Syndecan-4 Is a Key Facilitator of the SARS-CoV-2 Delta Variant’s Superior Transmission
Abstract
:1. Introduction
2. Results
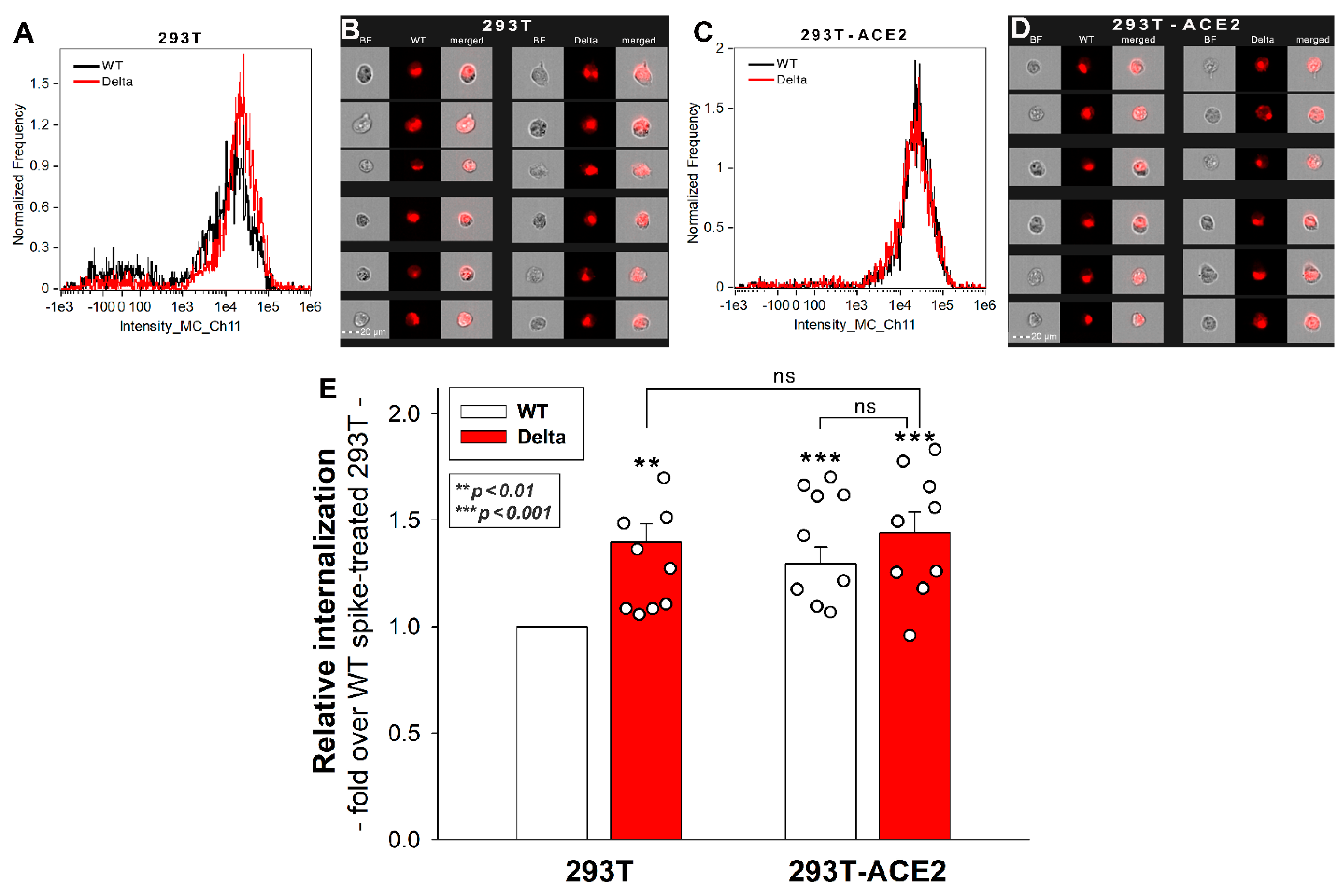
2.1. Effect of ACE2 Overexpression on the Cellular Internalization of the Spike Proteins
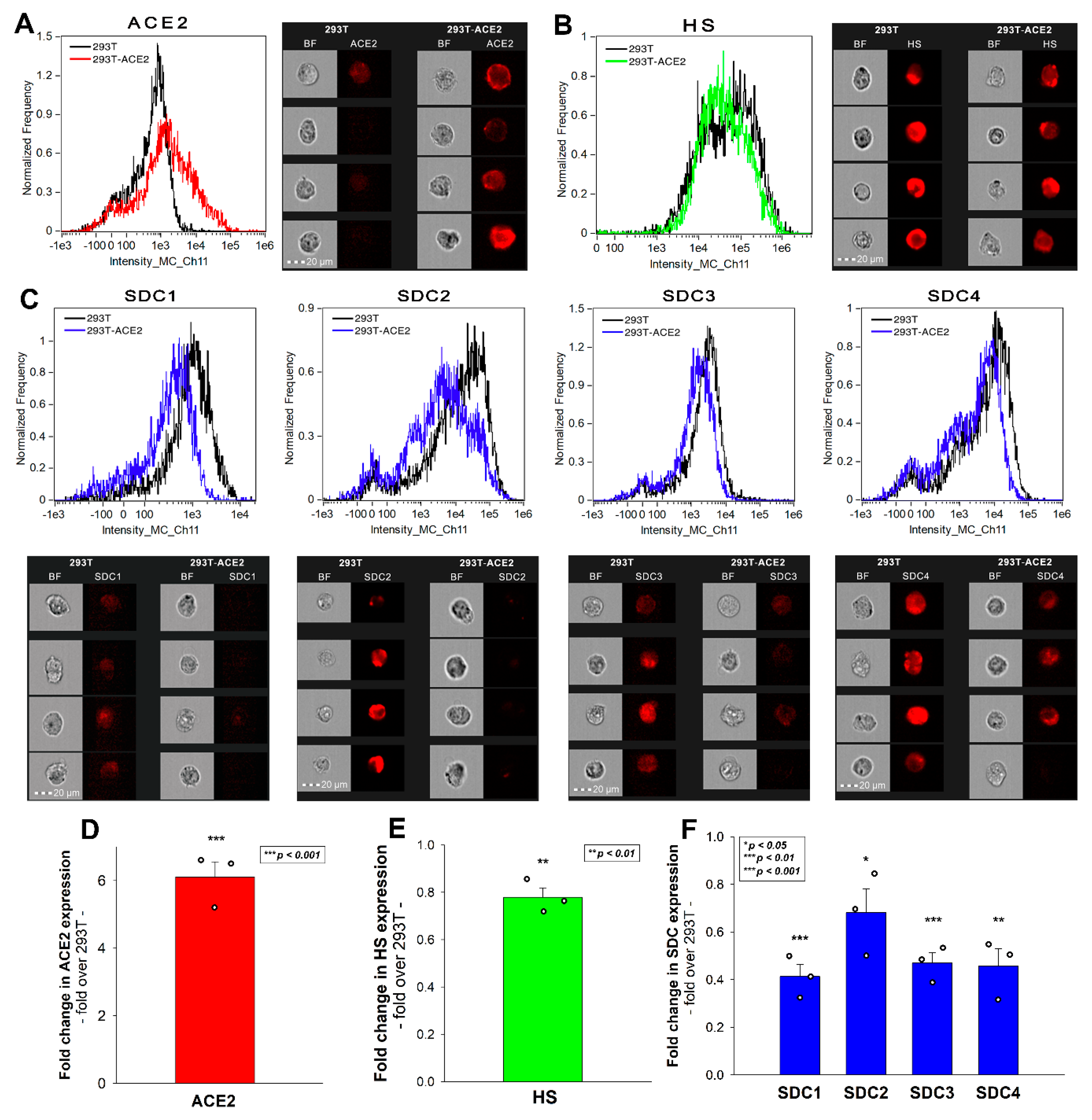
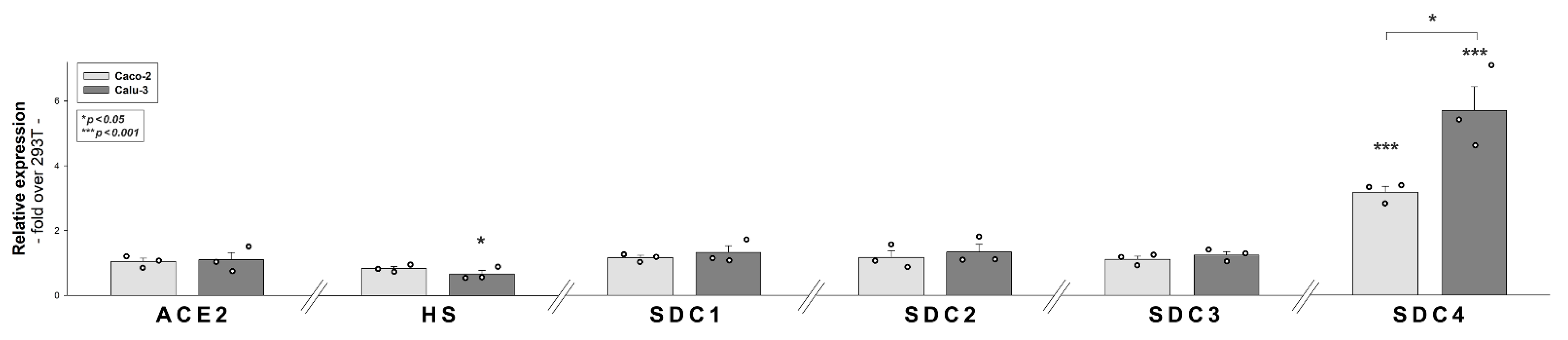
2.2. 293T-ACE2 Cells Exhibit Increased ACE2 Yet Reduced HS and SDC Expression
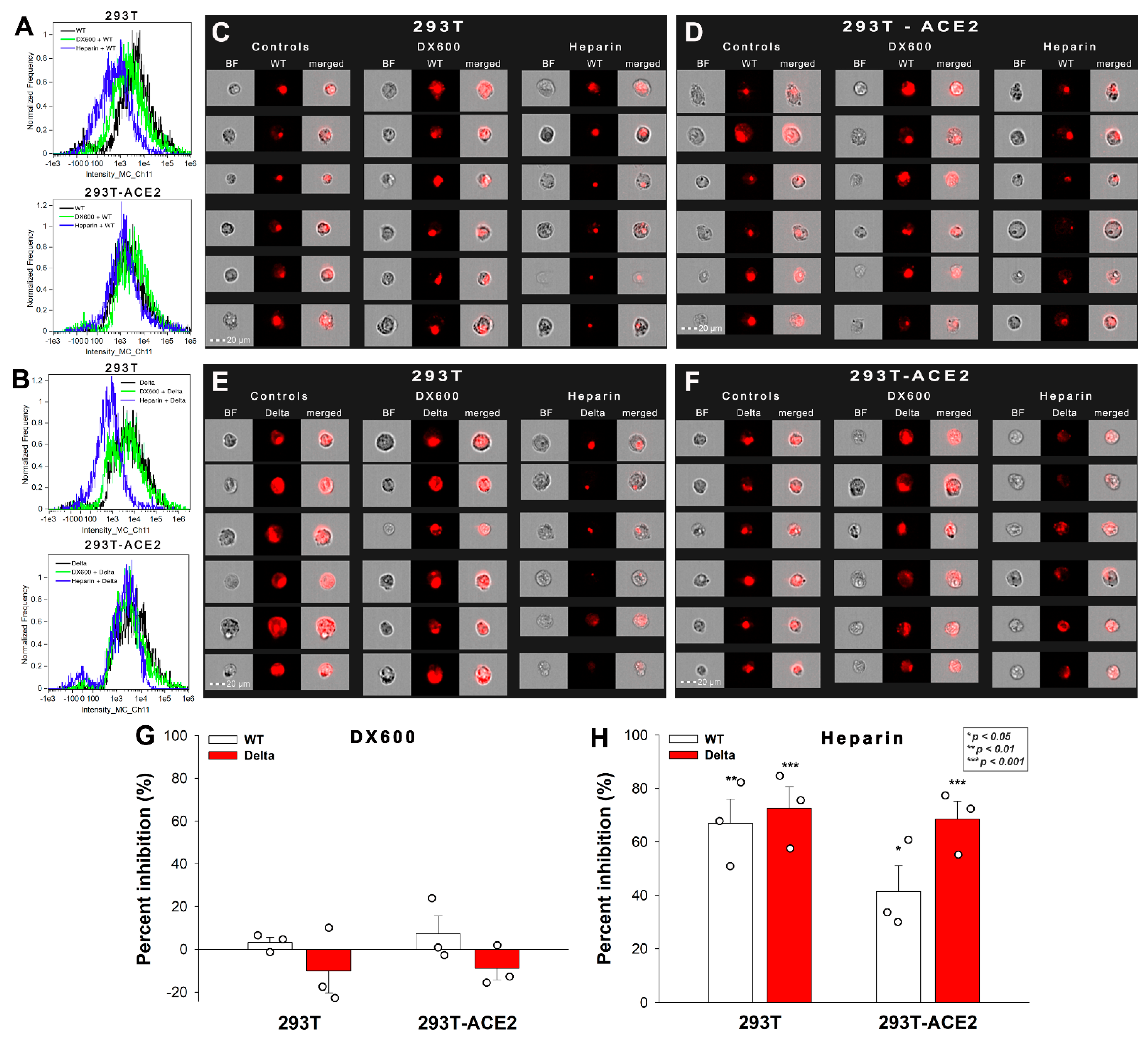
2.3. Effect of ACE2 and Proteoglycan Inhibition on Cellular Uptake
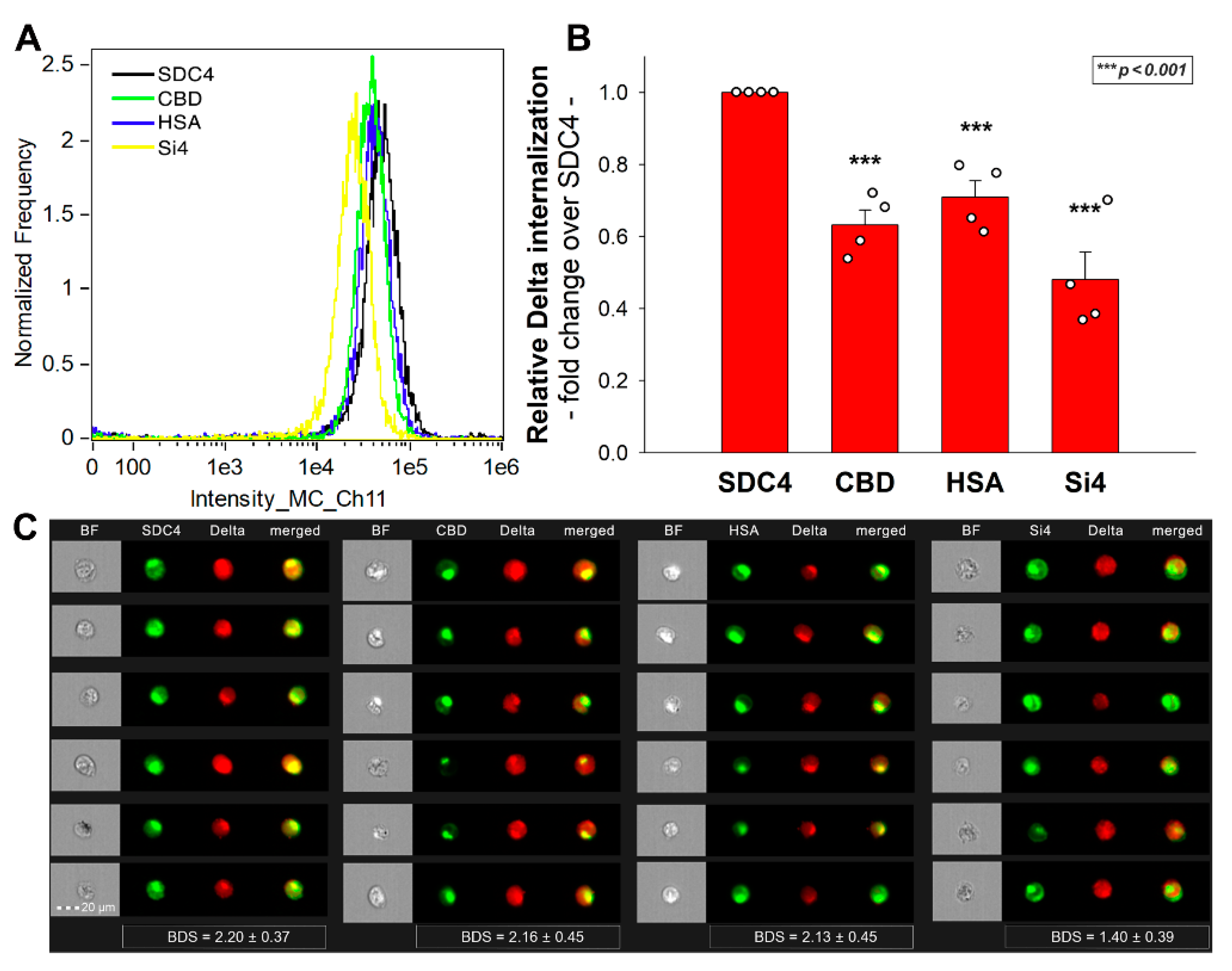
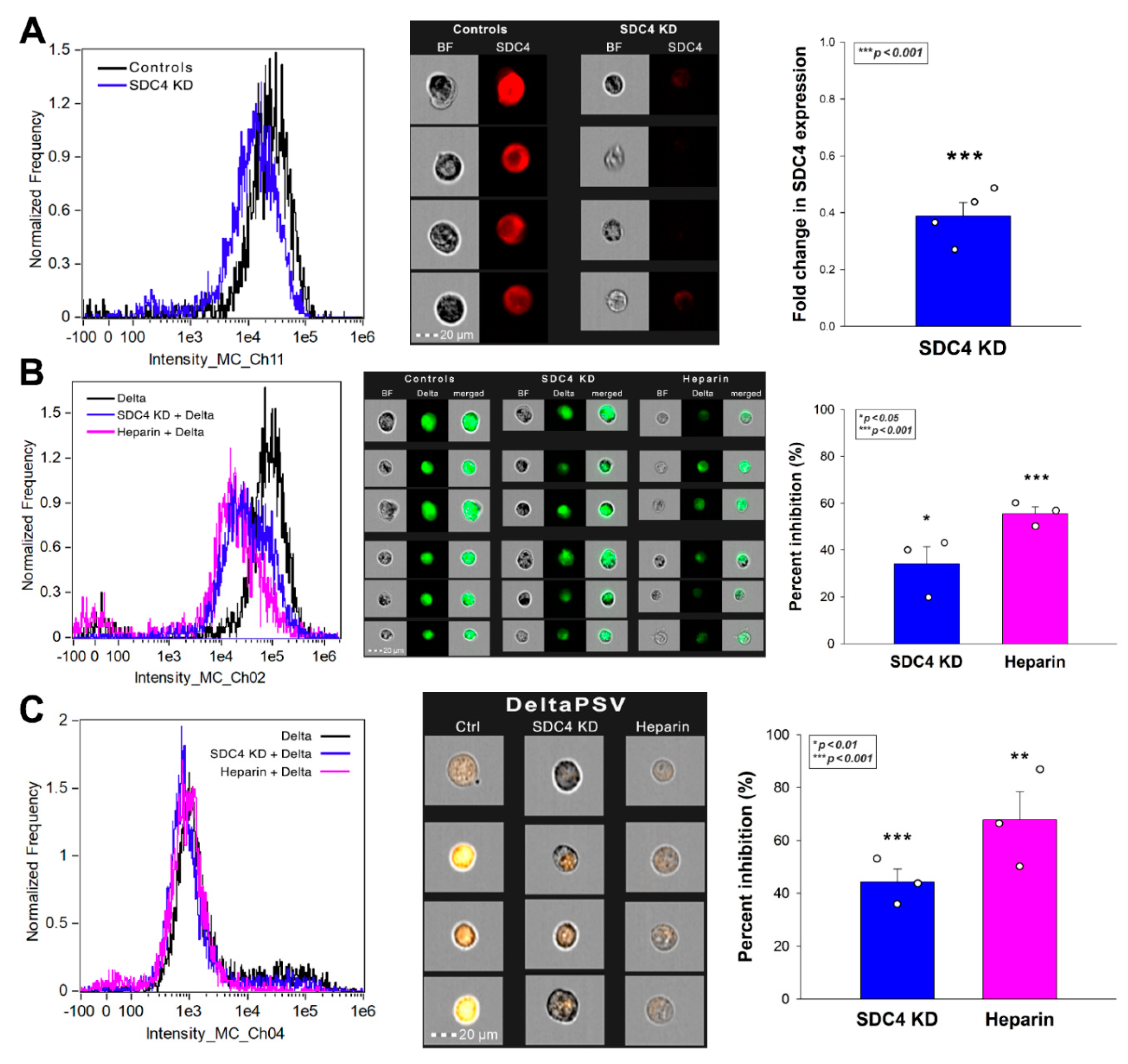
2.4. SDC4 Enhances the Cellular Uptake of the Delta Spike Protein
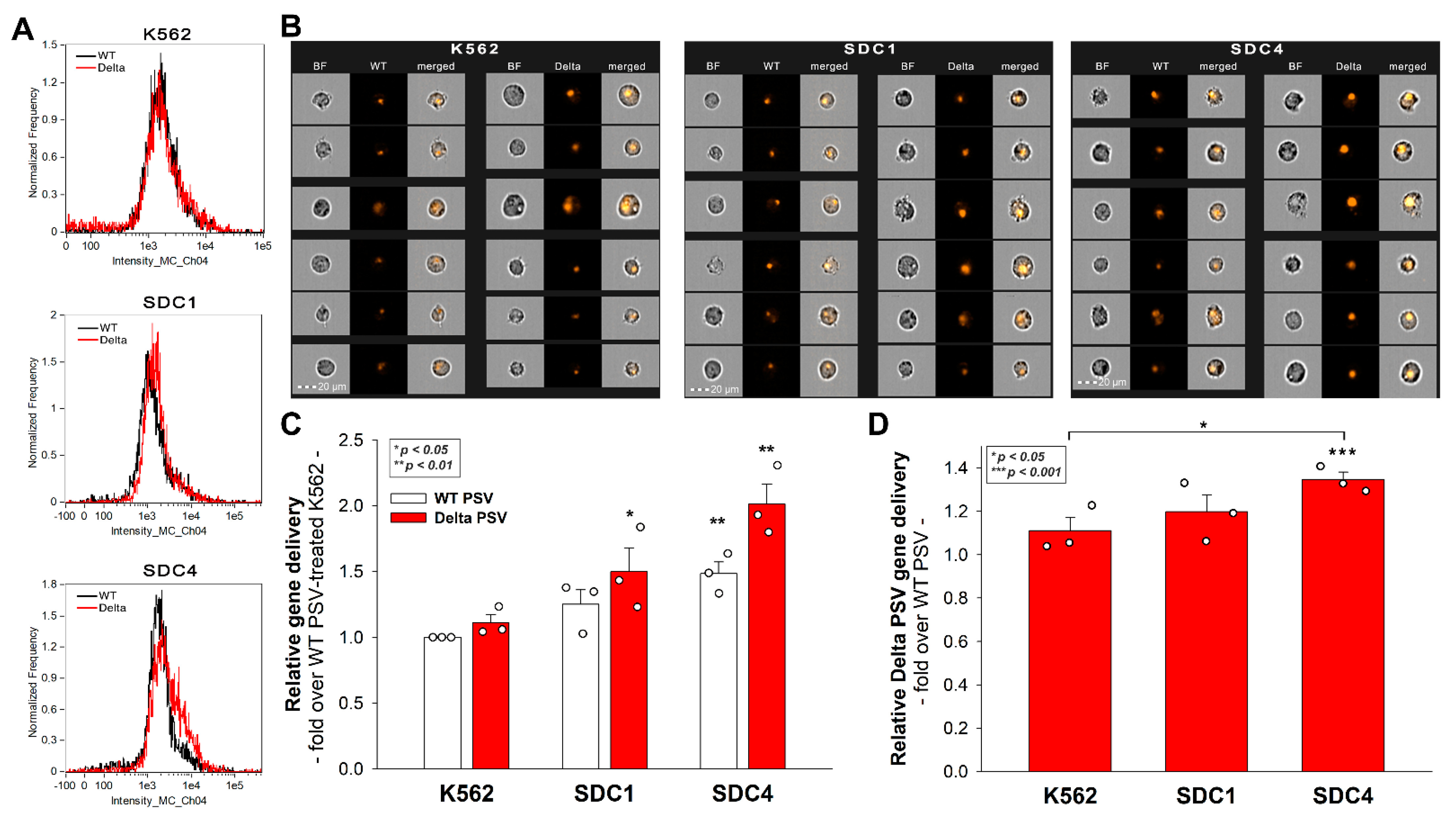
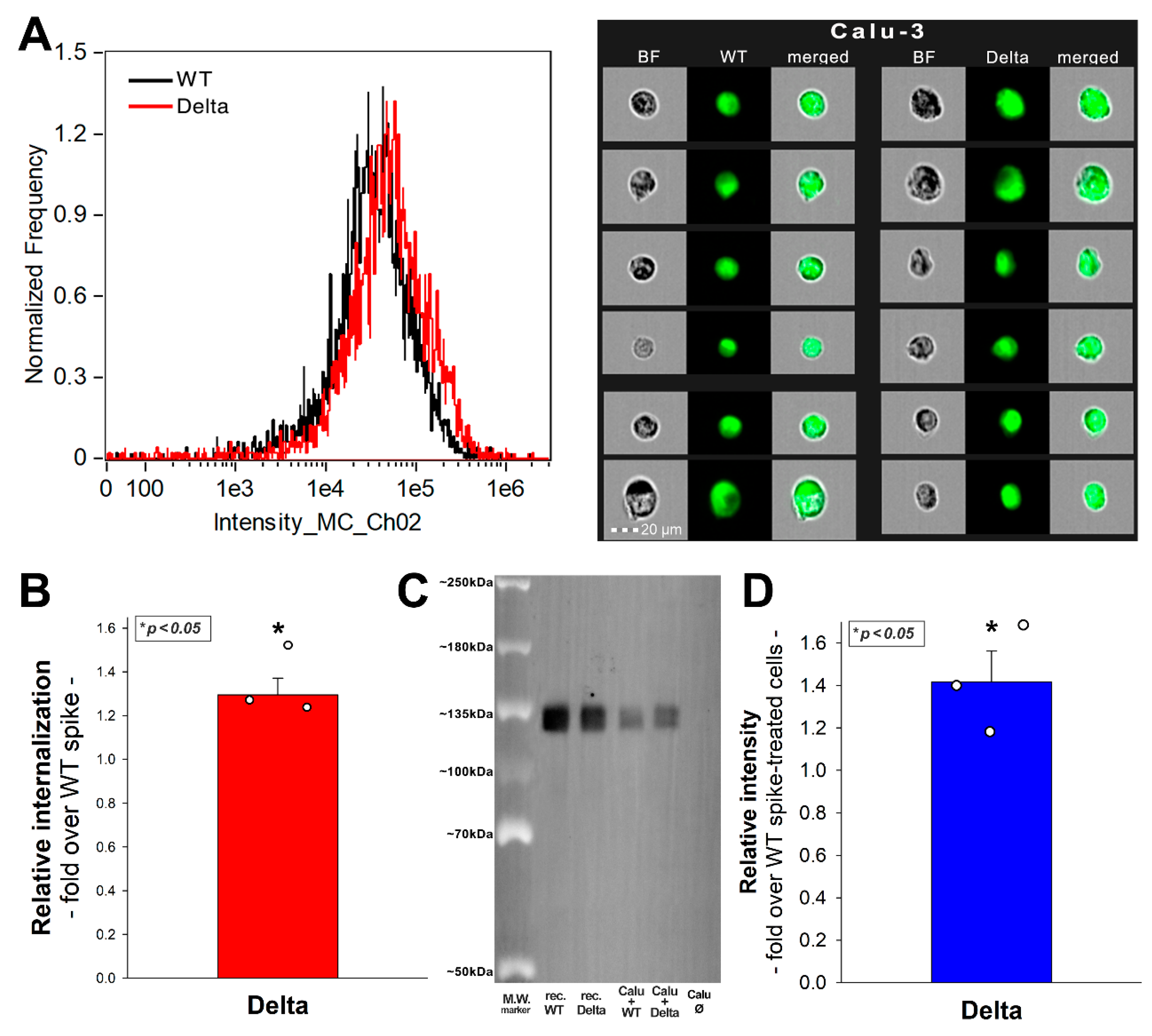
2.5. SDC4 Enhances the Cellular Uptake of the Delta PSV
2.6. Exploring the Delta Spike’s Interactions with the SDC4 Ectodomain
2.7. ACE2, HS and SDC Expression Profile of Calu-3 Cells
2.8. SDC4 Knockdown Decreases Delta’s Internalization into Calu-3 Cells
3. Discussion
4. Materials and Methods
4.1. Recombinant Proteins and Pseudoviruses
4.2. ACE2 and SDC Constructs, Cell Culture and Transfection
4.3. Flow Cytometry Analysis of HS, ACE2 and SDC Expression
4.4. Establishment of ACE2 or SDC4 KD Cell Lines
4.5. Pseudovirus (PSV) Studies
4.6. Flow Cytometry Analysis of Spike Uptake
4.7. Inhibitor Studies
4.8. Cell Viability Measurements
4.9. Co-Immunoprecipitation Experiments
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fara, A.; Mitrev, Z.; Rosalia, R.A.; Assas, B.M. Cytokine storm and COVID-19: A chronicle of pro-inflammatory cytokines. Open Biol. 2020, 10, 200160. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Khan, S.; Tiwari, R.; Sircar, S.; Bhat, S.; Malik, Y.S.; Singh, K.P.; Chaicumpa, W.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. Coronavirus Disease 2019-COVID-19. Clin. Microbiol. Rev. 2020, 33, e00028-20. [Google Scholar] [CrossRef] [PubMed]
