Mining the Wheat Grain Proteome
Abstract
:1. Introduction
2. Results and Discussion
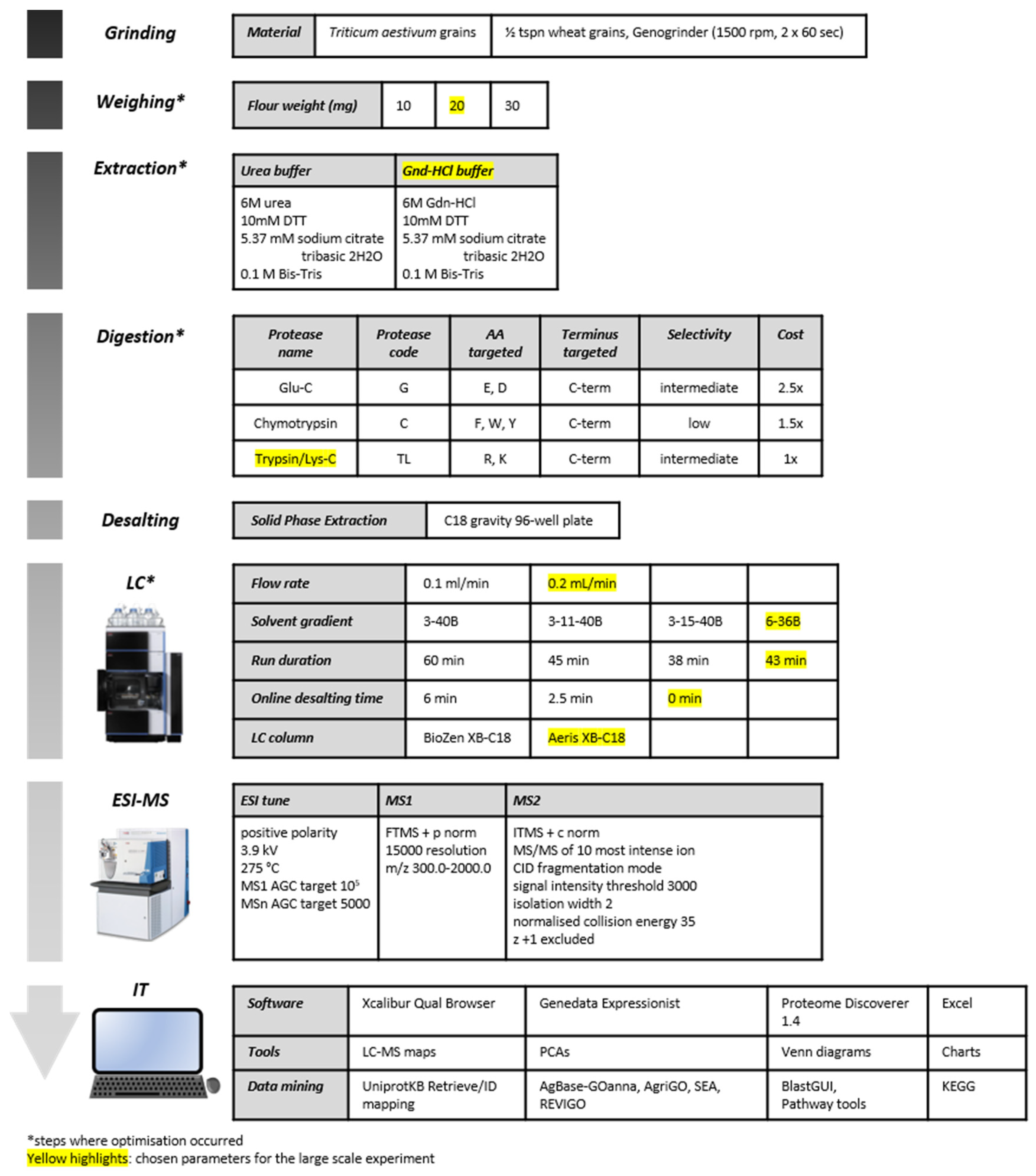
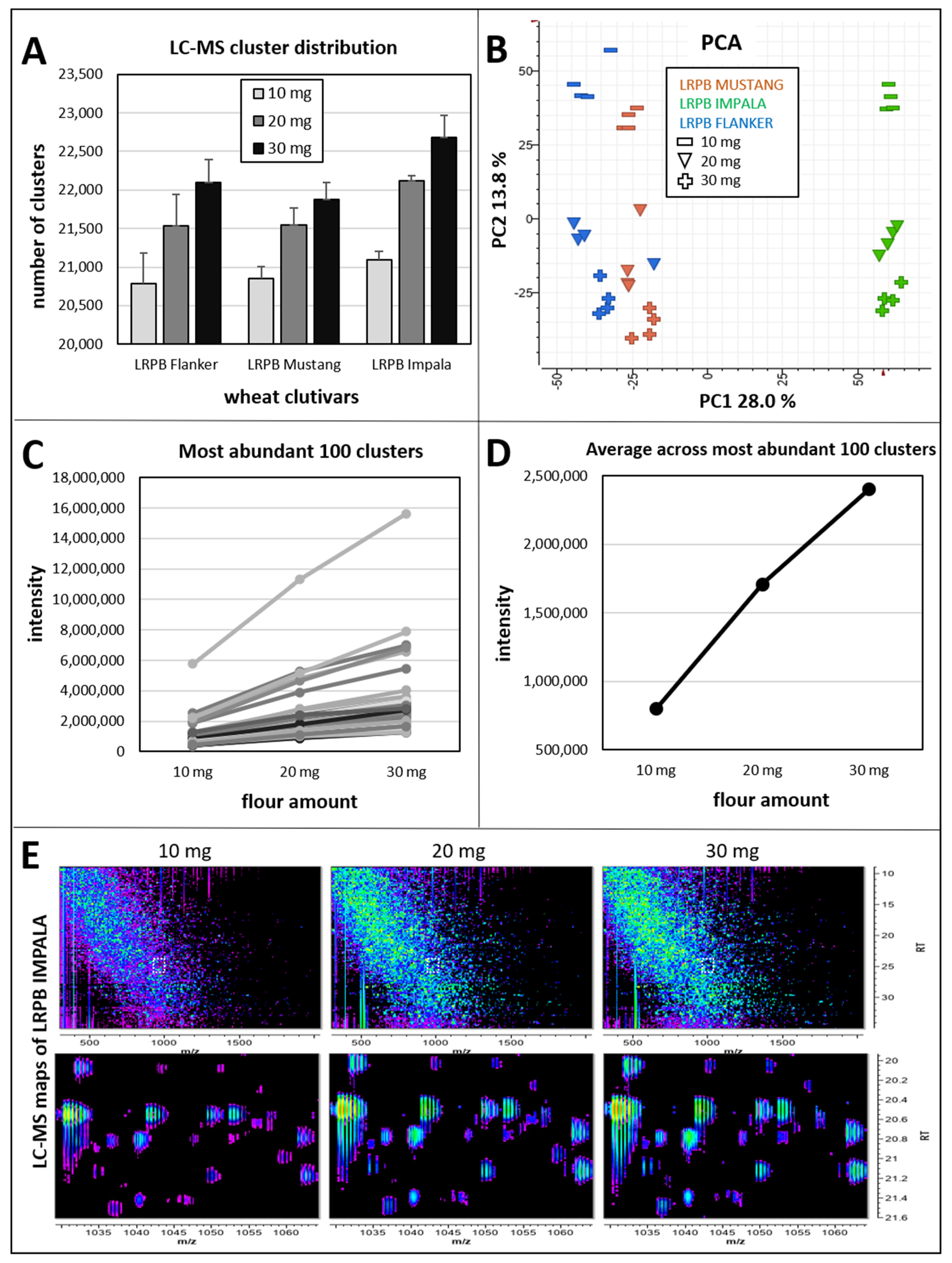
2.1. Testing Flour Weights
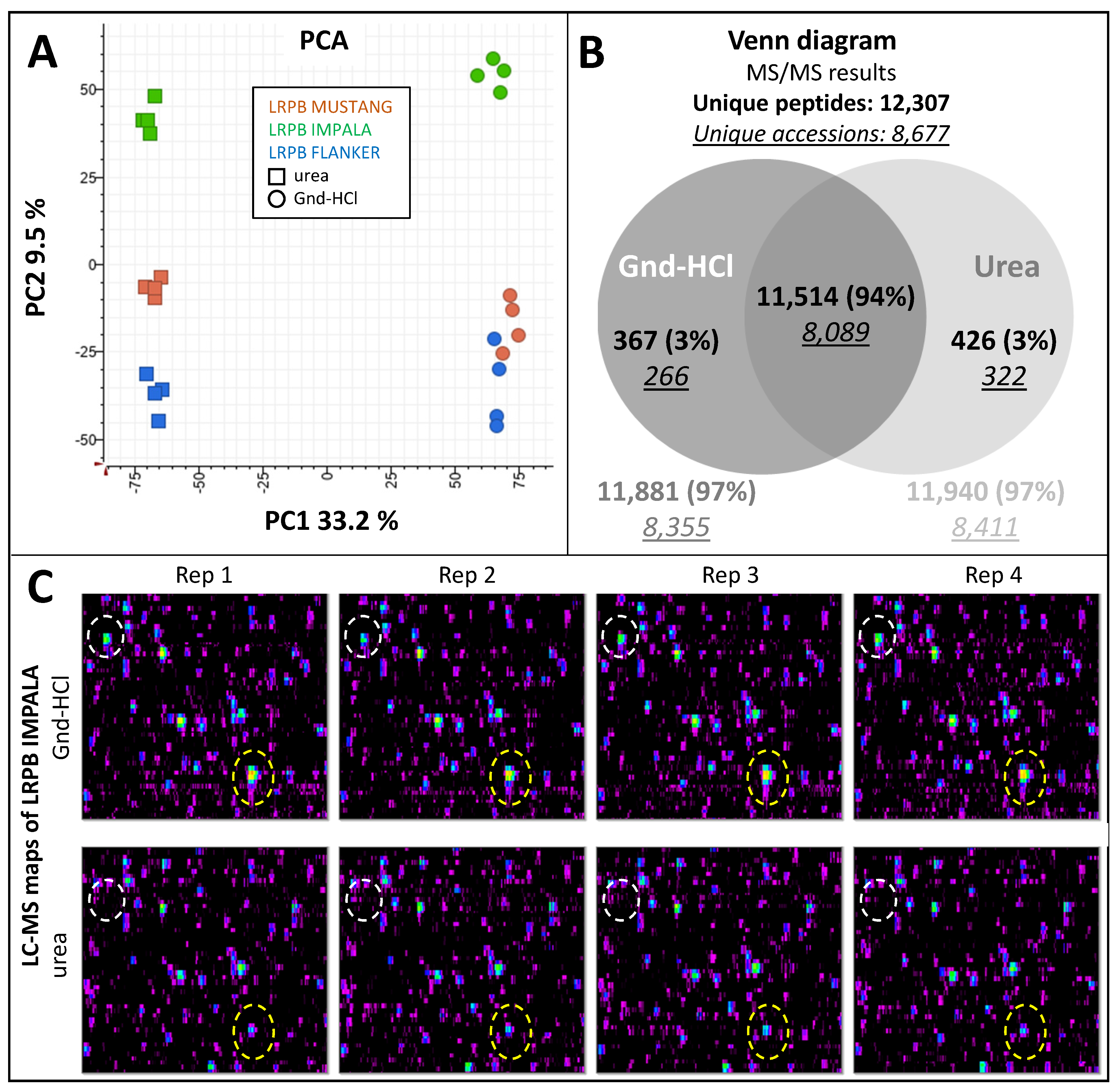
2.2. Testing Extraction Buffers
2.3. Testing Proteases
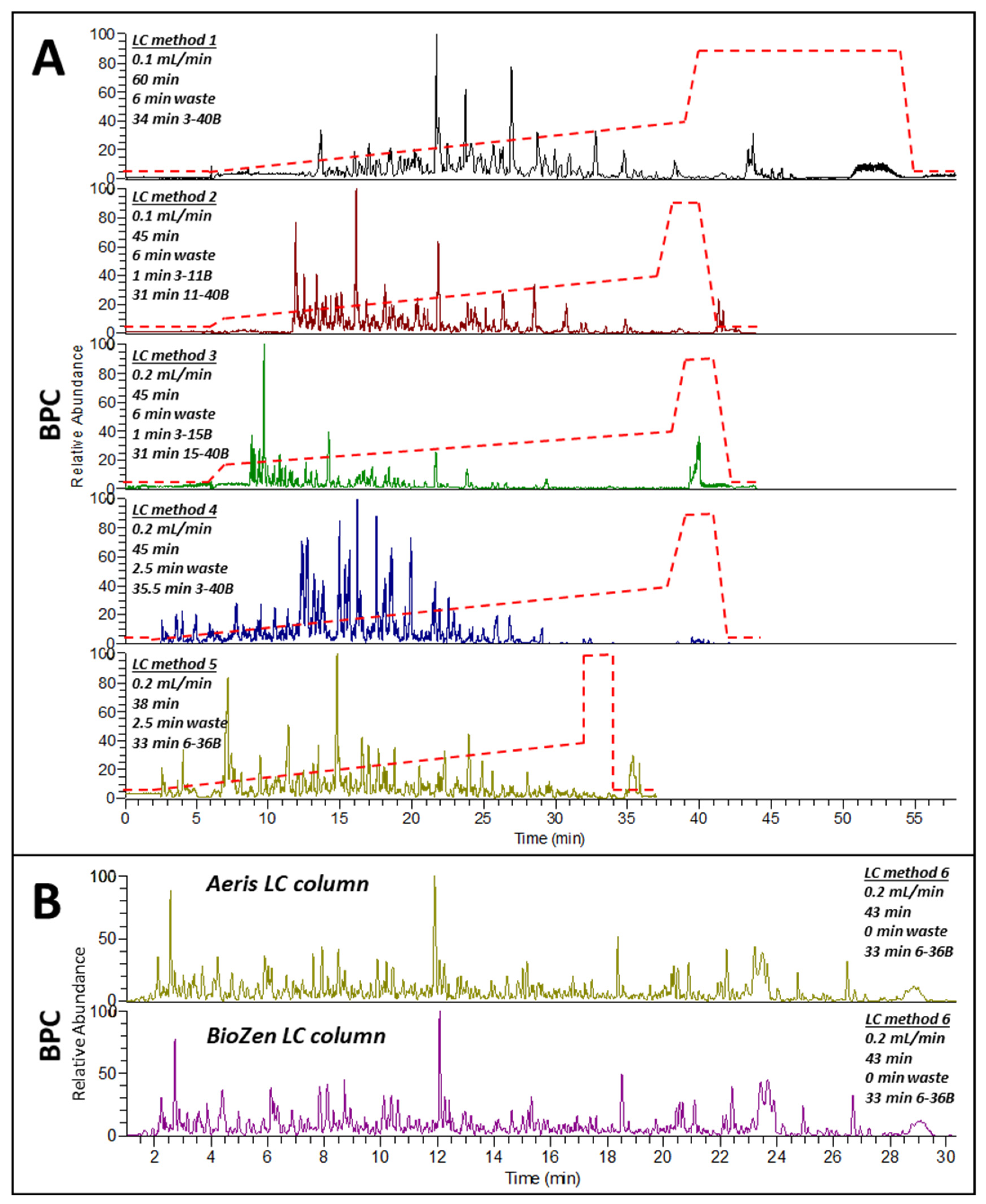
2.4. Testing LC Separation
2.5. Validating the Shotgun Proteomics Method
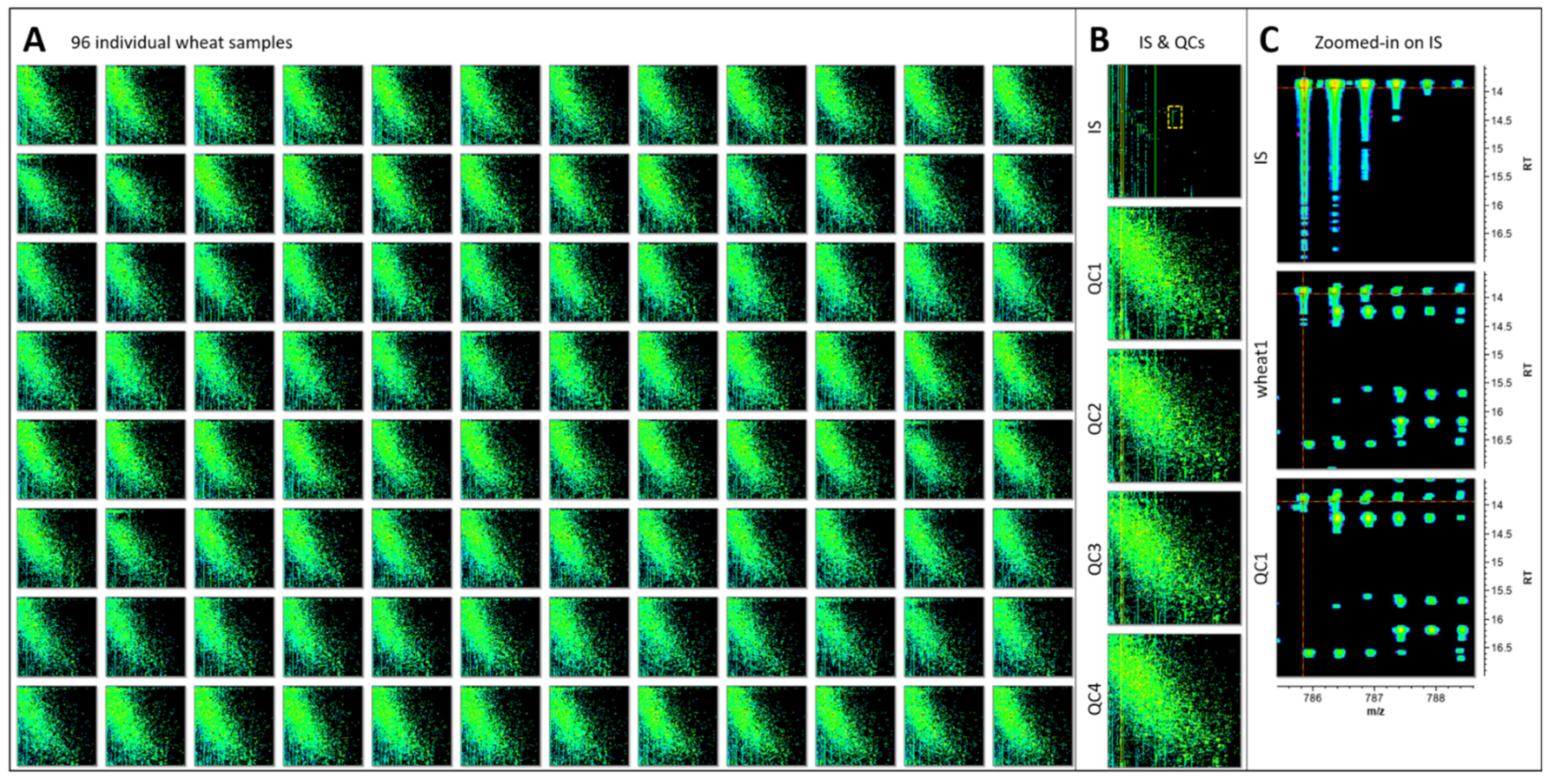
2.6. Data Mining of Protein Identification
3. Materials and Methods
3.1. Materials
3.1.1. Wheat Cultivation and Sampling
3.1.2. Wheat Grain Processing
3.2. Methods
3.2.1. Flour Weighing, Protein Extraction, and Protein Assay
3.2.2. Protein Digestion, Digest SPE Clean-Up, and Peptide Reconstitution
3.2.3. LC–MS and LC–MS/MS
LC Separation Columns
LC Methods
ESI–MS
ESI–MS/MS
LC–MS Validation Run
3.2.4. Data Processing, Database Search, and Statistical Analyses
Data File Processing
Protein Identification
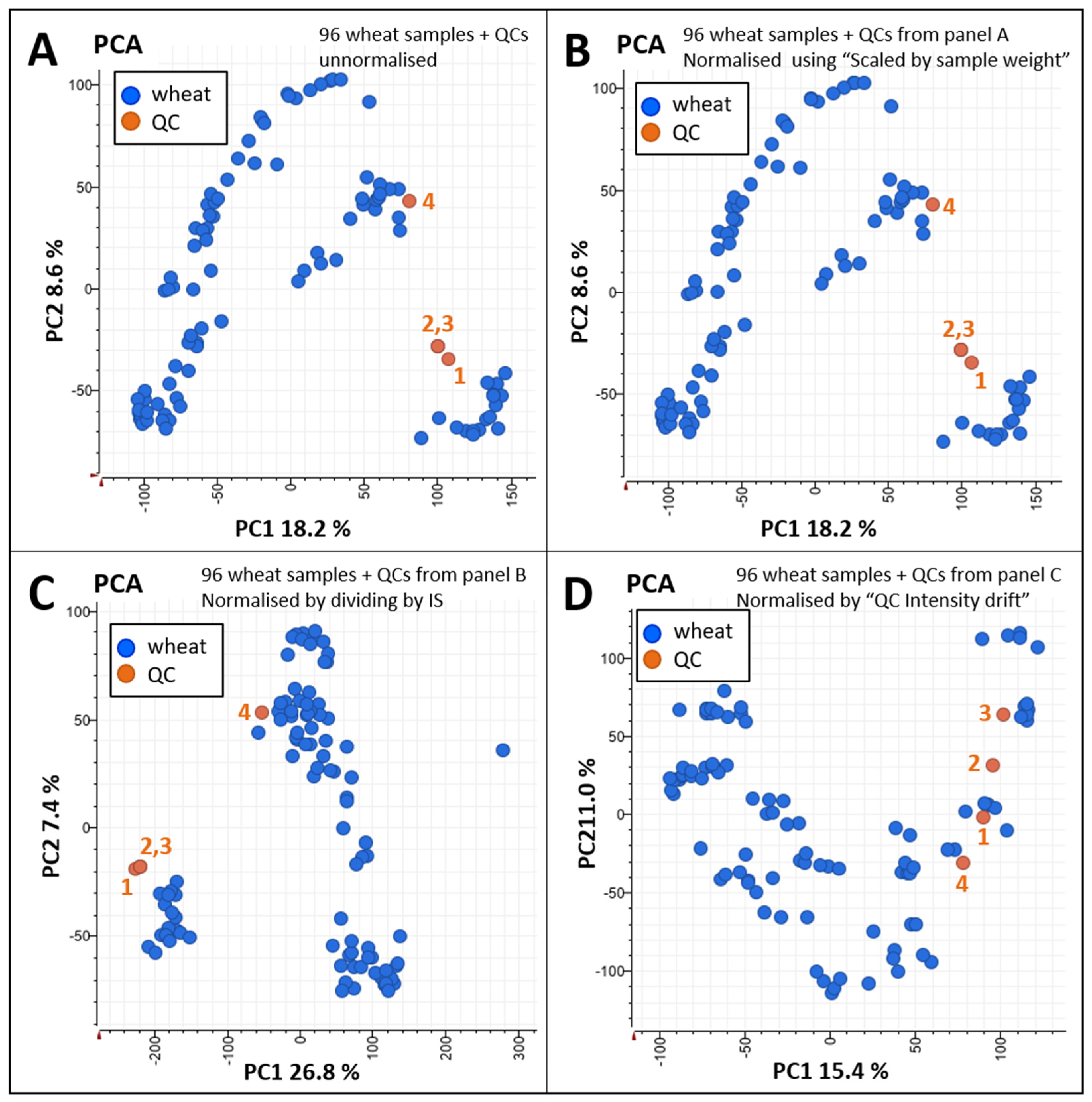
Data Normalisation and Statistical Analyses
Data Mining
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shewry, P.R. Wheat. J. Exp. Bot. 2009, 60, 1537–1553. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC); IWGSC RefSeq Principal Investigators; Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; iIWGSC Whole-Genome Assembly Principal Investigators; Pozniak, C.J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, J.; Garcia, D.F.; Zhou, Y.; Appels, R.; Li, A.; Mao, L. The Battle to Sequence the Bread Wheat Genome: A Tale of the Three Kingdoms. Genom. Proteom. Bioinform. 2020, 18, 221–229. [Google Scholar] [CrossRef]
- Bhalla, P.L.; Sharma, A.; Singh, M.B. Enabling Molecular Technologies for Trait Improvement in Wheat. Methods Mol. Biol. 2017, 1679, 3–24. [Google Scholar] [CrossRef]
- Bonomi, F.; Iametti, S.; Mamone, G.; Ferranti, P. The Performing Protein: Beyond Wheat Proteomics? Cereal Chem. 2013, 90, 358–366. [Google Scholar] [CrossRef]
