Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain
Abstract
1. Introduction
2. Results
2.1. mtHsp22 Preserves Spatial Reversal Learning and Memory in rTg4510 Mice
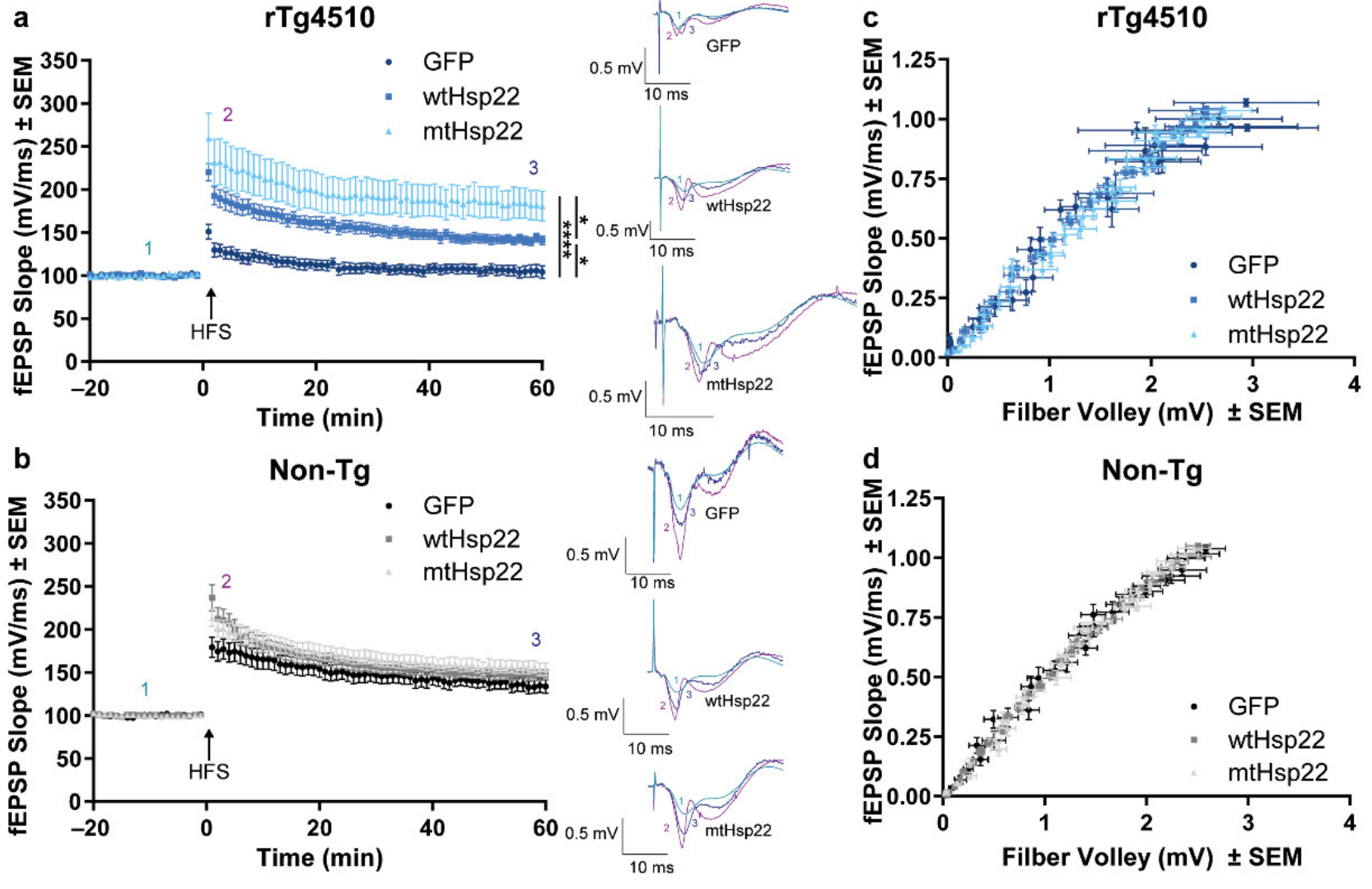
2.2. wtHsp22 and mtHsp22 Overexpression Rescue LTP Deficits in rTg4510 Mice
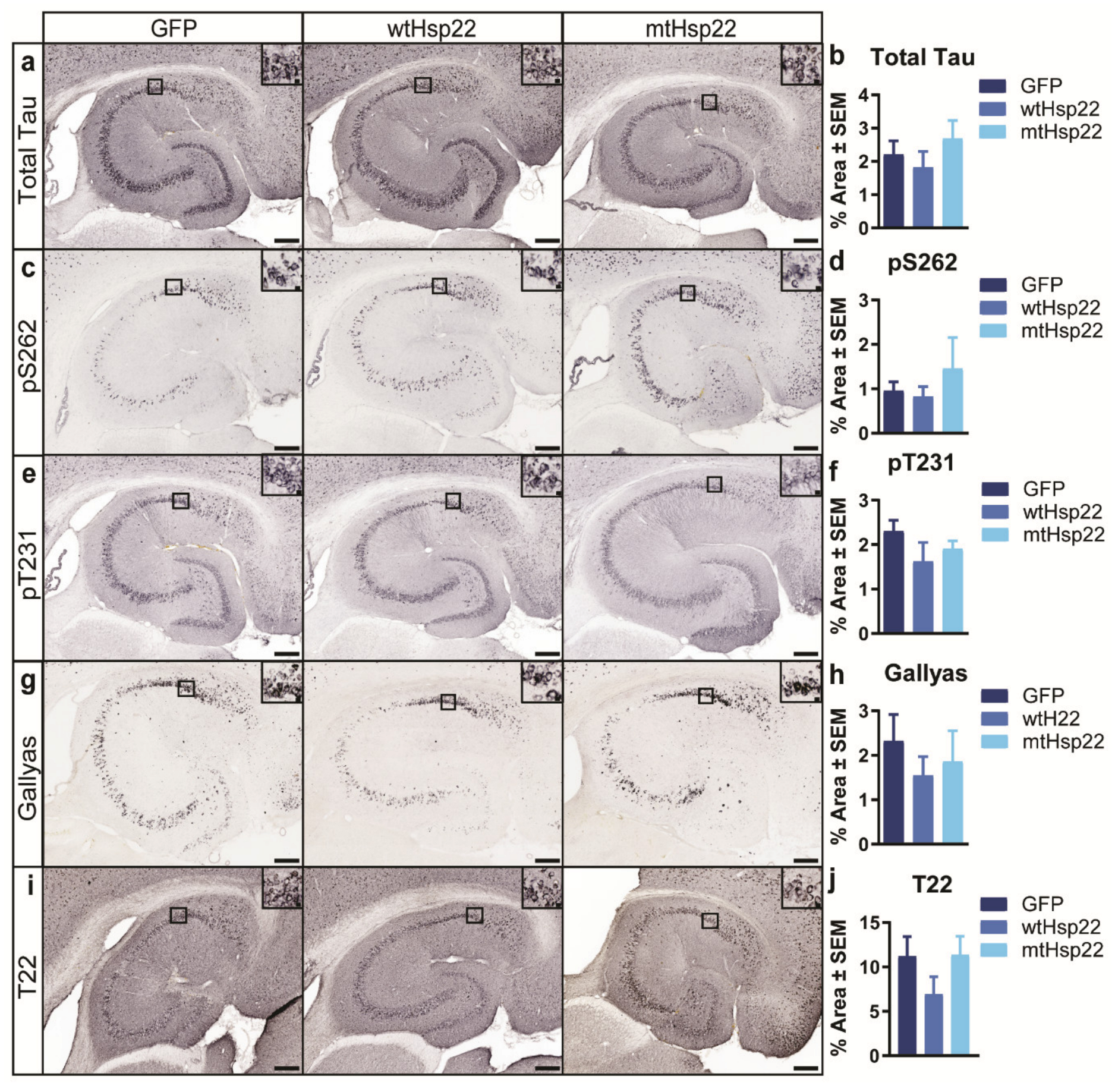
2.3. Overexpression of wtHsp22 and mtHsp22 Does Not Change Tau Levels or Phosphorylation Status
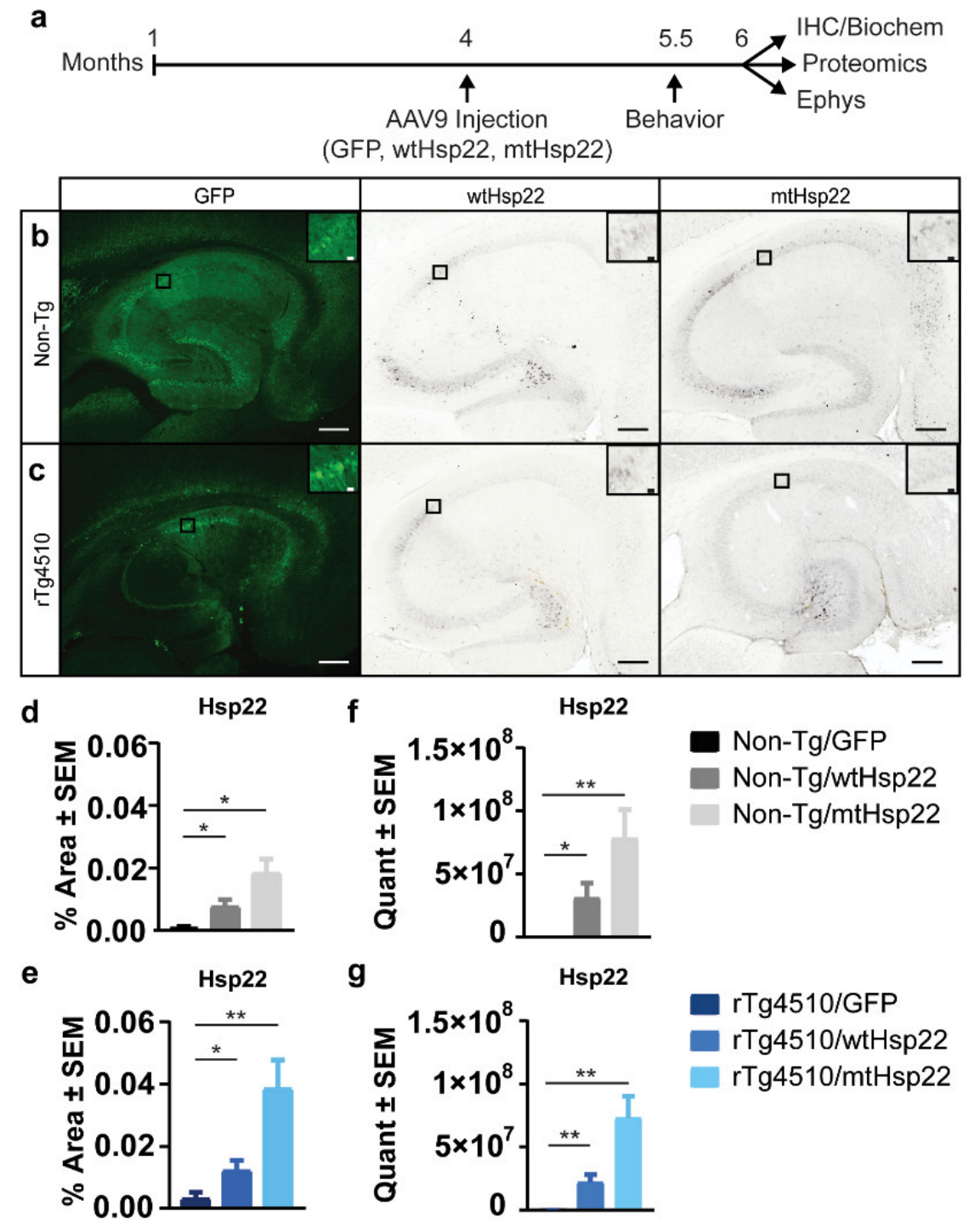
2.4. Hsp22 Overexpression Preserves Neurons in rTg4510 Mice
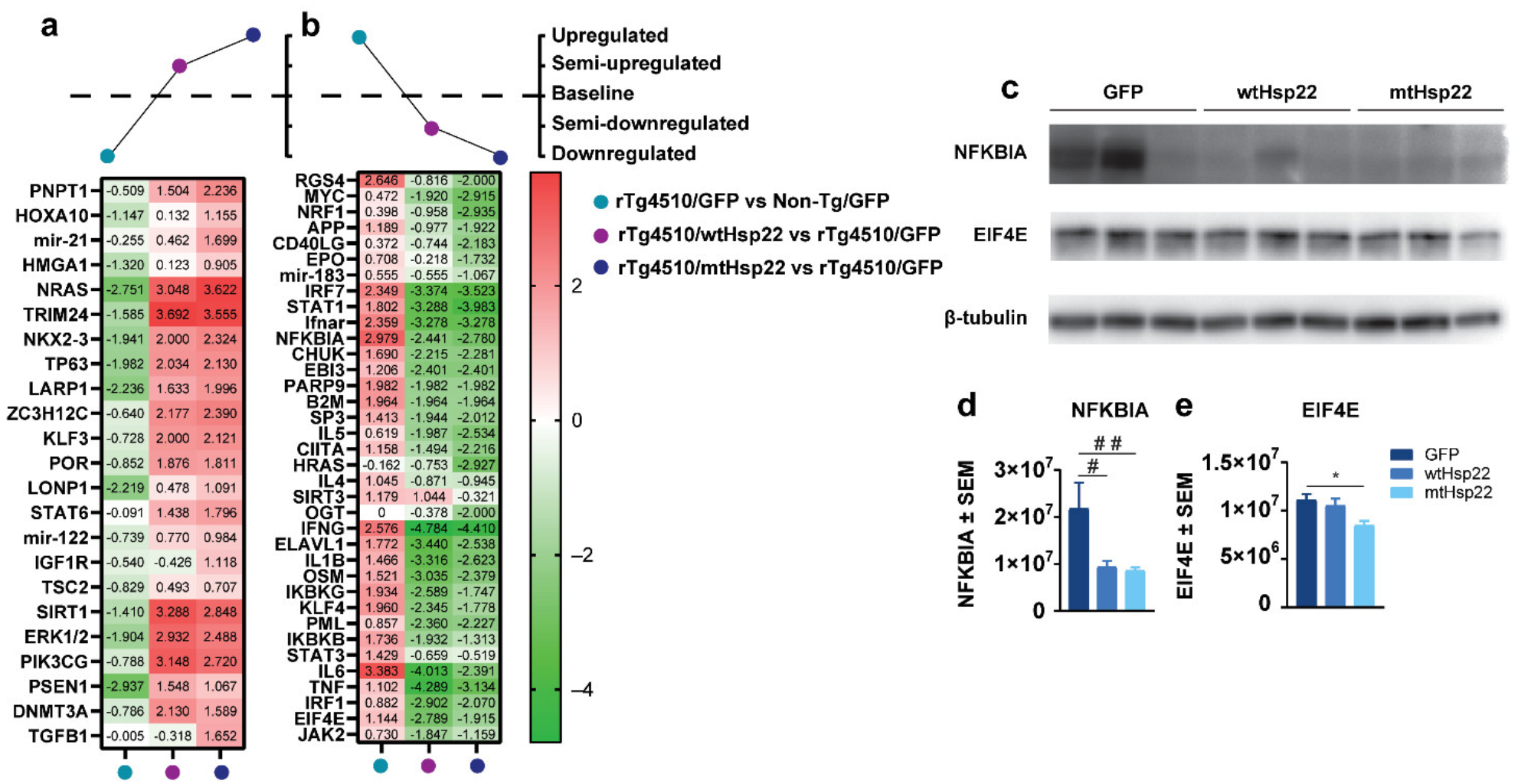
2.5. Overexpression of Hsp22 Changes Expression of Key Neuroprotective Proteins in rTg4510 Mice
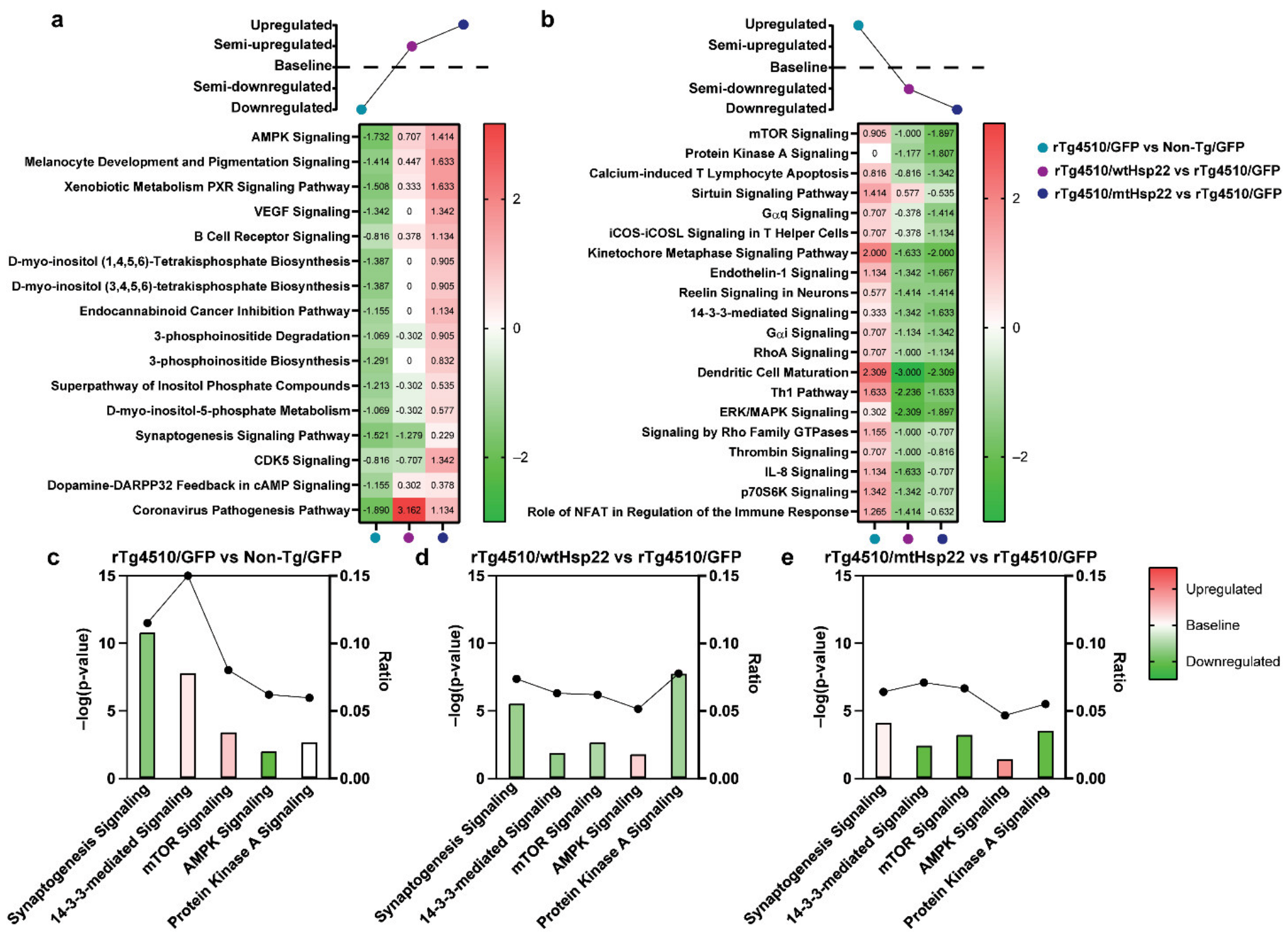
2.6. Hsp22 Overexpression Protects against Tau-Mediated Disruption of Cellular Metabolism, Health, and Integrity Pathways
3. Discussion
4. Materials and Methods
4.1. Virus Production
4.2. Animals
4.3. Radial-Arm Water Maze with Reversal and Open Field
4.4. Immunohistochemistry, Immunoblotting, and Gallyas Staining
4.5. Unbiased Stereology
4.6. Electrophysiology
4.7. Mass Spectrometry
4.7.1. Experimental Design and Statistical Rationale
4.7.2. Parallel Reaction Monitoring
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spillantini, M.G.; Goedert, M. Tau pathology and neurodegeneration. Lancet Neurol. 2013, 12, 609–622. [Google Scholar] [CrossRef]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Shao, E.; Mucke, L. Tau: Enabler of diverse brain disorders and target of rapidly evolving therapeutic strategies. Science 2021, 371, eabb8255. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef]
- Klaips, C.L.; Jayaraj, G.G.; Hartl, F.U. Pathways of cellular proteostasis in aging and disease. J. Cell Biol. 2018, 217, 51–63. [Google Scholar] [CrossRef]
- Carra, S.; Alberti, S.; Arrigo, P.A.; Benesch, J.L.; Benjamin, I.J.; Boelens, W.; Bartelt-Kirbach, B.; Brundel, B.; Buchner, J.; Bukau, B.; et al. The growing world of small heat shock proteins: From structure to functions. Cell Stress Chaperones 2017, 22, 601–611. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small heat shock proteins: Role in cellular functions and pathology. Biochim. Biophys. Acta 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Stokoe, D.; Engel, K.; Campbell, D.G.; Cohen, P.; Gaestel, M. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 1992, 313, 307–313. [Google Scholar] [CrossRef]
- Peschek, J.; Braun, N.; Rohrberg, J.; Back, K.C.; Kriehuber, T.; Kastenmuller, A.; Weinkauf, S.; Buchner, J. Regulated structural transitions unleash the chaperone activity of alphaB-crystallin. Proc. Natl. Acad. Sci. USA 2013, 110, E3780–E3789. [Google Scholar] [CrossRef]
