Abstract
Actin molecules are fundamental for embryonic structural and functional differentiation; γ-actin is specifically required for the maintenance and function of cytoskeletal structures in the ear, resulting in hearing. Baraitser–Winter Syndrome (B-WS, OMIM #243310, #614583) is a rare, multiple-anomaly genetic disorder caused by mutations in either cytoplasmically expressed actin gene, ACTB (β-actin) or ACTG1 (γ-actin). The resulting actinopathies cause characteristic cerebrofrontofacial and developmental traits, including progressive sensorineural deafness. Both ACTG1-related non-syndromic A20/A26 deafness and B-WS diagnoses are characterized by hypervariable penetrance in phenotype. Here, we identify a 28th patient worldwide carrying a mutated γ-actin ACTG1 allele, with mildly manifested cerebrofrontofacial B-WS traits, hypervariable penetrance of developmental traits and sensorineural hearing loss. This patient also displays brachycephaly and a complete absence of speech faculty, previously unreported for ACTG1-related B-WS or DFNA20/26 deafness, representing phenotypic expansion. The patient’s exome sequence analyses (ES) confirms a de novo ACTG1 variant previously unlinked to the pathology. Additional microarray analysis uncover no further mutational basis for dual molecular diagnosis in our patient. We conclude that γ-actin c.542C > T, p.Ala181Val is a dominant pathogenic variant, associated with mildly manifested facial and cerebral traits typical of B-WS, hypervariable penetrance of developmental traits and sensorineural deafness. We further posit and present argument and evidence suggesting ACTG1-related non-syndromic DFNA20/A26 deafness is a manifestation of undiagnosed ACTG1-related B-WS.
1. Introduction
Actin isoforms β and γ are identical except for four amino acid substitutions within the first 10 N-terminal residues. Both isoforms are vital to embryonic differentiation and interact in some tissues, with each having hundreds of first-level interactions with other cellular molecules [1]. Nonetheless, these forms have different specific expression patterns and are involved in multiple, largely separate processes, with little functional redundancy [1,2]. γ-Actin expression predominates in the auditory hair cells [3] and is required for the formation and maintenance of the structures involved in the mechanotransduction of sound waves to electrical signals which are relayed to the brain as hearing [4,5,6,7]. Consistent with differential expression patterns, the accumulated data support phenotype–genotype correlation for B-WS-associated mutations in ACTG1 versus ACTB (OMIM#102560 and #102630). Clinically, both ACTB-related and ACTG1-related B-WS share characteristic dysmorphic frontofacial traits: hypertelorism, ptosis, wide face and frequently, trigonocephaly. Both also present with a range of developmental traits, including sensorineural deafness, iris and retinal coloboma, growth retardation and intellectual disability. Cleft palate, hallux duplication and congenital heart problems are more common in ACTB-related BW-S. However, ACTG1-related B-WS patients show milder and highly variable trait manifestation [8,9,10] and varying degrees of deafness and time of onset, as well as other, predominantly neuromigration-related developmental traits. These include intellectual disability, microcephaly, coloboma of the iris and lens or/and optic nerve, optic nerve anomalies and structural/microstructural changes in the brain, such as pachygyria, myelination defects, lissencephaly and microlissencephaly [8]. Hypervariable penetrance associated with ACTG1-related disease means that any, all or possibly even none of these may be apparent in a patient carrying a pathogenic ACTG1 allele.
The correlation of different intragenic point mutations, or actin protein domains, with specific phenotypic features, however, remains unclear.
The majority (28/51) of confirmed pathogenic ACTG1 mutations in the Human Gene Mutation Database (HGMD) are reported to be associated with non-syndromal sensorineural hearing loss (NSHL) DFNA20/26 (Table 1).

Table 1.
All ACTG1 mutations listed in HGMD as of 30 November 2021. DM + red, disease-causing mutation; DM? + orange, possibly disease-causing mutation; B-WS + grey, Baraitser–Winter cerebrofrontofacial syndrome; green, mutations correlated with neither hearing loss nor B-WS. Nucleotide changes in the ACTG1 gene are reported according to RefSeq transcript NM_001614.5. Amino acid changes are reported according to RefSeq transcript NP_001605.1. (↓) and (↑) indicate two different nucleotide substitutions within the same codon resulting in two variants.
