Y.E.; Lee, S.J.; Chae, Y.S.; Jung, J.H.; Kim, I.S.; Park, H.Y.; Baek, M.C. Fibronectin on circulating extracellular vesicles as a liquid biopsy to detect breast cancer. Oncotarget 2016, 7, 40189–40199. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Kamohara, H.; Kinoshita, K.; Kurashige, J.; Ishimoto, T.; Iwatsuki, M.; Watanabe, M.; Baba, H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer-Am. Cancer Soc. 2013, 119, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Chiam, K.; Wang, T.T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215. [Google Scholar] [CrossRef] [Green Version]
- Tokuhisa, M.; Ichikawa, Y.; Kosaka, N.; Ochiya, T.; Yashiro, M.; Hirakawa, K.; Kosaka, T.; Makino, H.; Akiyama, H.; Kunisaki, C.; et al. Exosomal miRNAs from Peritoneum Lavage Fluid as Potential Prognostic Biomarkers of Peritoneal Metastasis in Gastric Cancer. PLoS ONE 2015, 10, e0130472. [Google Scholar] [CrossRef]
- Kumata, Y.; Iinuma, H.; Suzuki, Y.; Tsukahara, D.; Midorikawa, H.; Igarashi, Y.; Soeda, N.; Kiyokawa, T.; Horikawa, M.; Fukushima, R. Exosome-encapsulated microRNA-23b as a minimally invasive liquid biomarker for the prediction of recurrence and prognosis of gastric cancer patients in each tumor stage. Oncol. Rep. 2018, 40, 319–330. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, W.; Liu, J.B.; Zang, X.Y.; Gu, J.M.; Pan, L.; Shi, H.; Fu, M.; Huang, Z.H.; Zhang, Y.; et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J. Exp. Clin. Cancer Res. 2018, 37, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, H.L.; Yang, H.; Zhang, X.; Wang, B.; Mao, J.H.; Li, X.; Wang, M.; Zhang, B.; Sun, Z.X.; Qian, H.; et al. Exosomal TRIM3 is a novel marker and therapy target for gastric cancer. J. Exp. Clin. Cancer Res. 2018, 37, 162. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Lin, W.R.; Qi, P.; Xu, M.D.; Wu, X.B.; Ni, S.J.; Huang, D.; Weng, W.W.; Tan, C.; Sheng, W.Q.; et al. Circulating Long RNAs in Serum Extracellular Vesicles: Their Characterization and Potential Application as Biomarkers for Diagnosis of Colorectal Cancer. Cancer Epidemiol. Prev. Biomark. 2016, 25, 1158–1166. [Google Scholar] [CrossRef] [Green Version]
- Yan, S.S.; Jiang, Y.; Liang, C.H.; Cheng, M.; Jin, C.W.; Duan, Q.H.; Xu, D.H.; Yang, L.; Zhang, X.Y.; Ren, B.; et al. Exosomal miR-6803-5p as potential diagnostic and prognostic marker in colorectal cancer. J. Cell Biochem. 2018, 119, 4113–4119. [Google Scholar] [CrossRef]
- Fu, F.F.; Jiang, W.Q.; Zhou, L.F.; Chen, Z. Circulating Exosomal miR-17-5p and miR-92a-3p Predict Pathologic Stage and Grade of Colorectal Cancer. Transl. Oncol. 2018, 11, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.P.; Bao, C.Y.; Li, S.Y.; Guo, W.J.; Zhao, J.; Chen, D.; Gu, J.R.; He, X.H.; Huang, S.L. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Li, Y.; Yuan, Y.H.; Liu, B.; Pan, S.M.; Liu, Q.Q.; Qi, X.; Zhou, H.M.; Dong, W.J.; Jia, L. The potential of exosomes derived from colorectal cancer as a biomarker. Clin. Chim. Acta 2019, 490, 186–193. [Google Scholar] [CrossRef]
- Santasusagna, S.; Moreno, I.; Navarro, A.; Castellano, J.J.; Martinez, F.; Hernandez, R.; Munoz, C.; Monzo, M. Proteomic Analysis of Liquid Biopsy from Tumor-Draining Vein Indicates that High Expression of Exosomal ECM1 Is Associated with Relapse in Stage I-III Colon Cancer. Transl. Oncol. 2018, 11, 715–721. [Google Scholar] [CrossRef]
- Sun, B.; Li, Y.M.; Zhou, Y.M.; Ng, T.K.; Zhao, C.; Gan, Q.Q.; Gu, X.D.; Xiang, J.B. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J. Cell Physiol. 2019, 234, 1416–1425. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Jiang, Y.; Yang, L.; Yan, S.S.; Wang, Y.G.; Lu, X.J. Decreased levels of serum exosomal miR-638 predict poor prognosis in hepatocellular carcinoma. J. Cell Biochem. 2018, 119, 4711–4716. [Google Scholar] [CrossRef]