- Dickey, C.; Kraft, C.; Jinwal, U.; Koren, J.; Johnson, A.; Anderson, L.; Lebson, L.; Lee, D.; Dickson, D.; de Silva, R.; et al. Aging analysis reveals slowed tau turnover and enhanced stress response in a mouse model of tauopathy. Am. J. Pathol. 2009, 174, 228–238. [Google Scholar] [CrossRef]
- Webster, J.M.; Darling, A.L.; Sanders, T.A.; Blazier, D.M.; Vidal-Aguiar, Y.; Beaulieu-Abdelahad, D.; Plemmons, D.G.; Hill, S.E.; Uversky, V.N.; Bickford, P.C.; et al. Hsp22 with an N-Terminal Domain Truncation Mediates a Reduction in Tau Protein Levels. Int. J. Mol. Sci. 2020, 21, 5442. [Google Scholar] [CrossRef]
- Koopman, M.B.; Rüdiger, S.G.D. Alzheimer Cells on Their Way to Derailment Show Selective Changes in Protein Quality Control Network. Front. Mol. Biosci. 2020, 7, 214. [Google Scholar] [CrossRef]
- Ojha, J.; Masilamoni, G.; Dunlap, D.; Udoff, R.A.; Cashikar, A.G. Sequestration of toxic oligomers by HspB1 as a cytoprotective mechanism. Mol. Cell Biol. 2011, 31, 3146–3157. [Google Scholar] [CrossRef]
- Shimura, H.; Miura-Shimura, Y.; Kosik, K.S. Binding of tau to heat shock protein 27 leads to decreased concentration of hyperphosphorylated tau and enhanced cell survival. J. Biol. Chem. 2004, 279, 17957–17962. [Google Scholar] [CrossRef]
- Bruinsma, I.B.; Bruggink, K.A.; Kinast, K.; Versleijen, A.A.; Segers-Nolten, I.M.; Subramaniam, V.; Kuiperij, H.B.; Boelens, W.; de Waal, R.M.; Verbeek, M.M. Inhibition of alpha-synuclein aggregation by small heat shock proteins. Proteins 2011, 79, 2956–2967. [Google Scholar] [CrossRef]
- Baughman, H.E.R.; Pham, T.T.; Adams, C.S.; Nath, A.; Klevit, R.E. Release of a disordered domain enhances HspB1 chaperone activity toward tau. Proc. Natl. Acad. Sci. USA 2020, 117, 2923–2929. [Google Scholar] [CrossRef]
- Abisambra, J.F.; Blair, L.J.; Hill, S.E.; Jones, J.R.; Kraft, C.; Rogers, J.; Koren, J., 3rd; Jinwal, U.K.; Lawson, L.; Johnson, A.G.; et al. Phosphorylation dynamics regulate Hsp27-mediated rescue of neuronal plasticity deficits in tau transgenic mice. J. Neurosci. 2010, 30, 15374–15382. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; Boelens, W.C.; Otte-Holler, I.; Kamps, B.; Kusters, B.; Maat-Schieman, M.L.; de Waal, R.M.; Verbeek, M.M. Small heat shock protein HspB8: Its distribution in Alzheimer’s disease brains and its inhibition of amyloid-beta protein aggregation and cerebrovascular amyloid-beta toxicity. Acta Neuropathol. 2006, 111, 139–149. [Google Scholar] [CrossRef]
- Morrow, G.; Tanguay, R.M. Drosophila melanogaster Hsp22: A mitochondrial small heat shock protein influencing the aging process. Front. Genet. 2015, 6, 1026. [Google Scholar] [CrossRef]