In the literature to date, these variants do not overlap with the 17/51 confirmed B-WS-related ACTG1 variants [53]. The two conditions are therefore considered to be separate molecular and clinical diagnoses. Pathology that is neither B-WS nor deafness has been reported for 13 other listed ACTG1 variants (2, 12, 13, 14, 23, 27, 31, 37, 40, 46, 47, 49 and 69; Table 1). Pathogenic or possibly pathogenic variants listed for ACTG1 therefore currently fall into three non-overlapping subsets according to the reported phenotype, with causal implications. However, the accumulated literature [10,24,50] reveals a snowballing degree of wobble in the molecular classification of ACTG1 variants into subsets causing different pathologies. As individual ACTG1-related B-WS cases and familial DFNA20/26 studies accumulate, as well as ACTG1-linked pathologies reported as unassociated with either, the overlap of the symptoms and molecular etiology has become increasingly evident. A compilation of the data to date in Table 1 presents this information, which is taken up in our discussion. This has considerable diagnostic, clinical and genetic counseling impact, as well as biomedical interest. Here we present a 28th B-WS patient carrying a previously unassociated mutation in ACTG1 that encapsulates and informs this issue.
2. Results
Our patient is a boy born at term in the 39th week of the mother’s third pregnancy by natural delivery. Parents (father: 40 years, healthy; mother: 39 years at date of birth, treated with Plaquenil for lupus erythematosus diagnosed 3 years earlier) are non-consanguineous Caucasians of Polish descent. During pregnancy, nuchal translucency at the 95th percentile and a higher risk of trisomy 21 was determined by a non-invasive test (PAPP-A), and a cell-free fetal DNA (cffDNA) test was performed. This showed a low risk of Down syndrome.
2.1. Patient Parameters at Birth
Patient parameters at birth were as follows: weight, 3350 g (50 c); length, 56 cm (>95 c); occipital frontal circumference (OFC), 34 cm (25 c); Apgar score, 10; head circumference, 34 cm (−2 SD of the normal mean). Distinctive dysmorphic features and cryptorchidism were noted; an ultrasound examination revealed unilateral duplication of the pelvicalyceal system and echocardiography showed a membranous ventricular septal defect (VSD). Ultrasound images of all other internal organs were normal. The patient tested negative at birth for toxoplasmosis and cytomegaly.
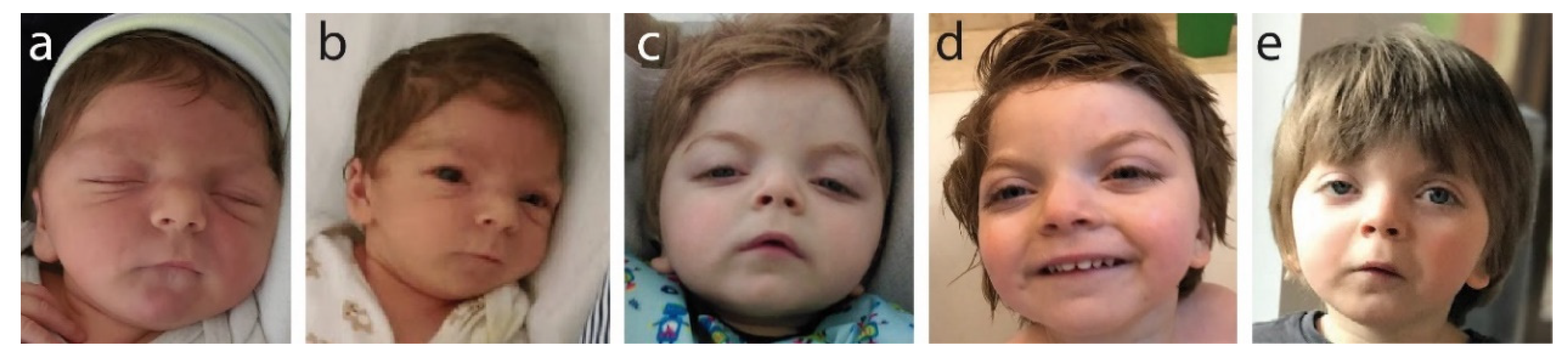
Facial features noted at birth and as the child grew (Figure 1) show wide-set eyes (hypertelorism), microcephaly and a wide face.