- Askitas, N.; Tatsiramos, K.; Verheyden, B. Estimating worldwide effects of non-pharmaceutical interventions on COVID-19 incidence and population mobility patterns using a multiple-event study. Sci. Rep. 2021, 11, 1972. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Morgenstern, C.; Kelly, J.; Lowe, R.; Group, C.C.-W.; Jit, M. The impact of non-pharmaceutical interventions on SARS-CoV-2 transmission across 130 countries and territories. BMC Med. 2021, 19, 40. [Google Scholar] [CrossRef]
- Forni, G.; Mantovani, A.; COVID-19 Commission of Accademia Nazionale dei Lincei, Rome. COVID-19 vaccines: Where we stand and challenges ahead. Cell Death Differ. 2021, 28, 626–639. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, W. Fast-spreading SARS-CoV-2 variants: Challenges to and new design strategies of COVID-19 vaccines. Signal Transduct. Target Ther. 2021, 6, 226. [Google Scholar] [CrossRef]
- Vaidyanathan, G. Coronavirus variants are spreading in India-what scientists know so far. Nature 2021, 593, 321–322. [Google Scholar] [CrossRef]
- Mishra, S.; Mindermann, S.; Sharma, M.; Whittaker, C.; Mellan, T.A.; Wilton, T.; Klapsa, D.; Mate, R.; Fritzsche, M.; Zambon, M.; et al. Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. Clin. Med. 2021, 39, 101064. [Google Scholar] [CrossRef]
- Callaway, E.; Ledford, H. How bad is Omicron? What scientists know so far. Nature 2021, 600, 197–199. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Dingens, A.S.; Bloom, J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021, 2, 100255. [Google Scholar] [CrossRef]
- Liu, Y.; Rocklov, J. The reproductive number of the Delta variant of SARS-CoV-2 is far higher compared to the ancestral SARS-CoV-2 virus. J. Travel Med. 2021, 28, 7. [Google Scholar] [CrossRef]