- Wang, Y.R.; Zhang, C.Y.; Zhang, P.J.; Guo, G.H.; Jiang, T.; Zhao, X.M.; Jiang, J.J.; Huang, X.L.; Tong, H.L.; Tian, Y.P. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.; Chen, Y.M.; Dong, X.Y.; Wang, X.J. Serum Exosomal Long Noncoding RNAs ENSG00000258332.1 and LINC00635 for the Diagnosis and Prognosis of Hepatocellular Carcinoma. Cancer Epidemiol. Prev. Biomark. 2018, 27, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Arbelaiz, A.; Azkargorta, M.; Krawczyk, M.; Santos-Laso, A.; Lapitz, A.; Perugorria, M.; Erice, O.; Gonzalez, E.; Jimenez-Aguero, R.; Lacasta, A.; et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis (PSC), cholangiocarcinoma (CCA) and hepatocellular carcinoma (HCC). Hepatology 2017, 66, 1125–1143. [Google Scholar] [CrossRef] [PubMed]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M.; et al. An elevated expression of serum exosomal microRNA-191,-21,-451a of pancreatic neoplasm is considered to be efficient diagnostic marker. BMC Cancer 2018, 18, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhan, Y.; Du, L.T.; Wang, L.S.; Jiang, X.M.; Zhang, S.J.; Li, J.; Yan, K.Q.; Duan, W.L.; Zhao, Y.H.; Wang, L.L.; et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol. Cancer 2018, 17, 142. [Google Scholar] [CrossRef]
- Capitanio, D.; Moriggi, M.; Torretta, E.; Barbacini, P.; De Palma, S.; Vigano, A.; Lochmuller, H.; Muntoni, F.; Ferlini, A.; Mora, M.; et al. Comparative proteomic analyses of Duchenne muscular dystrophy and Becker muscular dystrophy muscles: Changes contributing to preserve muscle function in Becker muscular dystrophy patients. J. Cachexia Sarcopenia Muscle 2020, 11, 547–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beckham, C.J.; Olsen, J.; Yin, P.N.; Wu, C.H.; Ting, H.J.; Hagen, F.K.; Scosyrev, E.; Messing, E.M.; Lee, Y.F. Bladder Cancer Exosomes Contain EDIL-3/Del1 and Facilitate Cancer Progression. J. Urol. 2014, 192, 583–592. [Google Scholar] [CrossRef]
- Zhang, W.; Ni, M.W.; Su, Y.; Wang, H.; Zhu, S.X.; Zhao, A.; Li, G.R. MicroRNAs in Serum Exosomes as Potential Biomarkers in Clear-cell Renal Cell Carcinoma. Eur. Urol. Focus 2018, 4, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Raimondo, F.; Morosi, L.; Corbetta, S.; Chinello, C.; Brambilla, P.; Della Mina, P.; Villa, A.; Albo, G.; Battaglia, C.; Bosari, S.; et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol. Biosyst. 2013, 9, 1220–1233. [Google Scholar] [CrossRef] [Green Version]
- Testa, A.; Venturelli, E.; Brizzi, M.F. Extracellular Vesicles as a Novel Liquid Biopsy-Based Diagnosis for the Central Nervous System, Head and Neck, Lung, and Gastrointestinal Cancers: Current and Future Perspectives. Cancers 2021, 13, 2792. [Google Scholar] [CrossRef]
- Testa, A.; Venturelli, E.; Brizzi, M.F. Extracellular Vesicles: New Tools for Early Diagnosis of Breast and Genitourinary Cancers. Int. J. Mol. Sci. 2021, 22, 8430. [Google Scholar] [CrossRef]
- Weston, W.W.; Ganey, T.; Temple, H.T. The Relationship between Exosomes and Cancer: Implications for Diagnostics and Therapeutics. Biodrugs 2019, 33, 137–158. [Google Scholar] [CrossRef] [PubMed]
- Sung, B.H.; Ketova, T.; Hoshino, D.; Zijlstra, A.; Weaver, A.M. Directional cell movement through tissues is controlled by exosome secretion. Nat. Commun. 2015, 6, 7164. [Google Scholar] [CrossRef] [Green Version]