- Komatsu, S.; Kamal, A.H.; Hossain, Z. Wheat proteomics: Proteome modulation and abiotic stress acclimation. Front. Plant Sci. 2014, 5, 684. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Liu, Y.; Dong, J.; Zhao, W.; Kashyap, S.; Gao, X.; Rustgi, S.; Wen, S. Probing early wheat grain development via transcriptomic and proteomic approaches. Funct. Integr. Genom. 2020, 20, 63–74. [Google Scholar] [CrossRef]
- Zhang, S.; Ghatak, A.; Bazargani, M.M.; Bajaj, P.; Varshney, R.K.; Chaturvedi, P.; Jiang, D.; Weckwerth, W. Spatial distribution of proteins and metabolites in developing wheat grain and their differential regulatory response during the grain filling process. Plant J. 2021, 107, 669–687. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar] [CrossRef]
- Garcia-Molina, M.D.; Muccilli, V.; Saletti, R.; Foti, S.; Masci, S.; Barro, F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J. Proteom. 2017, 165, 102–112. [Google Scholar] [CrossRef]
- Nadaud, I.; Girousse, C.; Debiton, C.; Chambon, C.; Bouzidi, M.F.; Martre, P.; Branlard, G. Proteomic and morphological analysis of early stages of wheat grain development. Proteomics 2010, 10, 2901–2910. [Google Scholar] [CrossRef] [PubMed]
- Uvackova, L.; Skultety, L.; Bekesova, S.; McClain, S.; Hajduch, M. MS(E) based multiplex protein analysis quantified important allergenic proteins and detected relevant peptides carrying known epitopes in wheat grain extracts. J. Proteome Res. 2013, 12, 4862–4869. [Google Scholar] [CrossRef]
- Wong, J.H.; Cai, N.; Tanaka, C.K.; Vensel, W.H.; Hurkman, W.J.; Buchanan, B.B. Thioredoxin reduction alters the solubility of proteins of wheat starchy endosperm: An early event in cereal germination. Plant Cell Physiol. 2004, 45, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Matros, A.; Mock, H.P.; Muhling, K.H. Protein Composition and Baking Quality of Wheat Flour as Affected by Split Nitrogen Application. Front. Plant Sci. 2019, 10, 642. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Jorgensen, A.D.; Li, H.; Sondergaard, I.; Finnie, C.; Svensson, B.; Jiang, D.; Wollenweber, B.; Jacobsen, S. Implications of high-temperature events and water deficits on protein profiles in wheat (Triticum aestivum L. cv. Vinjett) grain. Proteomics 2011, 11, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Lakhneko, O.; Danchenko, M.; Morgun, B.; Kovac, A.; Majerova, P.; Skultety, L. Comprehensive Comparison of Clinically Relevant Grain Proteins in Modern and Traditional Bread Wheat Cultivars. Int. J. Mol. Sci. 2020, 21, 3445. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, S.; Stenoien, D.L.; Pasa-Tolic, L. High-throughput proteomics. Annu. Rev. Anal. Chem. 2014, 7, 427–454. [Google Scholar] [CrossRef] [Green Version]
- Di Francesco, A.; Saletti, R.; Cunsolo, V.; Svensson, B.; Muccilli, V.; Vita, P.; Foti, S. Qualitative proteomic comparison of metabolic and CM-like protein fractions in old and modern wheat Italian genotypes by a shotgun approach. J. Proteom. 2020, 211, 103530. [Google Scholar] [CrossRef] [PubMed]
- Salplachta, J.; Marchetti, M.; Chmelik, J.; Allmaier, G. A new approach in proteomics of wheat gluten: Combining chymotrypsin cleavage and matrix-assisted laser desorption/ionization quadrupole ion trap reflectron tandem mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 2725–2728. [Google Scholar] [CrossRef] [PubMed]
- Tosi, P.; Gritsch, C.S.; He, J.; Shewry, P.R. Distribution of gluten proteins in bread wheat (Triticum aestivum) grain. Ann. Bot. 2011, 108, 23–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mughal, I.; Shah, Y.; Tahir, S.; Haider, W.; Fayyaz, M.; Yasmin, T.; Ilyas, M.; Farrakh, S. Protein quantification and enzyme activity estimation of Pakistani wheat landraces. PLoS ONE 2020, 15, e0239375. [Google Scholar] [CrossRef]
- Pilolli, R.; Gadaleta, A.; Di Stasio, L.; Lamonaca, A.; De Angelis, E.; Nigro, D.; De Angelis, M.; Mamone, G.; Monaci, L. A Comprehensive Peptidomic Approach to Characterize the Protein Profile of Selected Durum Wheat Genotypes: Implication for Coeliac Disease and Wheat Allergy. Nutrients 2019, 11, 2321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bose, U.; Broadbent, J.A.; Byrne, K.; Hasan, S.; Howitt, C.A.; Colgrave, M.L. Optimisation of protein extraction for in-depth profiling of the cereal grain proteome. J. Proteom. 2019, 197, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Pilolli, R.; Gadaleta, A.; Mamone, G.; Nigro, D.; De Angelis, E.; Montemurro, N.; Monaci, L. Scouting for Naturally Low-Toxicity Wheat Genotypes by a Multidisciplinary Approach. Sci. Rep. 2019, 9, 1646. [Google Scholar] [CrossRef] [PubMed]