- Salleh, M.Z.; Derrick, J.P.; Deris, Z.Z. Structural Evaluation of the Spike Glycoprotein Variants on SARS-CoV-2 Transmission and Immune Evasion. Int. J. Mol. Sci. 2021, 22, 7425. [Google Scholar] [CrossRef]
- Pascarella, S.; Ciccozzi, M.; Zella, D.; Bianchi, M.; Benedetti, F.; Benvenuto, D.; Broccolo, F.; Cauda, R.; Caruso, A.; Angeletti, S.; et al. SARS-CoV-2 B.1.617 Indian variants: Are electrostatic potential changes responsible for a higher transmission rate? J. Med. Virol. 2021, 93, 6551–6556. [Google Scholar] [CrossRef] [PubMed]
- McCallum, M.; Walls, A.C.; Sprouse, K.R.; Bowen, J.E.; Rosen, L.; Dang, H.V.; deMarco, A.; Franko, N.; Tilles, S.W.; Logue, J.; et al. Molecular basis of immune evasion by the delta and kappa SARS-CoV-2 variants. bioRxiv 2021, 374, 1621–1626. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Sidarovich, A.; Krüger, N.; Graichen, L.; Moldenhauer, A.-S.; Winkler, M.S.; Schulz, S.; Jäck, H.-M.; et al. Increased lung cell entry of B.1.617.2 and evasion of antibodies induced by infection and BNT162b2 vaccination. bioRxiv 2021. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Kruger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Karthika, T.; Joseph, J.; Das, V.R.A.; Nair, N.; Charulekha, P.; Roji, M.D.; Raj, V.S. SARS-CoV-2 Cellular Entry Is Independent of the ACE2 Cytoplasmic Domain Signaling. Cells 2021, 10, 1814. [Google Scholar] [CrossRef]
- Gadanec, L.K.; McSweeney, K.R.; Qaradakhi, T.; Ali, B.; Zulli, A.; Apostolopoulos, V. Can SARS-CoV-2 Virus Use Multiple Receptors to Enter Host Cells? Int. J. Mol. Sci. 2021, 22, 992. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, T.; Zhang, W.; Sun, Q.; Li, H.; Li, J.-p. Elucidating the Interactions Between Heparin/Heparan Sulfate and SARS-CoV-2-Related Proteins—An Important Strategy for Developing Novel Therapeutics for the COVID-19 Pandemic. Front. Mol. Biosci. 2021, 7, 490. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, C.Z.; Swaroop, M.; Xu, M.; Wang, L.; Lee, J.; Wang, A.Q.; Pradhan, M.; Hagen, N.; Chen, L.; et al. Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov. 2020, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chopra, P.; Li, X.; Bouwman, K.M.; Tompkins, S.M.; Wolfert, M.A.; de Vries, R.P.; Boons, G.-J. Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2. ACS Central Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Jambrina, M.; Eder, J.; Kaptein, T.M.; van Hamme, J.L.; Helgers, L.C.; Vlaming, K.E.; Brouwer, P.J.M.; van Nuenen, A.C.; Spaargaren, M.; de Bree, G.J.; et al. Infection and transmission of SARS-CoV-2 depend on heparan sulfate proteoglycans. EMBO J. 2021, 40, e106765. [Google Scholar] [CrossRef]
- Hudák, A.; Letoha, A.; Szilák, L.; Letoha, T. Contribution of Syndecans to the Cellular Entry of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 5336. [Google Scholar]
- Wu, C.; Jin, X.; Tsueng, G.; Afrasiabi, C.; Su, A.I. BioGPS: Building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 2016, 44, D313–D316. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Macleod, I.; Su, A.I. BioGPS and MyGene.info: Organizing online, gene-centric information. Nucleic Acids Res. 2013, 41, D561–D565. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W., 3rd; et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009, 10, R130. [Google Scholar] [CrossRef]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef]
- Nakase, I.; Tadokoro, A.; Kawabata, N.; Takeuchi, T.; Katoh, H.; Hiramoto, K.; Negishi, M.; Nomizu, M.; Sugiura, Y.; Futaki, S. Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry 2007, 46, 492–501. [Google Scholar] [CrossRef]
- Sorkin, A.; von Zastrow, M. Endocytosis and signalling: Intertwining molecular networks. Nat. Rev. Mol. Cell Biol. 2009, 10, 609–622. [Google Scholar] [CrossRef] [Green Version]