- Lopatina, T.; Grange, C.; Cavallari, C.; Navarro-Tableros, V.; Lombardo, G.; Rosso, A.; Cedrino, M.; Pomatto, M.; Alba, C.; Koni, M.; et al. Targeting IL-3Rα on tumor-derived endothelial cells blunts metastatic spread of triple-negative breast cancer via extracellular vesicle reprogramming. Oncogenesis 2020, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21, Erratum in Gynecol. Oncol. 2010, 116, 153. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gong, M.F.; Hu, Y.Y.; Liu, H.S.; Zhang, W.Q.; Zhang, M.M.; Hu, X.X.; Aubert, D.; Zhu, S.B.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef]
- Fang, X.; Chen, C.; Liu, B.; Ma, Z.; Hu, F.; Li, H.; Gu, H.; Xu, H. A magnetic bead-mediated selective adsorption strategy for extracellular vesicle separation and purification. Acta Biomater. 2021, 124, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Brett, S.I.; Lucien, F.; Guo, C.; Williams, K.C.; Kim, Y.; Durfee, P.N.; Brinker, C.J.; Chin, J.I.; Yang, J.; Leong, H.S. Immunoaffinity based methods are superior to kits for purification of prostate derived extracellular vesicles from plasma samples. Prostate 2017, 77, 1335–1343. [Google Scholar] [CrossRef]
- Pandey, C.M.; Augustine, S.; Kumar, S.; Kumar, S.; Nara, S.; Srivastava, S.; Malhotra, B.D. Microfluidics Based Point-of-Care Diagnostics. Biotechnol. J. 2018, 13, 1700047. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, D.; Yang, L.; Liu, Y.; Wu, Y. Application of Microfluidics in Molecular Diagnostics. Zhongguo Yi Liao Qi Xie Za Zhi 2020, 44, 520–524. [Google Scholar]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Gamez-Valero, A.; Monguio-Tortajada, M.; Carreras-Planella, L.; Franquesa, M.-L.; Beyer, K.; Borras, F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, M.R.; Wu, J.; Zhu, J.H.; Lubman, D.M. Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. J. Proteome Res. 2018, 17, 3599–3605. [Google Scholar] [CrossRef] [PubMed]
- Stranska, R.; Gysbrechts, L.; Wouters, J.; Vermeersch, P.; Bloch, K.; Dierickx, D.; Andrei, G.; Snoeck, R. Comparison of membrane affinity-based method with size-exclusion chromatography for isolation of exosome-like vesicles from human plasma. J. Transl. Med. 2018, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.Y.; Jiang, F.; Ma, Y.F.; Wang, J.Q.; Li, H.J.; Zhang, J.J. Isolation and Detection Technologies of Extracellular Vesicles and Application on Cancer Diagnostic. Dose-Response 2019, 17, 1559325819891004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, D.B.; Zhang, W.H.; Zhang, H.Y.; Zhang, F.Q.; Chen, L.M.; Ma, L.X.; Larcher, L.M.; Chen, S.X.; Liu, N.; Zhao, Q.X.; et al. Progress, opportunity, and perspective on exosome isolation-efforts for efficient exosome-based theranostics. Theranostics 2020, 10, 3684–3707. [Google Scholar] [CrossRef]
- Taly, V.; Pekin, D.; Benhaim, L.; Kotsopoulos, S.K.; Le Corre, D.; Li, X.Y.; Atochin, I.; Link, D.R.; Griffiths, A.D.; Pallier, K.; et al. Multiplex Picodroplet Digital PCR to Detect KRAS Mutations in Circulating DNA from the Plasma of Colorectal Cancer Patients. Clin. Chem. 2013, 59, 1722–1731. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Bhattacharya, S.; Kotsopoulos, S.; Olson, J.; Taly, V.; Griffiths, A.D.; Link, D.R.; Larson, J.W. Multiplex digital PCR: Breaking the one target per color barrier of quantitative PCR. Lab Chip 2011, 11, 2167–2174. [Google Scholar] [CrossRef] [Green Version]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef]
- Williams, R.; Peisajovich, S.G.; Miller, O.J.; Magdassi, S.; Tawfik, D.S.; Griffiths, A.D. Amplification of complex gene libraries by emulsion PCR. Nat. Methods 2006, 3, 545–550. [Google Scholar] [CrossRef]