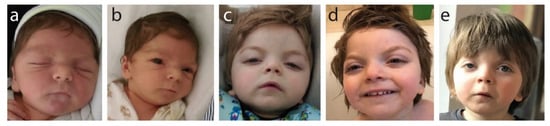
Figure 1.
Photographic documentation of frontofaciocerebral features: (a) 3 months old (hypertelorism, short nose, round face); (b) 8 months old (small chin, retrognathia, posteriorly rotated ears); (c) 12 months old (arched eyebrows, elongated palpebral fissures, short nose, mouth corners directed down, round face); (d) 2 years and 2 months old (high forehead and frontal bossing); (e) 4 years old (unilateral ptosis, visible changing of the face’s shape with age, which is becoming less round with more visible arched eyebrows).
Clinically, these specific facial features presenting together are strongly indicative of B-WS. Brachycephaly, a short nose and a long philtrum were also noted at birth and were present at all later examination age points. Progressively, we saw the following: at age 3 months: hypertelorism, short nose and round face; at age 8 months: small chin, retrognathia and posteriorly rotated ears; at age 12 months: progressive and more pronounced arching of the eyebrows, elongated palpebral fissures, mouth corners directed down, face becoming less round and more elongated, and unilateral ptosis; at age 2 years 2 months: high forehead with frontal bossing evident; at age 4years: strong unilateral ptosis, more pronounced arching of the eyebrows and face shape changing with age, becoming increasingly less round.
2.2. Sensorineural Deafness
Screening tests after birth suggested deafness. Hearing tests between the ages of 2 to 4 months were inconclusive. At 4 months, severe deafness was diagnosed using the Auditory Brainstem Response (ABR) test, with 70 dB (left) and 80–90 dB (right) auditory deficits. At age 3 and a half months, CT imaging revealed shadowing of the entire pneumatic structures of both pyramids, suggesting inner ear inflammation. His middle ear canals were drained, anti-inflammatory treatment was applied and bilateral hearing aids were used from 6 months. Brainstem auditory evoked potentials (BAEP) at 5 months and at 16 months were positive. Hearing tests at 3 years 4 months showed sensorineural hearing loss of medium degree with a reaction to sound at 35 dB and 40–45 dB with and without hearing aids, respectively. This reflects an appreciable regression of deafness, out of character for B-WS or isolated ACTG1-related deafness, which are both normally progressive. We conclude that early infection of the ear canals and subsequent healing were responsible for the regressive nature of deafness in our patient, since hearing improved after drainage and anti-inflammatory treatment. We note that congenital malformations of the skull (microcephaly, brachycephaly) may contribute to an anomalous inner ear structure causing secretion and fluid accumulation, and a predisposition to ear canal infections exacerbating hearing loss.