- Dupont, F.M.; Vensel, W.H.; Tanaka, C.K.; Hurkman, W.J.; Altenbach, S.B. Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry. Proteome Sci. 2011, 9, 10. [Google Scholar] [CrossRef] [Green Version]
- Zilic, S.; Barac, M.; Pesic, M.; Dodig, D.; Ignjatovic-Micic, D. Characterization of proteins from grain of different bread and durum wheat genotypes. Int. J. Mol. Sci. 2011, 12, 5878–5894. [Google Scholar] [CrossRef]
- Yu, Z.; Han, C.; Yan, X.; Li, X.; Jiang, G.; Yan, Y. Rapid characterization of wheat low molecular weight glutenin subunits by ultraperformance liquid chromatography (UPLC). J. Agric. Food Chem. 2013, 61, 4026–4034. [Google Scholar] [CrossRef]
- Prandi, B.; Bencivenni, M.; Tedeschi, T.; Marchelli, R.; Dossena, A.; Galaverna, G.; Sforza, S. Common wheat determination in durum wheat samples through LC/MS analysis of gluten peptides. Anal. Bioanal. Chem. 2012, 403, 2909–2914. [Google Scholar] [CrossRef]
- Cho, K.; Jang, Y.R.; Lim, S.H.; Altenbach, S.B.; Gu, Y.Q.; Simon-Buss, A.; Lee, J.Y. Proteomic Determination of Low-Molecular-Weight Glutenin Subunit Composition in Aroona Near-Isogenic Lines and Standard Wheat Cultivars. Int. J. Mol. Sci. 2021, 22, 7709. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, K.L.; McGrath, S.C.; Callahan, J.H.; Ross, M.M. Characterization of grain-specific peptide markers for the detection of gluten by mass spectrometry. J. Agric. Food Chem. 2014, 62, 5835–5844. [Google Scholar] [CrossRef]
- Osborne, T.B. The Vegetable Proteins, 2nd ed.; Longmans, Green and Company: London, UK, 1924; Volume 2. [Google Scholar]
- Zhao, J.; Li, Z.; Khan, M.U.; Gao, X.; Yu, M.; Gao, H.; Li, Y.; Zhang, H.; Dasanayaka, B.P.; Lin, H. Extraction of total wheat (Triticum aestivum) protein fractions and cross-reactivity of wheat allergens with other cereals. Food Chem. 2021, 347, 129064. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Gao, X.; Dong, J.; Gandhi, N.; Cai, H.; von Wettstein, D.H.; Rustgi, S.; Wen, S. Pattern of Protein Expression in Developing Wheat Grains Identified through Proteomic Analysis. Front. Plant Sci. 2017, 8, 962. [Google Scholar] [CrossRef] [Green Version]