- Gholizadeh, S.; Draz, M.S.; Zarghooni, M.; Sanati-Nezhad, A.; Ghavami, S.; Shafiee, H.; Akbari, M. Microfluidic approaches for isolation, detection, and characterization of extracellular vesicles: Current status and future directions. Biosens. Bioelectron. 2017, 91, 588–605. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Naranjo, J.C.; Wu, H.J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, X.; He, M.; Shang, Y.; Tetlow, A.L.; Godwin, A.K.; Zeng, Y. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat. Biomed. Eng. 2019, 3, 438–451. [Google Scholar] [CrossRef]
- Schlucker, S. SERS Microscopy: Nanoparticle Probes and Biomedical Applications. Chemphyschem 2009, 10, 1344–1354. [Google Scholar] [CrossRef]
- Kneipp, J.; Kneipp, H.; Kneipp, K. SERS—A single-molecule and nanoscale tool for bioanalytics. Chem. Soc. Rev. 2008, 37, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Q.; Yan, B.; Chen, L.X. SERS Tags: Novel Optical Nanoprobes for Bioanalysis. Chem. Rev. 2013, 113, 1391–1428. [Google Scholar] [CrossRef]
- Zamay, G.S.; Kolovskaya, O.S.; Zamay, T.N.; Glazyrin, Y.E.; Krat, A.V.; Zubkova, O.; Spivak, E.; Wehbe, M.; Gargaun, A.; Muharemagic, D.; et al. Aptamers Selected to Postoperative Lung Adenocarcinoma Detect Circulating Tumor Cells in Human Blood. Mol. Ther. 2015, 23, 1486–1496. [Google Scholar] [CrossRef] [Green Version]
- Boriachek, K.; Islam, M.N.; Gopalan, V.; Lam, A.K.; Nguyen, N.T.; Shiddiky, M.J.A. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst 2017, 142, 2211–2219. [Google Scholar] [CrossRef] [PubMed]
- Schure, M.; Moran, R.; Schuster, S.; Wagner, B.; Luo, C.P. Size exclusion chromatography with superficially porous particles. Abstr. Pap. Am. Chem. S. 2017, 1480, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Zheng, L.; Li, B.; Liu, C.C.; Pan, W.L. New technology for analysis of extracellular vesicles towards clinical diagnosis. Cancer Sci. 2021, 112, 985. [Google Scholar]
- Boing, A.N.; van der Pol, E.; Grootemaat, A.E.; Coumans, F.A.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef]
- Zhang, Y.; Deng, Z.; Lou, D.; Wang, Y.; Wang, R.; Hu, R.; Zhang, X.; Zhu, Q.; Chen, Y.; Liu, F. High-Efficiency Separation of Extracellular Vesicles from Lipoproteins in Plasma by Agarose Gel Electrophoresis. Anal. Chem. 2020, 92, 7493–7499. [Google Scholar] [CrossRef] [PubMed]
- Gilboa, T.; Maley, A.M.; Ogata, A.F.; Wu, C.; Walt, D.R. Sequential Protein Capture in Multiplex Single Molecule Arrays: A Strategy for Eliminating Assay Cross-Reactivity. Adv. Healthc. Mater. 2021, 10, e2001111. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Jing, F.; Li, G.; Wu, Z.; Cheng, Z.; Zhang, J.; Zhang, H.; Jia, C.; Jin, Q.; Mao, H.; et al. Absolute quantification of lung cancer related microRNA by droplet digital PCR. Biosens. Bioelectron. 2015, 74, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; He, C.; Liu, G.; Zhao, Y.; Hui, L.; Mu, Y.; Tang, R.K.; Luo, Y.; Zheng, S.; Wang, B. Nanoparticle Counting by Microscopic Digital Detection: Selective Quantitative Analysis of Exosomes via Surface-Anchored Nucleic Acid Amplification. Anal. Chem. 2018, 90, 6556–6562. [Google Scholar] [CrossRef]
- Cohen, L.; Cui, N.W.; Cai, Y.M.; Garden, P.M.; Li, X.; Weitz, D.A.; Walt, D.R. Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay. ACS Nano 2020, 14, 9491–9501. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]
- Cho, S.M.; Shin, S.; Kim, Y.; Song, W.; Hong, S.G.; Jeong, S.H.; Kang, M.S.; Lee, K.A. A novel approach for tuberculosis diagnosis using exosomal DNA and droplet digital PCR. Clin. Microbiol. Infect. 2020, 26, 942.e1–942.e5. [Google Scholar] [CrossRef]