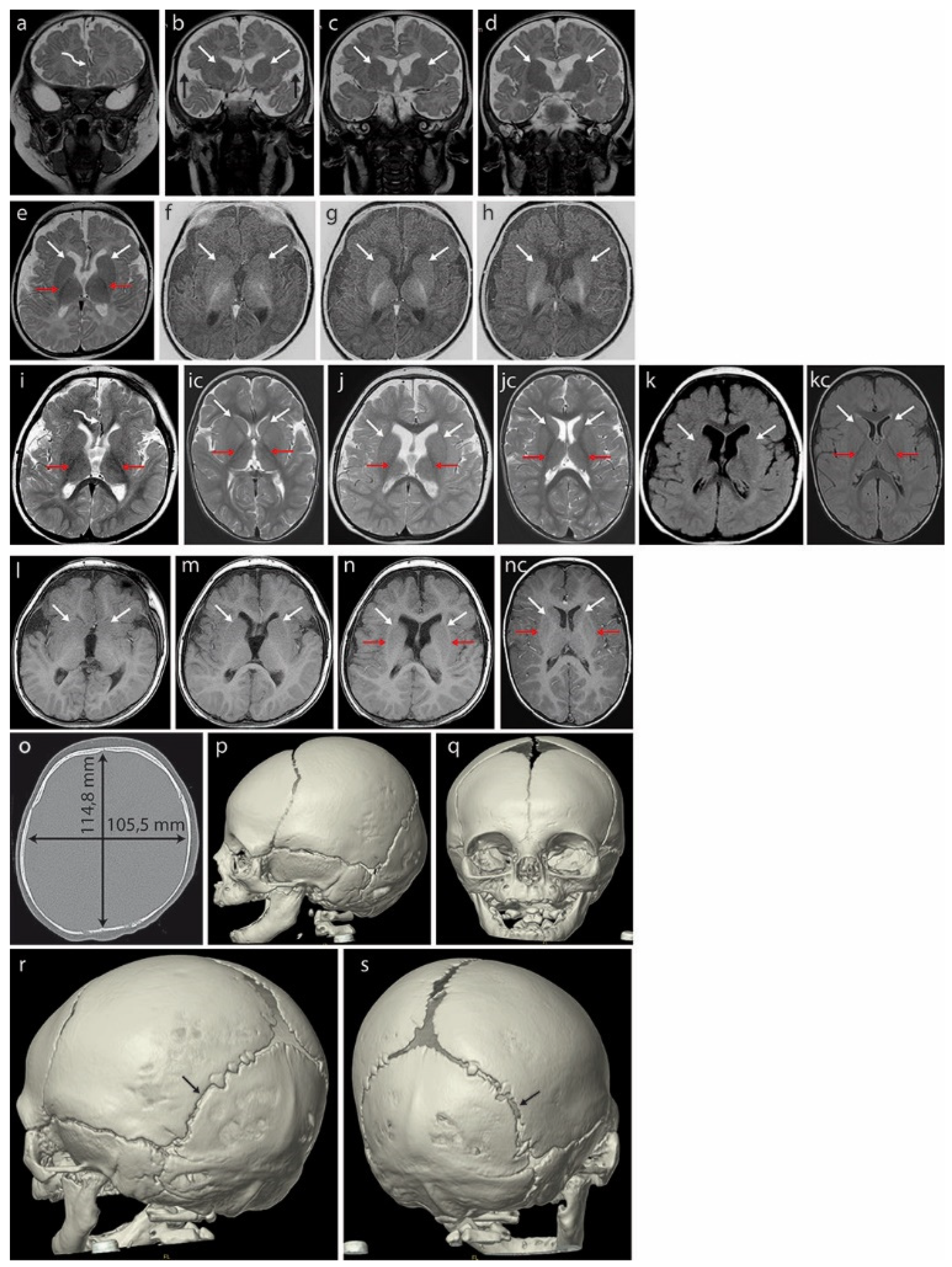
Magnetic resonance imaging (MRI) of the brain at 3 months (Figure 2a–h) revealed hypoplasia/fenestration of the anterior falx cerebri with the gyri of the right cerebral hemisphere crossing the brain midline (Figure 2a—curved arrow), as well as incomplete opercularization (Figure 2b—black arrows).
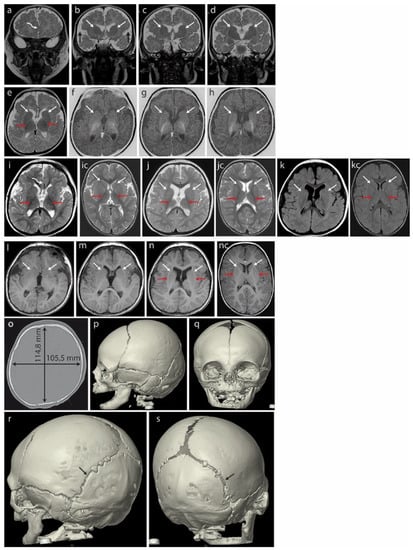
Figure 2.