- Shemetov, A.A.; Seit-Nebi, A.S.; Bukach, O.V.; Gusev, N.B. Phosphorylation by cyclic AMP-dependent protein kinase inhibits chaperone-like activity of human HSP22 in vitro. Biochemistry 2008, 73, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Mymrikov, E.V.; Seit-Nebi, A.S.; Gusev, N.B. Heterooligomeric complexes of human small heat shock proteins. Cell Stress Chaperones 2012, 17, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.L.; Dahrendorff, J.; Creodore, S.G.; Dickey, C.A.; Blair, L.J.; Uversky, V.N. Small heat shock protein 22 kDa can modulate the aggregation and liquid-liquid phase separation behavior of tau. Protein Sci. 2021, 30, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.D.; Shelton, L.B.; Zheng, D.; Favretto, F.; Nordhues, B.A.; Darling, A.; Sullivan, L.E.; Sun, Z.; Solanki, P.K.; Martin, M.D.; et al. Human cyclophilin 40 unravels neurotoxic amyloids. PLoS Biol. 2017, 15, e2001336. [Google Scholar] [CrossRef] [PubMed]
- Selenica, M.L.; Benner, L.; Housley, S.B.; Manchec, B.; Lee, D.C.; Nash, K.R.; Kalin, J.; Bergman, J.A.; Kozikowski, A.; Gordon, M.N.; et al. Histone deacetylase 6 inhibition improves memory and reduces total tau levels in a mouse model of tau deposition. Alzheimer’s Res. Ther. 2014, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, G.; Laczo, J.; Hort, J.; Minihane, A.M.; Hornberger, M. Spatial navigation deficits—Overlooked cognitive marker for preclinical Alzheimer disease? Nat. Rev. Neurol. 2018, 14, 496–506. [Google Scholar] [CrossRef]
- Wang, X.; Smith, K.; Pearson, M.; Hughes, A.; Cosden, M.L.; Marcus, J.; Hess, J.F.; Savage, M.J.; Rosahl, T.; Smith, S.M.; et al. Early intervention of tau pathology prevents behavioral changes in the rTg4510 mouse model of tauopathy. PLoS ONE 2018, 13, e0195486. [Google Scholar] [CrossRef]
- Spires, T.L.; Orne, J.D.; SantaCruz, K.; Pitstick, R.; Carlson, G.A.; Ashe, K.H.; Hyman, B.T. Region-specific dissociation of neuronal loss and neurofibrillary pathology in a mouse model of tauopathy. Am. J. Pathol. 2006, 168, 1598–1607. [Google Scholar] [CrossRef]
- Shen, J.; Wang, D.; Wang, X.; Gupta, S.; Ayloo, B.; Wu, S.; Prasad, P.; Xiong, Q.; Xia, J.; Ge, S. Neurovascular Coupling in the Dentate Gyrus Regulates Adult Hippocampal Neurogenesis. Neuron 2019, 103, 878–890. [Google Scholar] [CrossRef]
- Ramirez-Rodriguez, G.; Babu, H.; Klempin, F.; Krylyshkina, O.; Baekelandt, V.; Gijsbers, R.; Debyser, Z.; Overall, R.W.; Nicola, Z.; Fabel, K.; et al. The alpha crystallin domain of small heat shock protein b8 (Hspb8) acts as survival and differentiation factor in adult hippocampal neurogenesis. J. Neurosci. 2013, 33, 5785–5796. [Google Scholar] [CrossRef]
- Kwok, A.S.; Phadwal, K.; Turner, B.J.; Oliver, P.L.; Raw, A.; Simon, A.K.; Talbot, K.; Agashe, V.R. HspB8 mutation causing hereditary distal motor neuropathy impairs lysosomal delivery of autophagosomes. J. Neurochem. 2011, 119, 1155–1161. [Google Scholar] [CrossRef]