- Schutz, D.; Ruiz-Blanco, Y.B.; Munch, J.; Kirchhoff, F.; Sanchez-Garcia, E.; Muller, J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry. Adv. Drug Deliv. Rev. 2020, 167, 47–65. [Google Scholar] [CrossRef]
- Fukushi, S.; Mizutani, T.; Saijo, M.; Matsuyama, S.; Miyajima, N.; Taguchi, F.; Itamura, S.; Kurane, I.; Morikawa, S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005, 86, 2269–2274. [Google Scholar] [CrossRef] [PubMed]
- Paiardi, G.; Richter, S.; Oreste, P.; Urbinati, C.; Rusnati, M.; Wade, R.C. The binding of heparin to spike glycoprotein inhibits SARS-CoV-2 infection by three mechanisms. J. Biol. Chem. 2021, 101507. [Google Scholar] [CrossRef]
- De Pasquale, V.; Quiccione, M.S.; Tafuri, S.; Avallone, L.; Pavone, L.M. Heparan Sulfate Proteoglycans in Viral Infection and Treatment: A Special Focus on SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 6574. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Fernandez, E.; Egia-Mendikute, L.; Vila-Vecilla, L.; Bosch, A.; Barreira-Manrique, A.; Lee, S.Y.; Garcia-Del Rio, A.; Antonana-Vildosola, A.; Jimenez-Lasheras, B.; Moreno-Cugnon, L.; et al. Hypoxia reduces cell attachment of SARS-CoV-2 spike protein by modulating the expression of ACE2, neuropilin-1, syndecan-1 and cellular heparan sulfate. Emerg. Microbes Infect. 2021, 10, 1065–1076. [Google Scholar] [CrossRef]
- Suzuki, K.; Okada, H.; Tomita, H.; Sumi, K.; Kakino, Y.; Yasuda, R.; Kitagawa, Y.; Fukuta, T.; Miyake, T.; Yoshida, S.; et al. Possible involvement of Syndecan-1 in the state of COVID-19 related to endothelial injury. Thromb. J. 2021, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Jambrina, M.; Eder, J.; Kaptein, T.M.; van Hamme, J.L.; Helgers, L.C.; Vlaming, K.E.; Brouwer, P.J.M.; Vlaar, A.P.J.; van Baarle, F.E.H.P.; Spaargaren, M.; et al. Infection and transmission of SARS-CoV-2 depends on heparan sulfate proteoglycans. bioRxiv 2021. [Google Scholar] [CrossRef]