- Cherkaoui, M.; Geairon, A.; Lollier, V.; Clemente, H.S.; Larre, C.; Rogniaux, H.; Jamet, E.; Guillon, F.; Francin-Allami, M. Cell Wall Proteome Investigation of Bread Wheat (Triticum Aestivum) Developing Grain in Endosperm and Outer Layers. Proteomics 2018, 18, e1800286. [Google Scholar] [CrossRef] [PubMed]
- Cherkaoui, M.; Lollier, V.; Geairon, A.; Bouder, A.; Larre, C.; Rogniaux, H.; Jamet, E.; Guillon, F.; Francin-Allami, M. Cell Wall Proteome of Wheat Grain Endosperm and Outer Layers at Two Key Stages of Early Development. Int. J. Mol. Sci. 2019, 21, 239. [Google Scholar] [CrossRef] [Green Version]
- Daba, S.D.; Liu, X.; Aryal, U.; Mohammadi, M. A proteomic analysis of grain yield-related traits in wheat. AoB Plants 2020, 12, plaa042. [Google Scholar] [CrossRef]
- Raynes, J.K.; Vincent, D.; Zawadzki, J.L.; Savin, K.; Mertens, D.; Logan, A.; Williams, R.P.W. Investigation of Age Gelation in UHT Milk. Beverages 2018, 4, 95. [Google Scholar] [CrossRef] [Green Version]
- Vincent, D.; Binos, S.; Rochfort, S.; Spangenberg, G. Top-down proteomics of medicinal cannabis. Proteomes 2019, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Vincent, D.; Elkins, A.; Condina, M.R.; Ezernieks, V.; Rochfort, S. Quantitation and Identification of Intact Major Milk Proteins for High-Throughput LC-ESI-Q-TOF MS Analyses. PLoS ONE 2016, 11, e0163471. [Google Scholar] [CrossRef]
- Vincent, D.; Ezernieks, V.; Rochfort, S.; Spangenberg, G. A Multiple Protease Strategy to Optimise the Shotgun Proteomics of Mature Medicinal Cannabis Buds. Int. J. Mol. Sci. 2019, 20, 5630. [Google Scholar] [CrossRef] [Green Version]
- Vincent, D.; Mertens, D.; Rochfort, S. Optimisation of Milk Protein Top-Down Sequencing Using In-Source Collision-Induced Dissociation in the Maxis Quadrupole Time-of-Flight Mass Spectrometer. Molecules 2018, 23, 2777. [Google Scholar] [CrossRef] [Green Version]