- Lin, B.Q.; Tian, T.; Lu, Y.Z.; Liu, D.; Huang, M.J.; Zhu, L.; Zhu, Z.; Song, Y.L.; Yang, C.Y. Tracing Tumor-Derived Exosomal PD-L1 by Dual-Aptamer Activated Proximity-Induced Droplet Digital PCR. Angew. Chem. Int. Ed. 2021, 60, 7582–7586. [Google Scholar] [CrossRef]
- He, M.; Zeng, Y. Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. J. Lab. Autom. 2016, 21, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Feng, Q.; Sun, J.S. Lipid Nanovesicles by Microfluidics: Manipulation, Synthesis, and Drug Delivery. Adv. Mater. 2019, 31, 1804788. [Google Scholar] [CrossRef]
- Wu, M.X.; Ouyang, Y.S.; Wang, Z.Y.; Zhang, R.; Huang, P.H.; Chen, C.Y.; Li, H.; Li, P.; Quinn, D.; Dao, M.; et al. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589, Erratum in Proc. Natl. Acad. Sci. USA 2020, 117, 28525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruus, H.; Dual, J.; Hawkes, J.; Hill, M.; Laurell, T.; Nilsson, J.; Radel, S.; Sadhal, S.; Wiklund, M. Forthcoming Lab on a Chip tutorial series on acoustofluidics: Acoustofluidics-exploiting ultrasonic standing wave forces and acoustic streaming in microfluidic systems for cell and particle manipulation. Lab Chip 2011, 11, 3579–3580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, L.Y.; Friend, J.R. Surface Acoustic Wave Microfluidics. Annu. Rev. Fluid Mech. 2014, 46, 379–406. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Shao, H.L.; Weissleder, R.; Lee, H. Acoustic Purification of Extracellular Microvesicles. ACS Nano 2015, 9, 2321–2327. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.C.; Xu, X.N.; Li, B.; Situ, B.; Pan, W.L.; Hu, Y.; An, T.X.; Yao, S.H.; Zheng, L. Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 2018, 18, 4226–4232. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Gill, D.; Kilponen, R.G.; Rimai, L. Resonance Raman scattering of laser radiation by vibrational modes of carotenoid pigment molecules in intact plant tissues. Nature 1970, 227, 743–744. [Google Scholar] [CrossRef]
- Xu, H.Y.; Liao, C.; Zuo, P.; Liu, Z.W.; Ye, B.C. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem. 2018, 90, 13451–13458. [Google Scholar] [CrossRef]
- Stremersch, S.; Marro, M.; Pinchasik, B.E.; Baatsen, P.; Hendrix, A.; De Smedt, S.C.; Loza-Alvarez, P.; Skirtach, A.G.; Raemdonck, K.; Braeckmans, K. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016, 12, 3292–3301. [Google Scholar] [CrossRef]
- Lee, C.; Carney, R.P.; Hazari, S.; Smith, Z.J.; Knudson, A.; Robertson, C.S.; Lam, K.S.; Wachsmann-Hogiu, S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 2015, 7, 9290–9297. [Google Scholar] [CrossRef]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.H.; Kim, H.K.; Hong, S.; Park, J.H.; Choi, Y. Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Shi, H.; Tang, K.; Qiao, L.; Yu, G.; Ding, C.; Yu, S. Microfluidic Raman biochip detection of exosomes: A promising tool for prostate cancer diagnosis. Lab Chip 2020, 20, 4632–4637. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.G.; Choi, B.H.; Kang, K.W.; Jeong, H.; Park, Y.; et al. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef] [PubMed]
- Hassan, E.M.; Willmore, W.G.; DeRosa, M.C. Aptamers: Promising Tools for the Detection of Circulating Tumor Cells. Nucleic Acid Ther. 2016, 26, 335–347. [Google Scholar] [CrossRef]
- Yoshida, M.; Hibino, K.; Yamamoto, S.; Matsumura, S.; Yajima, Y.; Shiba, K. Preferential capture of EpCAM-expressing extracellular vesicles on solid surfaces coated with an aptamer-conjugated zwitterionic polymer. Biotechnol. Bioeng. 2018, 115, 536–544. [Google Scholar] [CrossRef]