- Letoha, T.; Hudak, A.; Kusz, E.; Pettko-Szandtner, A.; Domonkos, I.; Josvay, K.; Hofmann-Apitius, M.; Szilak, L. Contribution of syndecans to cellular internalization and fibrillation of amyloid-beta(1-42). Sci. Rep. 2019, 9, 1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudak, A.; Kusz, E.; Domonkos, I.; Josvay, K.; Kodamullil, A.T.; Szilak, L.; Hofmann-Apitius, M.; Letoha, T. Contribution of syndecans to cellular uptake and fibrillation of alpha-synuclein and tau. Sci. Rep. 2019, 9, 16543. [Google Scholar] [CrossRef] [Green Version]
- Lang, J.; Yang, N.; Deng, J.; Liu, K.; Yang, P.; Zhang, G.; Jiang, C. Inhibition of SARS pseudovirus cell entry by lactoferrin binding to heparan sulfate proteoglycans. PLoS ONE 2011, 6, e23710. [Google Scholar] [CrossRef]
- Lei, C.; Qian, K.; Li, T.; Zhang, S.; Fu, W.; Ding, M.; Hu, S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun 2020, 11, 2070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millet, J.K.; Tang, T.; Nathan, L.; Jaimes, J.A.; Hsu, H.L.; Daniel, S.; Whittaker, G.R. Production of Pseudotyped Particles to Study Highly Pathogenic Coronaviruses in a Biosafety Level 2 Setting. J. Vis. Exp. 2019, 145. [Google Scholar] [CrossRef] [Green Version]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020, 9, 680–686. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [Green Version]
- Yue, J.; Jin, W.; Yang, H.; Faulkner, J.; Song, X.; Qiu, H.; Teng, M.; Azadi, P.; Zhang, F.; Linhardt, R.J.; et al. Heparan Sulfate Facilitates Spike Protein-Mediated SARS-CoV-2 Host Cell Invasion and Contributes to Increased Infection of SARS-CoV-2 G614 Mutant and in Lung Cancer. Front. Mol. Biosci. 2021, 8, 649575. [Google Scholar] [CrossRef] [PubMed]
- Krause, P.R.; Fleming, T.R.; Longini, I.M.; Peto, R.; Briand, S.; Heymann, D.L.; Beral, V.; Snape, M.D.; Rees, H.; Ropero, A.M.; et al. SARS-CoV-2 Variants and Vaccines. N. Engl. J. Med. 2021, 385, 179–186. [Google Scholar] [CrossRef]