Brain MRI of the patient at 3 months (a–h) revealed hypoplasia/fenestration of the anterior falx cerebri with the gyri of the right cerebral hemisphere crossing the midline ((a)—curved arrow) and incomplete opercularization, which is normally complete at term ((b)—black arrows). Roundish basal ganglia in the coronal plane in T2-weighted images (T2WI) ((b–d)—white arrows). At this age, the posterior limbs of the internal capsules (PLICs—red arrows) are well myelinated: black in T2WI ((e)—axial plane) and white in the inversion recovery (IR) sequence, best depicting the myelinated structures (f–h). There is no trace of even the unmyelinated lines separating the caudate and lentiform nuclei that would represent the anterior limbs of the internal capsules (ALICs) ((b–h)—white arrows). A further brain MRI scan at 18 months (i–n) did not detect the ALICs which are normally appreciable at 10–11 months. The basal ganglia appear to be fused bilaterally in both hemispheres ((j–n) white arrows in the axial plane: (j)—T2WI, (k)—FLAIR, (l–n)—T1WI); opercularization remained incomplete (i). Normal PLICs ((i)—red arrows). (ic) Corresponding sections in the normal brain of an 18-month-old boy, T2WI. PLICs—red arrows; normal ALICs separating the caudate and lentiform nuclei—white arrows. (jc) Corresponding sections in the normal brain of an 18-month-old boy, T2WI. PLICs—red arrows; normal ALICs separating the caudate and lentiform nuclei—white arrows. (kc) Corresponding sections in the normal brain of an 18-month-old boy, FLAIR. PLICs—red arrows; normal ALICs separating the caudate and lentiform nuclei—white arrows. (nc) Corresponding sections in the normal brain of an 18-month-old boy, T1WI. PLICs—red arrows; normal ALICs separating the caudate and lentiform nuclei—white arrows. Computed tomography (CT) of the skull at age 4 months (o–s) showed a brachycephalic skull (the cephalic index of 91.9 exceeded the range of 76 to 81 compatible with a mesaticephalic skull in normal males). Asymmetric lambdoid suture: narrower on the left (r—black arrow) compared with the wider right one (s—black arrow).
Unusually rounded basal ganglia are visible in the coronal plane in T2-weighted images (T2WI) (Figure 2b–d, white arrows). The posterior limbs of the internal capsules (PLICs, red arrows) are well myelinated: these structures appear black in T2WI on the axial plane (Figure 2e) and white in inversion recovery (IR) sequences highlighting the myelinated structures (Figure 2f–h). The anatomical line normally separating the caudate and lentiform nuclei from the anterior limbs of the internal capsules (ALICs) remains undetectable at the structural and myelination levels (Figure 2b–h, white arrows), indicating severely disrupted/delayed myelination. This is currently considered rare in B-WS patients [8].
A brain MRI at 18 months (Figure 2i–n) showed that opercularization remained incomplete (Figure 2i, red arrows). The anterior limbs of the internal capsules (ALICs) that are normally appreciable at 10–11 months were not detected. The basal ganglia appear to be fused bilaterally (Figure 2j–n, arrows showing fusion lines).
A CT of the skull at age 4 months (Figure 2o–q) showed a brachycephalic skull with a cephalic index of 91.9, indicating pathologically disturbed proportions of the skull. This exceeded the range of 76 to 81 compatible with a mesaticephalic skull in normal males. A SD greater than 3.5 between the length and width of the skull was noted at 33 months (−2.29 SD vs. 1.32 SD). We note that the initial physical examination suggested positional brachycephaly with a discrete asymmetry of the occiput (plagiocephaly), but a later CT examination ruled out positional skull deformation because there were clear differences in the width of the lambdoid suture: it was narrow on the left side and wide on the right (Figure 2r,s).
A chest X-ray at 2 months showed an abnormal shape of the ribs (third to eighth).