- Kaushik, A.M.; Hsieh, K.; Wang, T.H. Droplet microfluidics for high-sensitivity and high-throughput detection and screening of disease biomarkers. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2018, 10, e1522. [Google Scholar] [CrossRef] [PubMed]
- Darmanis, S.; Nong, R.Y.; Hammond, M.; Gu, J.J.; Alderborn, A.; Vanelid, J.; Siegbahn, A.; Gustafsdottir, S.; Ericsson, O.; Landegren, U.; et al. Sensitive Plasma Protein Analysis by Microparticle-based Proximity Ligation Assays. Mol. Cell Proteom. 2010, 9, 327–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, S.; Zhang, L.Q.; Wang, S.; Liu, Y.; Wu, C.C.; Cui, C.; Sun, H.; Shi, M.L.; Jiang, Y.; Li, L.; et al. Molecular Recognition-Based DNA Nanoassemblies on the Surfaces of Nanosized Exosomes. J. Am. Chem. Soc. 2017, 139, 5289–5292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Zu, Y. A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viswanathan, S.; Rani, C.; Ho, J.A.A. Electrochemical immunosensor for multiplexed detection of food-borne pathogens using nanocrystal bioconjugates and MWCNT screen-printed electrode. Talanta 2012, 94, 315–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, F.Y.; Xu, B.Y.; Xu, J.J.; Chen, H.Y. Simultaneous electrochemical immunoassay using CdS/DNA and PbS/DNA nanochains as labels. Biosens. Bioelectron. 2013, 39, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Shiddiky, M.J.A.; Rauf, S.; Kithva, P.H.; Trau, M. Graphene/quantum dot bionanoconjugates as signal amplifiers in stripping voltammetric detection of EpCAM biomarkers. Biosens. Bioelectron. 2012, 35, 251–257. [Google Scholar] [CrossRef]
- Shiddiky, M.J.A.; Kithva, P.H.; Kozak, D.; Trau, M. An electrochemical immunosensor to minimize the nonspecific adsorption and to improve sensitivity of protein assays in human serum. Biosens. Bioelectron. 2012, 38, 132–137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Rahimian, A.; Son, K.; Shin, D.S.; Patel, T.; Revzin, A. Development of an aptasensor for electrochemical detection of exosomes. Methods 2016, 97, 88–93. [Google Scholar] [CrossRef] [Green Version]
- Doldan, X.; Fagundez, P.; Cayota, A.; Laiz, J.; Tosar, J.P. Electrochemical Sandwich Immunosensor for Determination of Exosomes Based on Surface Marker-Mediated Signal Amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef]
- Wu, M.; Chen, Z.; Xie, Q.; Xiao, B.; Zhou, G.; Chen, G.; Bian, Z. One-step quantification of salivary exosomes based on combined aptamer recognition and quantum dot signal amplification. Biosens. Bioelectron. 2021, 171, 112733. [Google Scholar] [CrossRef]
- Wang, M.; Pan, Y.; Wu, S.; Sun, Z.; Wang, L.; Yang, J.; Yin, Y.; Li, G. Detection of colorectal cancer-derived exosomes based on covalent organic frameworks. Biosens. Bioelectron. 2020, 169, 112638. [Google Scholar] [CrossRef]
- Wang, Z.X.; Wu, H.J.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.J.; Liu, X.W. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.; Choi, M.; Lee, H.; Kim, Y.H.; Han, J.Y.; Lee, E.S.; Cho, Y. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J. Nanobiotechnol. 2019, 17, 1. [Google Scholar] [CrossRef] [PubMed]
- Scherr, S.M.; Daaboul, G.G.; Trueb, J.; Sevenler, D.; Fawcett, H.; Goldberg, B.; Connor, J.H.; Unlu, M.S. Real-Time Capture and Visualization of Individual Viruses in Complex Media. ACS Nano 2016, 10, 2827–2833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Crescitelli, R.; Lasser, C.; Jang, S.C.; Cvjetkovic, A.; Malmhall, C.; Karimi, N.; Hoog, J.L.; Johansson, I.; Fuchs, J.; Thorsell, A.; et al. Subpopulations of extracellular vesicles from human metastatic melanoma tissue identified by quantitative proteomics after optimized isolation. J. Extracell. Vesicles 2020, 9, 1722433. [Google Scholar] [CrossRef] [PubMed]