- Khateeb, J.; Li, Y.; Zhang, H. Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Crit. Care 2021, 25, 244. [Google Scholar] [CrossRef]
- Planas, D.; Veyer, D.; Baidaliuk, A.; Staropoli, I.; Guivel-Benhassine, F.; Rajah, M.M.; Planchais, C.; Porrot, F.; Robillard, N.; Puech, J.; et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature 2021, 596, 276–280. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sapkal, G.N.; Abraham, P.; Ella, R.; Deshpande, G.; Patil, D.Y.; Nyayanit, D.A.; Gupta, N.; Sahay, R.R.; Shete, A.M.; et al. Neutralization of variant under investigation B.1.617 with sera of BBV152 vaccinees. Clin. Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hofmann-Winkler, H.; Kruger, N.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Arora, P.; Sidarovich, A.; Moldenhauer, A.S.; Winkler, M.S.; et al. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021, 36, 109415. [Google Scholar] [CrossRef] [PubMed]
- Edara, V.V.; Pinsky, B.A.; Suthar, M.S.; Lai, L.; Davis-Gardner, M.E.; Floyd, K.; Flowers, M.W.; Wrammert, J.; Hussaini, L.; Ciric, C.R.; et al. Infection and Vaccine-Induced Neutralizing-Antibody Responses to the SARS-CoV-2 B.1.617 Variants. N. Engl. J. Med. 2021, 385, 664–666. [Google Scholar] [CrossRef]
- Irvin, S.C.; Herold, B.C. Molecular mechanisms linking high dose medroxyprogesterone with HIV-1 risk. PLoS ONE 2015, 10, e0121135. [Google Scholar] [CrossRef] [Green Version]
- Connor, J.P.; Felder, M.; Kapur, A.; Onujiogu, N. DcR3 binds to ovarian cancer via heparan sulfate proteoglycans and modulates tumor cells response to platinum with corresponding alteration in the expression of BRCA1. BMC Cancer 2012, 12, 176. [Google Scholar] [CrossRef] [Green Version]