- Crippa, V.; Carra, S.; Rusmini, P.; Sau, D.; Bolzoni, E.; Bendotti, C.; De Biasi, S.; Poletti, A. A role of small heat shock protein B8 (HspB8) in the autophagic removal of misfolded proteins responsible for neurodegenerative diseases. Autophagy 2010, 6, 958–960. [Google Scholar] [CrossRef]
- Chowdary, T.K.; Raman, B.; Ramakrishna, T.; Rao, C.M. Mammalian Hsp22 is a heat-inducible small heat-shock protein with chaperone-like activity. Biochem. J. 2004, 381 Pt 2, 379–387. [Google Scholar] [CrossRef]
- Yue, M.; Hanna, A.; Wilson, J.; Roder, H.; Janus, C. Sex difference in pathology and memory decline in rTg4510 mouse model of tauopathy. Neurobiol. Aging 2011, 32, 590–603. [Google Scholar] [CrossRef]
- Latchman, D.S. Protective effect of heat shock proteins in the nervous system. Curr. Neurovasc. Res. 2004, 1, 21–27. [Google Scholar] [CrossRef]
- Sharp, F.R.; Zhan, X.; Liu, D.Z. Heat shock proteins in the brain: Role of Hsp70, Hsp 27, and HO-1 (Hsp32) and their therapeutic potential. Transl. Stroke Res. 2013, 4, 685–692. [Google Scholar] [CrossRef]
- Han, H.J.; Allen, C.C.; Buchovecky, C.M.; Yetman, M.J.; Born, H.A.; Marin, M.A.; Rodgers, S.P.; Song, B.J.; Lu, H.C.; Justice, M.J.; et al. Strain background influences neurotoxicity and behavioral abnormalities in mice expressing the tetracycline transactivator. J. Neurosci. 2012, 32, 10574–10586. [Google Scholar] [CrossRef]
- Santacruz, K.; Lewis, J.; Spires, T.; Paulson, J.; Kotilinek, L.; Ingelsson, M.; Guimaraes, A.; DeTure, M.; Ramsden, M.; McGowan, E.; et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science 2005, 309, 476–481. [Google Scholar] [CrossRef]
- Arendt, T.; Stieler, J.T.; Holzer, M. Tau and tauopathies. Brain Res. Bull. 2016, 126 Pt 3, 238–292. [Google Scholar] [CrossRef]
- Hoover, B.R.; Reed, M.N.; Su, J.; Penrod, R.D.; Kotilinek, L.A.; Grant, M.K.; Pitstick, R.; Carlson, G.A.; Lanier, L.M.; Yuan, L.L.; et al. Tau mislocalization to dendritic spines mediates synaptic dysfunction independently of neurodegeneration. Neuron 2010, 68, 1067–1081. [Google Scholar] [CrossRef]
- Wang, B.Z.; Yang, J.J.; Zhang, H.; Smith, C.A.; Jin, K. AMPK Signaling Regulates the Age-Related Decline of Hippocampal Neurogenesis. Aging Dis. 2019, 10, 1058–1074. [Google Scholar] [CrossRef] [PubMed]
- Buonarati, O.R.; Hammes, E.A.; Watson, J.F.; Greger, I.H.; Hell, J.W. Mechanisms of postsynaptic localization of AMPA-type glutamate receptors and their regulation during long-term potentiation. Sci. Signal. 2019, 12, eaar6889. [Google Scholar] [CrossRef] [PubMed]