- Quaglia, F.; Krishn, S.R.; Daaboul, G.G.; Sarker, S.; Pippa, R.; Domingo-Domenech, J.; Kumar, G.; Fortina, P.; McCue, P.; Kelly, W.K.; et al. Small extracellular vesicles modulated by alphaVbeta3 integrin induce neuroendocrine differentiation in recipient cancer cells. J. Extracell. Vesicles 2020, 9, 1761072. [Google Scholar] [CrossRef] [PubMed]
- Harkonen, K.; Oikari, S.; Kyykallio, H.; Capra, J.; Hakkola, S.; Ketola, K.; Thanigai Arasu, U.; Daaboul, G.; Malloy, A.; Oliveira, C.; et al. CD44s Assembles Hyaluronan Coat on Filopodia and Extracellular Vesicles and Induces Tumorigenicity of MKN74 Gastric Carcinoma Cells. Cells 2019, 8, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Scheme | Cancer Type | Marker Type | Name | Expression | Source | Clinical Value | Ref. |
|---|---|---|---|---|---|---|---|
| Central nervous | Glioma | DNA | IDH1 mutation | ↑ | Serum | Diagnosis, Prognosis | [35] |
| mRNA | EGFRvIII | cerebrospinal fluid | Diagnosis | [43,44] | |||
| miRNA | miR-454-3P/miR-320/miR-574-3P | Serum | Diagnosis, Prognosis judgment | [45,46] | |||
| snRNA | RNU6-1 | Serum | Diagnosis, Prognosis judgment | [46] | |||
| Thoracic | Lung cancer | mRNA | BRAF/EGFR/FRS2/GREB1/LZTS1 | ↑ | saliva | Diagnosis | [47] |
| miRNA | miR-451a/miR-425-3p/miR-4257 | Serum plasma | Recurrence/Resistance/Prognosis judgment | [48,49,50] | |||
| Protein | Tim-3/LBP/LRG1 | Serum/plasma/Urine | Transferrer/Staging/Diagnosis | [51,52,53] | |||
| Breast tumors | mRNA | hTERT | Serum | Early diagnosis/Recurrence | [54] | ||
| miRNA | miR-223-3P | Plasma | [32] | ||||
| Protein | FN | Plasma | [55] | ||||
| Digestive | Esophageal cancer | miRNA | miR-21/RUN6-1/miR-16-5p | ↑ | Serum | Diagnosis | [56,57] |
| Gastric carcinoma | miRNA | miR-423-5p/miR-21,miR-1225-5p/miR-23b | ↑/-/↓ | Serum/Peritoneal lavage fluid/Plasma | Diagnosis, Prognosis judgment/Recurrence/Recurrence, Prognosis judgment | [31,58,59] | |
| LncRNA | UFC1 | ↑ | Serum | Diagnosis, Prognosis judgment | [60] | ||
| Protein | TRIM3 | ↓ | Serum | Diagnosis | [61] | ||
| Colorectal cancer | mRNA | KRTAP5/MAGEA3 | ↑ | Serum | - | [62] | |
| LncRNA | BCAR4 | Serum | - | ||||
| miRNA | miR-6803-5P, miR-548c-5p, miR-92a-3p | Serum | Transferrer/Staging/Diagnosis | [63,64] | |||
| Diagnosis/Transferrer | |||||||
| circRNA | Circ-KLDHC10 | Serum | - | [65] | |||
| Protein | TAG72/CA125/CPNE3 | Plasma | Resistance/Transferrer/Diagnosis, Prognosis judgment | [66,67,68] | |||
| Liver cancer | mRNA | hnRNPH1 | ↑ | Serum | Diagnosis | [29] | |
| miRNA | miR-638 | ↓ | Serum | Diagnosis | [69] | ||
| miR-122 | ↑ | [70] | |||||
| miR-148a | ↑ | ||||||
| LncRNA | LINC00635 | ↑ | Serum | Diagnosis, Prognosis judgment | [71] | ||
| Protein | LG3BP/PIGR | ↑ | Serum | Diagnosis | [72] | ||
| Pancreatic carcinoma | miRNA | miR-191/miR-21/miR-451a | ↑ | Serum | Diagnosis | [73] | |
| Protein | GPC1+ | [36] | |||||
| Urinary | Bladder cancer | miRNA | miR-615-3p | ↑ | Urine | Diagnosis, Prognosis judgment | [30] |
| LncRNA | MALAT1/PCAT-1 | Urine | [74] | ||||
| Protein | TACSTD2/EDIL-3 | Urine | [75,76] | ||||
| Kidney cancer | miRNA | miR-210/miR-1233 | Serum | [77] | |||
| Protein | MMP9/PODXL/DKK4 | Urine | [78] |
| Methods | Mechanism | Cancer | Advantage | Limitation | Significance | Ref. |
|---|---|---|---|---|---|---|
| Size exclusion chromatography | Substances eluted out in accordance with their particle size or charge difference |
|
|
| This method is not only suitable for processing trace amounts of liquid samples but also easily scalable and automated for high-throughput exosomal preparation, which allows fast, precise, scalable, and automated exosomal isolation. | [91,92,93,94] |