- Dieudonne, F.X.; Marion, A.; Marie, P.J.; Modrowski, D. Targeted inhibition of T-cell factor activity promotes syndecan-2 expression and sensitization to doxorubicin in osteosarcoma cells and bone tumors in mice. J. Bone Miner Res. 2012, 27, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Eustace, A.D.; McNaughton, E.F.; King, S.; Kehoe, O.; Kungl, A.; Mattey, D.; Nobbs, A.H.; Williams, N.; Middleton, J. Soluble syndecan-3 binds chemokines, reduces leukocyte migration in vitro and ameliorates disease severity in models of rheumatoid arthritis. Arthritis Res. Ther. 2019, 21, 172. [Google Scholar] [CrossRef] [Green Version]
- Tomoda, H.; Kishimoto, Y.; Lee, Y.C. Temperature effect on endocytosis and exocytosis by rabbit alveolar macrophages. J. Biol. Chem. 1989, 264, 15445–15450. [Google Scholar] [PubMed]
- Chang, C.C.; Wu, M.; Yuan, F. Role of specific endocytic pathways in electrotransfection of cells. Mol. Ther. Methods Clin. Dev. 2014, 1, 14058. [Google Scholar] [CrossRef]
- Haridas, V.; Ranjbar, S.; Vorobjev, I.A.; Goldfeld, A.E.; Barteneva, N.S. Imaging flow cytometry analysis of intracellular pathogens. Methods 2017, 112, 91–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudák, A.; Veres, G.; Letoha, A.; Szilák, L.; Letoha, T. Syndecan-4 Is a Key Facilitator of the SARS-CoV-2 Delta Variant’s Superior Transmission. Int. J. Mol. Sci. 2022, 23, 796. https://doi.org/10.3390/ijms23020796
Hudák A, Veres G, Letoha A, Szilák L, Letoha T. Syndecan-4 Is a Key Facilitator of the SARS-CoV-2 Delta Variant’s Superior Transmission. International Journal of Molecular Sciences. 2022; 23(2):796. https://doi.org/10.3390/ijms23020796
Chicago/Turabian StyleHudák, Anett, Gábor Veres, Annamária Letoha, László Szilák, and Tamás Letoha. 2022. "Syndecan-4 Is a Key Facilitator of the SARS-CoV-2 Delta Variant’s Superior Transmission" International Journal of Molecular Sciences 23, no. 2: 796. https://doi.org/10.3390/ijms23020796