- Esteban, J.A.; Shi, S.H.; Wilson, C.; Nuriya, M.; Huganir, R.L.; Malinow, R. PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat. Neurosci. 2003, 6, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Hoeffer, C.A.; Klann, E. mTOR signaling: At the crossroads of plasticity, memory and disease. Trends Neurosci. 2010, 33, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Bell, C.M.; Rothbart, S.B.; Moran, R.G. AMP-activated Protein Kinase (AMPK) Control of mTORC1 Is p53- and TSC2-independent in Pemetrexed-treated Carcinoma Cells. J. Biol. Chem. 2015, 290, 27473–27486. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Park, J.Y. Emerging roles of 14-3-3gamma in the brain disorder. BMB Rep. 2020, 53, 500–511. [Google Scholar] [CrossRef] [PubMed]
- Kundu, A.; Shelar, S.; Ghosh, A.P.; Ballestas, M.; Kirkman, R.; Nam, H.; Brinkley, G.J.; Karki, S.; Mobley, J.A.; Bae, S.; et al. 14-3-3 proteins protect AMPK-phosphorylated ten-eleven translocation-2 (TET2) from PP2A-mediated dephosphorylation. J. Biol. Chem. 2020, 295, 1754–1766. [Google Scholar] [CrossRef]
- Carra, S.; Seguin, S.J.; Landry, J. HspB8 and Bag3: A new chaperone complex targeting misfolded proteins to macroautophagy. Autophagy 2008, 4, 237–239. [Google Scholar] [CrossRef]
- Inoue, K.; Rispoli, J.; Kaphzan, H.; Klann, E.; Chen, E.I.; Kim, J.; Komatsu, M.; Abeliovich, A. Macroautophagy deficiency mediates age-dependent neurodegeneration through a phospho-tau pathway. Mol. Neurodegener. 2012, 7, 48. [Google Scholar] [CrossRef]
- Kenny, A.; Jimenez-Mateos, E.M.; Zea-Sevilla, M.A.; Rabano, A.; Gili-Manzanaro, P.; Prehn, J.H.M.; Henshall, D.C.; Avila, J.; Engel, T.; Hernandez, F. Proteins and microRNAs are differentially expressed in tear fluid from patients with Alzheimer’s disease. Sci. Rep. 2019, 9, 15437. [Google Scholar] [CrossRef]
- Li, X.; An, W.L.; Alafuzoff, I.; Soininen, H.; Winblad, B.; Pei, J.J. Phosphorylated eukaryotic translation factor 4E is elevated in Alzheimer brain. Neuroreport 2004, 15, 2237–2240. [Google Scholar] [CrossRef]
- Duncan, R.; Milburn, S.C.; Hershey, J.W. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 1987, 262, 380–388. [Google Scholar] [CrossRef]
- Uttam, S.; Wong, C.; Price, T.J.; Khoutorsky, A. eIF4E-Dependent Translational Control: A Central Mechanism for Regulation of Pain Plasticity. Front. Genet. 2018, 9, 470. [Google Scholar] [CrossRef]
- Amorim, I.S.; Kedia, S.; Kouloulia, S.; Simbriger, K.; Gantois, I.; Jafarnejad, S.M.; Li, Y.; Kampaite, A.; Pooters, T.; Romano, N.; et al. Loss of eIF4E Phosphorylation Engenders Depression-like Behaviors via Selective mRNA Translation. J. Neurosci. 2018, 38, 2118–2133. [Google Scholar] [CrossRef]