2.3. Molecular Analyses
We used array comparative genomic hybridization (aCGH) to compare the patient’s and reference human genomes, and assess the genomic content variation and gene copy number aberrations. Our patient shows a normal genomic copy number and genomic content.
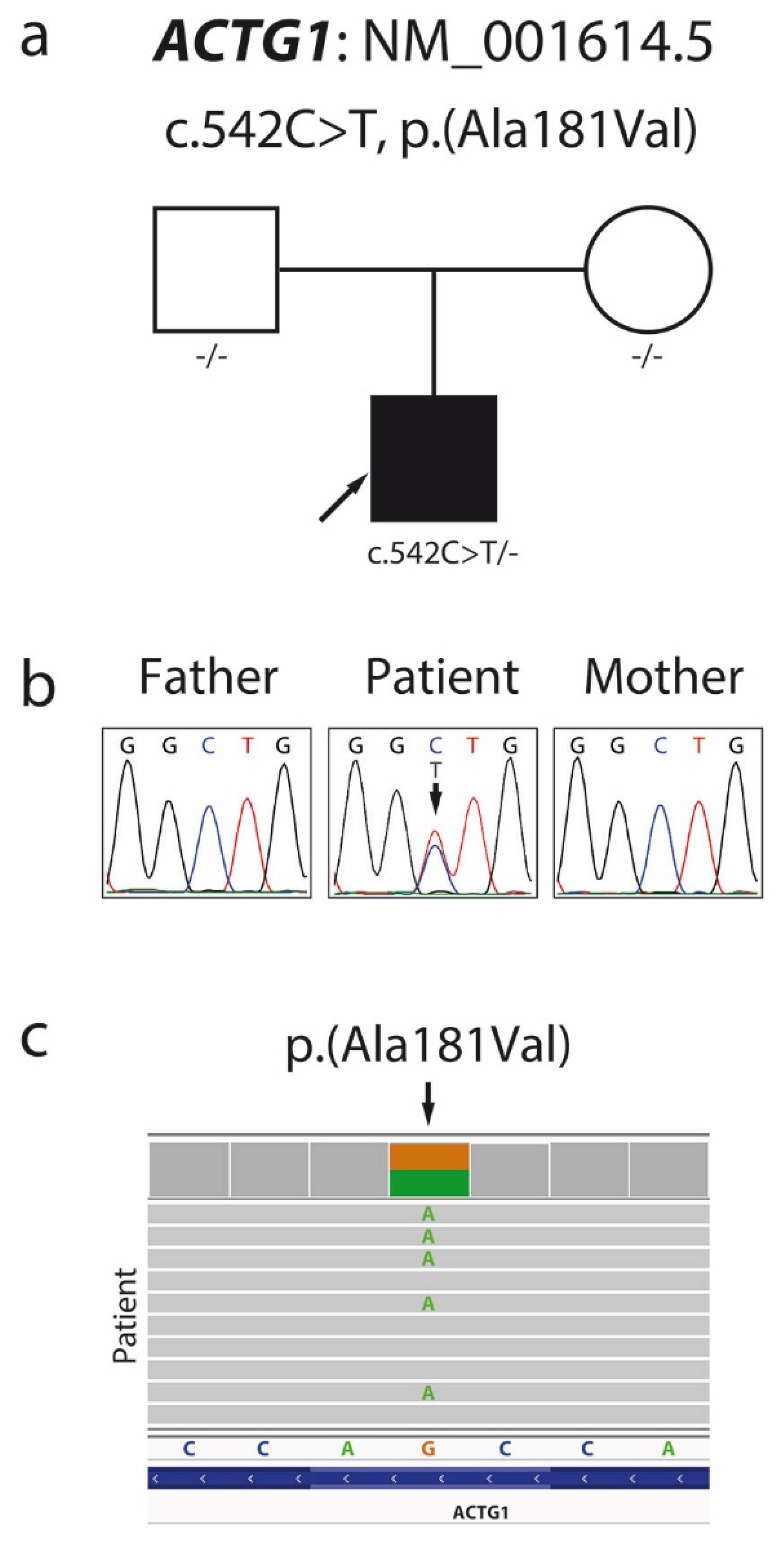
Patient exome screening for possible pathological mutations using exome sequencing (ES) identified a missense variant c.542C > T (NM_001614.5), p.Ala181Val (NP_001605.1) in the patient’s ACTG1 gene (Figure 3c), confirmed by local Sanger sequencing.