- Vincent, D.; Rochfort, S.; Spangenberg, G. Optimisation of Protein Extraction from Medicinal Cannabis Mature Buds for Bottom-Up Proteomics. Molecules 2019, 24, 659. [Google Scholar] [CrossRef] [Green Version]
- Vincent, D.; Savin, K.; Rochfort, S.; Spangenberg, G. The Power of Three in Cannabis Shotgun Proteomics: Proteases, Databases and Search Engines. Proteomes 2020, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Drapeau, G.R.; Boily, Y.; Houmard, J. Purification and properties of an extracellular protease of Staphylococcus aureus. J. Biol. Chem. 1972, 247, 6720–6726. [Google Scholar] [CrossRef]
- Tsiatsiani, L.; Heck, A.J. Proteomics beyond trypsin. FEBS J. 2015, 282, 2612–2626. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Vensel, W.H.; Dupont, F.M. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Res. Notes 2011, 4, 242. [Google Scholar] [CrossRef] [Green Version]
- Endo, S. Studies on protease produced by thermophilic bacteria. J. Ferment. Technol. 1962, 40, 346–353. [Google Scholar]
- Bhatt, D.K.; Prasad, B. Critical Issues and Optimized Practices in Quantification of Protein Abundance Level to Determine Interindividual Variability in DMET Proteins by LC-MS/MS Proteomics. Clin. Pharmacol. Ther. 2018, 103, 619–630. [Google Scholar] [CrossRef]
- Neilson, K.A.; Ali, N.A.; Muralidharan, S.; Mirzaei, M.; Mariani, M.; Assadourian, G.; Lee, A.; van Sluyter, S.C.; Haynes, P.A. Less label, more free: Approaches in label-free quantitative mass spectrometry. Proteomics 2011, 11, 535–553. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.E.; Mann, M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005, 1, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Bindschedler, L.V.; Cramer, R. Quantitative plant proteomics. Proteomics 2011, 11, 756–775. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, L. Sample normalization methods in quantitative metabolomics. J. Chromatogr. A 2016, 1430, 80–95. [Google Scholar] [CrossRef]
- Mizuno, H.; Ueda, K.; Kobayashi, Y.; Tsuyama, N.; Todoroki, K.; Min, J.Z.; Toyo’oka, T. The great importance of normalization of LC-MS data for highly-accurate non-targeted metabolomics. Biomed. Chromatogr. 2017, 31, e3864. [Google Scholar] [CrossRef]