- Herdy, B.; Jaramillo, M.; Svitkin, Y.V.; Rosenfeld, A.B.; Kobayashi, M.; Walsh, D.; Alain, T.; Sean, P.; Robichaud, N.; Topisirovic, I.; et al. Translational control of the activation of transcription factor NF-kappaB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 2012, 13, 543–550. [Google Scholar] [CrossRef]
- Liu, L.; Wu, Q.; Zhong, W.; Chen, Y.; Zhang, W.; Ren, H.; Sun, L.; Sun, J. Microarray Analysis of Differential Gene Expression in Alzheimer’s Disease Identifies Potential Biomarkers with Diagnostic Value. Med. Sci. Monit. 2020, 26, e919249. [Google Scholar] [CrossRef]
- Palomino-Alonso, M.; Lachen-Montes, M.; Gonzalez-Morales, A.; Ausin, K.; Perez-Mediavilla, A.; Fernandez-Irigoyen, J.; Santamaria, E. Network-Driven Proteogenomics Unveils an Aging-Related Imbalance in the Olfactory IkappaBalpha-NFkappaB p65 Complex Functionality in Tg2576 Alzheimer’s Disease Mouse Model. Int. J. Mol. Sci. 2017, 18, 2260. [Google Scholar] [CrossRef]
- O’Neill, L.A.; Kaltschmidt, C. NF-kappa B: A crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997, 20, 252–258. [Google Scholar] [CrossRef]
- Oikawa, K.; Odero, G.L.; Platt, E.; Neuendorff, M.; Hatherell, A.; Bernstein, M.J.; Albensi, B.C. NF-kappaB p50 subunit knockout impairs late LTP and alters long term memory in the mouse hippocampus. BMC Neurosci. 2012, 13, 45. [Google Scholar] [CrossRef]
- Boersma, M.C.; Dresselhaus, E.C.; De Biase, L.M.; Mihalas, A.B.; Bergles, D.E.; Meffert, M.K. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J. Neurosci. 2011, 31, 5414–5425. [Google Scholar] [CrossRef]
- Luo, S.; Kang, S.S.; Wang, Z.H.; Liu, X.; Day, J.X.; Wu, Z.; Peng, J.; Xiang, D.; Springer, W.; Ye, K. Akt Phosphorylates NQO1 and Triggers its Degradation, Abolishing Its Antioxidative Activities in Parkinson’s Disease. J. Neurosci. 2019, 39, 7291–7305. [Google Scholar] [CrossRef] [PubMed]
- Poppinga, W.J.; Munoz-Llancao, P.; Gonzalez-Billault, C.; Schmidt, M. A-kinase anchoring proteins: cAMP compartmentalization in neurodegenerative and obstructive pulmonary diseases. Br. J. Pharmacol. 2014, 171, 5603–5623. [Google Scholar] [CrossRef] [PubMed]
- Gorski, J.A.; Gomez, L.L.; Scott, J.D.; Dell’Acqua, M.L. Association of an A-kinase-anchoring protein signaling scaffold with cadherin adhesion molecules in neurons and epithelial cells. Mol. Biol. Cell 2005, 16, 3574–3590. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kudo, W.; Lee, H.P.; Smith, M.A.; Zhu, X.; Matsuyama, S.; Lee, H.G. Inhibition of Bax protects neuronal cells from oligomeric Abeta neurotoxicity. Cell Death Dis. 2012, 3, e309. [Google Scholar] [CrossRef] [PubMed]
- Orre, M.; Kamphuis, W.; Dooves, S.; Kooijman, L.; Chan, E.T.; Kirk, C.J.; Dimayuga Smith, V.; Koot, S.; Mamber, C.; Jansen, A.H.; et al. Reactive glia show increased immunoproteasome activity in Alzheimer’s disease. Brain 2013, 136 Pt 5 Pt 5, 1415–1431. [Google Scholar] [CrossRef]