| Droplet digital enzyme-linked immunosorbent assay | Based on the specific binding between exosome biomarkers and immobilized antibodies (ligands) |
|
|
| Collecting exosomes of specific origin not only facilitates the study of their parental cells but also provides essential indicators for disease diagnosis (for example, via detecting EpCAM positive exosomes to assess the existence of EpCAM related cancers). | [95] |
| droplet digital PCR | ddPCR technology uses a combination of microfluidics and proprietary surfactant chemistries to divide PCR samples into water-in-oil droplets |
|
|
| ddPCR uses aqueous droplets with volumes ranging from a few femtoliters to nanoliters dispersed in oil for compartmentalization of PCR reactions, opening up the possibility of having a theoretically unlimited number of compartments, thus largely increasing detection sensitivity. | [96,97,98,99] |
| Microfluidic technology | Based on different principles, including immunoaffinity, size, and density |
|
|
| Microfluidic techniques are dramatically innovating the landscape of exosome-based diagnosis by transferring the traditional two-step procedure (exosome isolation and characterization) to an integrated one-step process, which is especially valuable for non-invasive disease detection, such as early-stage cancer screening. | [100,101,102] |
| Surface-enhanced Raman scattering technology | SERS is a spectroscopic phenomenon that enhances the Raman signal by absorbing the molecule on the rough surface or nanometal materials. |
|
|
| Multiple analytes (or biomarkers) in one sample could be detected in a single cycle/run when using more than one SERS tag, which results in an accurate, efficient, and simplified diagnosis of diseases. | [103,104,105] |
| Aptamer-based separation method | The aptamer is combined with oligonucleotides or peptides as a detection probe, linked to a biological vector or emerging nanomaterials to achieve target monitoring |
|
|
| The biomarkers and corresponding aptamers can be exploited to improve cancer diagnostics and therapies. | [106] |
| Quantum dot-based exosome quantification | Using quantum dots as signal amplifiers |
|
|
| The approach could potentially represent an effective bioassay for the quantification of disease-specific exosomes in clinical samples. | [107] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.; Yang, Q.; Wang, R.; Luo, R.; Zhu, S.; Li, M.; Li, W.; Chen, C.; Zou, Y.; Huang, Z.; et al. Emerging Advances of Detection Strategies for Tumor-Derived Exosomes. Int. J. Mol. Sci. 2022, 23, 868. https://doi.org/10.3390/ijms23020868
Cheng H, Yang Q, Wang R, Luo R, Zhu S, Li M, Li W, Chen C, Zou Y, Huang Z, et al. Emerging Advances of Detection Strategies for Tumor-Derived Exosomes. International Journal of Molecular Sciences. 2022; 23(2):868. https://doi.org/10.3390/ijms23020868
Chicago/Turabian StyleCheng, Huijuan, Qian Yang, Rongrong Wang, Ruhua Luo, Shanshan Zhu, Minhui Li, Wenqi Li, Cheng Chen, Yuqing Zou, Zhihua Huang, and et al. 2022. "Emerging Advances of Detection Strategies for Tumor-Derived Exosomes" International Journal of Molecular Sciences 23, no. 2: 868. https://doi.org/10.3390/ijms23020868
APA StyleCheng, H., Yang, Q., Wang, R., Luo, R., Zhu, S., Li, M., Li, W., Chen, C., Zou, Y., Huang, Z., Xie, T., Wang, S., Zhang, H., & Tian, Q. (2022). Emerging Advances of Detection Strategies for Tumor-Derived Exosomes. International Journal of Molecular Sciences, 23(2), 868. https://doi.org/10.3390/ijms23020868