- Mitra, V.; Smilde, A.K.; Bischoff, R.; Horvatovich, P. Tutorial: Correction of shifts in single-stage LC-MS(/MS) data. Anal. Chim. Acta 2018, 999, 37–53. [Google Scholar] [CrossRef]
- Li, H.; Han, J.; Pan, J.; Liu, T.; Parker, C.E.; Borchers, C.H. Current trends in quantitative proteomics—An update. J. Mass Spectrom. 2017, 52, 319–341. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Kanehisa, M. The KEGG database. Novartis Found. Symp. 2002, 247, 91–101. [Google Scholar] [PubMed]
- Okuda, S.; Yamada, T.; Hamajima, M.; Itoh, M.; Katayama, T.; Bork, P.; Goto, S.; Kanehisa, M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008, 36, W423–W426. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Karp, P.D.; Latendresse, M.; Paley, S.M.; Krummenacker, M.; Ong, Q.D.; Billington, R.; Kothari, A.; Weaver, D.; Lee, T.; Subhraveti, P.; et al. Pathway Tools version 19.0 update: Software for pathway/genome informatics and systems biology. Brief. Bioinform. 2016, 17, 877–890. [Google Scholar] [CrossRef]
- Du, Z.; Wu, Q.; Wang, T.; Chen, D.; Huang, X.; Yang, W.; Luo, W. BlastGUI: A Python-based Cross-platform Local BLAST Visualization Software. Mol. Inform. 2020, 39, e1900120. [Google Scholar] [CrossRef] [PubMed]
- Choura, M.; Rebai, A.; Hanin, M. Proteome-wide analysis of protein disorder in Triticum aestivum and Hordeum vulgare. Comput. Biol. Chem. 2020, 84, 107138. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Ezernieks, V.; Elkins, A.; Nguyen, N.; Moate, P.J.; Cocks, B.G.; Rochfort, S. Milk Bottom-Up Proteomics: Method Optimization. Front. Genet. 2015, 6, 360. [Google Scholar] [CrossRef] [Green Version]
- Hopfgartner, G. Can MS fully exploit the benefits of fast chromatography? Bioanalysis 2011, 3, 121–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolmachev, A.V.; Monroe, M.E.; Purvine, S.O.; Moore, R.J.; Jaitly, N.; Adkins, J.N.; Anderson, G.A.; Smith, R.D. Characterization of strategies for obtaining confident identifications in bottom-up proteomics measurements using hybrid FTMS instruments. Anal. Chem. 2008, 