- Heneka, M.T.; O’Banion, M.K.; Terwel, D.; Kummer, M.P. Neuroinflammatory processes in Alzheimer’s disease. J. Neural Transm (Vienna). 2010, 117, 919–947. [Google Scholar] [CrossRef] [PubMed]
- Daily, J.L.; Nash, K.; Jinwal, U.; Golde, T.; Rogers, J.; Peters, M.M.; Burdine, R.D.; Dickey, C.; Banko, J.L.; Weeber, E.J. Adeno-associated virus-mediated rescue of the cognitive defects in a mouse model for Angelman syndrome. PLoS ONE 2011, 6, e27221. [Google Scholar] [CrossRef]
- Criado-Marrero, M.; Gebru, N.T.; Blazier, D.M.; Gould, L.A.; Baker, J.D.; Beaulieu-Abdelahad, D.; Blair, L.J. Hsp90 co-chaperones, FKBP52 and Aha1, promote tau pathogenesis in aged wild-type mice. Acta Neuropathol. Commun. 2021, 9, 65. [Google Scholar] [CrossRef]
- Carty, N.C.; Nash, K.; Lee, D.; Mercer, M.; Gottschall, P.E.; Meyers, C.; Muzyczka, N.; Gordon, M.N.; Morgan, D. Adeno-associated Viral (AAV) Serotype 5 Vector Mediated Gene Delivery of Endothelin-converting Enzyme Reduces Abeta Deposits in APP + PS1 Transgenic Mice. Mol. Ther. 2008, 16, 1580–1586. [Google Scholar] [CrossRef]
- Bobo, R.H.; Laske, D.W.; Akbasak, A.; Morrison, P.F.; Dedrick, R.L.; Oldfield, E.H. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl. Acad. Sci. USA 1994, 91, 2076–2080. [Google Scholar] [CrossRef]
- Alamed, J.; Wilcock, D.M.; Diamond, D.M.; Gordon, M.N.; Morgan, D. Two-day radial-arm water maze learning and memory task; robust resolution of amyloid-related memory deficits in transgenic mice. Nat. Protoc. 2006, 1, 1671–1679. [Google Scholar] [CrossRef]
- Blair, L.J.; Nordhues, B.A.; Hill, S.E.; Scaglione, K.M.; O’Leary, J.C., 3rd; Fontaine, S.N.; Breydo, L.; Zhang, B.; Li, P.; Wang, L.; et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. J. Clin. Investig. 2013, 123, 4158–4169. [Google Scholar] [CrossRef]
- Shelton, L.B.; Baker, J.D.; Zheng, D.; Sullivan, L.E.; Solanki, P.K.; Webster, J.M.; Sun, Z.; Sabbagh, J.J.; Nordhues, B.A.; Koren, J., 3rd; et al. Hsp90 activator Aha1 drives production of pathological tau aggregates. Proc. Natl. Acad. Sci. USA 2017, 114, 9707–9712. [Google Scholar] [CrossRef]
- Criado-Marrero, M.; Gebru, N.T.; Gould, L.A.; Blazier, D.M.; Vidal-Aguiar, Y.; Smith, T.M.; Abdelmaboud, S.S.; Shelton, L.B.; Wang, X.; Dahrendorff, J.; et al. FKBP52 overexpression accelerates hippocampal-dependent memory impairments in a tau transgenic mouse model. NPJ Aging Mech. Dis. 2021, 7, 9. [Google Scholar] [CrossRef]
- Blair, L.J.; Criado-Marrero, M.; Zheng, D.; Wang, X.; Kamath, S.; Nordhues, B.A.; Weeber, E.J.; Dickey, C.A. The Disease-Associated Chaperone FKBP51 Impairs Cognitive Function by Accelerating AMPA Receptor Recycling. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef]