80, 8514–8525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.; Zhou, X.; Ling, Y.; Zhang, Z.; Su, Z. agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010, 38, W64–W70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, F.M.; Wang, N.; Magee, G.B.; Nanduri, B.; Lawrence, M.L.; Camon, E.B.; Barrell, D.G.; Hill, D.P.; Dolan, M.E.; Williams, W.P.; et al. AgBase: A functional genomics resource for agriculture. BMC Genom. 2006, 7, 229. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Items Quantified | Quantities |
|---|---|
| Number of LC–MS peaks | 60,473 |
| Number of LC–MS clusters | 20,254 |
| Cluster size range | 2–11 |
| Cluster charge range | 2–10 |
| Cluster m/z range | 300.17–1996.52 |
| Cluster mass range | 598.34–8989.81 |
| Base peak range | 9–137,721 |
| Number of clusters with peptide identity | 13,165 |
| Number of identified unique peptides | 12,404 |
| Number of identified accessions | 8738 |
| Number of identified annotated proteins | 1390 |
| Range of peptides/accession | 1–65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vincent, D.; Bui, A.; Ram, D.; Ezernieks, V.; Bedon, F.; Panozzo, J.; Maharjan, P.; Rochfort, S.; Daetwyler, H.; Hayden, M. Mining the Wheat Grain Proteome. Int. J. Mol. Sci. 2022, 23, 713. https://doi.org/10.3390/ijms23020713
Vincent D, Bui A, Ram D, Ezernieks V, Bedon F, Panozzo J, Maharjan P, Rochfort S, Daetwyler H, Hayden M. Mining the Wheat Grain Proteome. International Journal of Molecular Sciences. 2022; 23(2):713. https://doi.org/10.3390/ijms23020713
Chicago/Turabian StyleVincent, Delphine, AnhDuyen Bui, Doris Ram, Vilnis Ezernieks, Frank Bedon, Joe Panozzo, Pankaj Maharjan, Simone Rochfort, Hans Daetwyler, and Matthew Hayden. 2022. "Mining the Wheat Grain Proteome" International Journal of Molecular Sciences 23, no. 2: 713. https://doi.org/10.3390/ijms23020713
APA StyleVincent, D., Bui, A., Ram, D., Ezernieks, V., Bedon, F., Panozzo, J., Maharjan, P., Rochfort, S., Daetwyler, H., & Hayden, M. (2022). Mining the Wheat Grain Proteome. International Journal of Molecular Sciences, 23(2), 713. https://doi.org/10.3390/ijms23020713