- Maclean, B.; Tomazela, D.M.; Abbatiello, S.E.; Zhang, S.; Whiteaker, J.R.; Paulovich, A.G.; Carr, S.A.; Maccoss, M.J. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 2010, 82, 10116–10124. [Google Scholar] [CrossRef]



| Experiment | Non-Tg | rTg4510 |
|---|---|---|
| Radial-Arm Water Maze | n = 31; wtHsp22 = 11 (6M/5F), mtHsp22 = 10 (5M/5F), GFP = 10 (5M/5F) | n = 30; wtHsp22 = 10 (5M/5F), mtHsp22 = 10 (5M/5F), GFP = 10 (5M/5F) |
| Open-Field Testing | n = 31; wtHsp22 = 11 (6M/5F), mtHsp22 = 10 (5M/5F), GFP = 10 (5M/5F) | n = 30; wtHsp22 = 10 (5M/5F), mtHsp22 = 10 (5M/5F), GFP = 10 (5M/5F) |
| Immunohistochemistry | N/A | n = 18; wtHsp22 = 6 (3M/3F), mtHsp22 = 6 (3M/3F), GFP = 6 (3M/3F) |
| Immunoblotting | N/A | n = 9; wtHsp22 = 3 (2M/1F), mtHsp22 = 3 (1M/2F), GFP = 3 (2M/1F) |
| Electrophysiology | n = 12; wtHsp22 = 10 slices from 4 (2M/2F), mtHsp22 = 31 slices from 4 (2M/2F), GFP = 31 slices from 4 (2M/2F) | n = 12; wtHsp22 = 12 slices from 4 (2M/2F), mtHsp22 = 18 slices from 4 (2M/2F), GFP = 13 slices from 4 (2M/2F) |
| Mass Spectrometry | n = 19; wtHsp22 = 7 (4M/3F), mtHsp22 = 6 (3M/3F), GFP = 6 (3M/3F) | n = 18; wtHsp22 = 6 (3M/3F), mtHsp22 = 6 (3M/3F), GFP = 6 (3M/3F) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez Ospina, S.; Blazier, D.M.; Criado-Marrero, M.; Gould, L.A.; Gebru, N.T.; Beaulieu-Abdelahad, D.; Wang, X.; Remily-Wood, E.; Chaput, D.; Stevens, S., Jr.; et al. Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain. Int. J. Mol. Sci. 2022, 23, 851. https://doi.org/10.3390/ijms23020851
Rodriguez Ospina S, Blazier DM, Criado-Marrero M, Gould LA, Gebru NT, Beaulieu-Abdelahad D, Wang X, Remily-Wood E, Chaput D, Stevens S Jr., et al. Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain. International Journal of Molecular Sciences. 2022; 23(2):851. https://doi.org/10.3390/ijms23020851
Chicago/Turabian StyleRodriguez Ospina, Santiago, Danielle M. Blazier, Marangelie Criado-Marrero, Lauren A. Gould, Niat T. Gebru, David Beaulieu-Abdelahad, Xinming Wang, Elizabeth Remily-Wood, Dale Chaput, Stanley Stevens, Jr., and et al. 2022. "Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain" International Journal of Molecular Sciences 23, no. 2: 851. https://doi.org/10.3390/ijms23020851
APA StyleRodriguez Ospina, S., Blazier, D. M., Criado-Marrero, M., Gould, L. A., Gebru, N. T., Beaulieu-Abdelahad, D., Wang, X., Remily-Wood, E., Chaput, D., Stevens, S., Jr., Uversky, V. N., Bickford, P. C., Dickey, C. A., & Blair, L. J. (2022). Small Heat Shock Protein 22 Improves Cognition and Learning in the Tauopathic Brain. International Journal of Molecular Sciences, 23(2), 851. https://doi.org/10.3390/ijms23020851