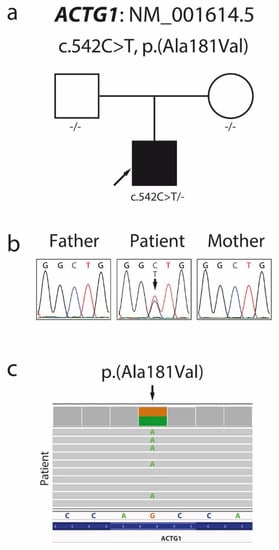
Figure 3.
Pedigree (a) and Sanger sequencing chromatogram (b) showing segregation within the family and confirming the de novo character of the c.542C > T p.Ala181Val mutation in the ACTG1 gene. Visual presentation of exome sequencing data made by the Integrative Genomics Viewer (c).
Family segregation analyses (Figure 3a,b) showed that the variant c.542C > T, p.Ala181Val arose de novo in one allele in our patient. In silico pathogenic prediction analysis resulted in 23 out of 25 prediction algorithms classifying this variant as deleterious, with high scores produced by the CADD and DANN algorithms (Table 2). The patient’s ES data revealed no additional pathogenic or likely pathogenic mutations to support dual molecular diagnoses for the patient’s phenotype.

Table 2.
Results of in silico variant analysis using various prediction algorithms.
3. Discussion
Our patient displays cranial, facial and developmental traits which together are clinically typical for B-WS [8]. These include microcephaly, specific dysmorphic facial features, intellectual disability, developmental defects of speech and brain microstructure, myelination defects and sensorineural hearing loss (Table 3, #14).

Table 3.
List of all 28 published patients with deleterious mutations in the ACTG1 gene and B-WS syndrome. Nd—no data; na—not applicable; R—regressive; CC–corpus callosum; F—female; M—male; F*—female proband in the study; F∆—mother of F*; M□—father of F*; M1 and M2—first and second patients from one study, red and bold—patient first described in this study.
He also carries a de novo point mutation in the γ-actin gene ATCG1 but no other mutation that could support a dual molecular diagnosis [14]. The specific point mutation identified here, c.542C > T, p.Ala181Val, was previously recorded in the dbSNP database (rs797044730) and the ClinVar clinical database (VCV000197198.4) as a variant with conflicting interpretations of pathogenicity, with no specific phenotype described; it is currently unlinked to any pathology. There is no further mention in the general literature, and it is not present in population genomic databases gnomAD v2 and v3. A pathogenic likelihood score of “potentially deleterious” was obtained for this variant when assessed by 23/25 predictive algorithms (Table 2). According to the American College of Medical Genetics and Genomics (ACMG) classification, this variant is classified as pathogenic (PS2, PM2, PM5, PP2, PP3). Furthermore, basal rate germline nucleotide point mutations per generation result in only one or two coding sequence mutations in a given exome [14], and our exome analyses detected only c.542C > T, p.Ala181Val, a coding missense point mutation in ACTG1. All B-WS-associated alleles for ACTG1 reported to date are missense point mutations (Table 1), and are usually de novo, although an inherited ACTG1 mutation linked to non-syndromic DFNA20/A26 deafness and B-WS in the same family has been reported [10]. Taken together, these data indicate that the variant identified here is responsible for the characteristic B-WS traits present, including deafness, and contributes to the phenotype expansion observed. Neither brachycephaly nor a complete absence of speech faculty have been previously described in ACTG1-related B-WS patients, nor for ACTG1-related isolated sensorineural deafness cases.
We therefore identify and reclassify the variant c.542C > T, p.Ala181Val as a dominant pathogenic allele of the ACTG1 gene that is causative for BW-S and also sensorineural deafness.
Our patient is the third person documented as carrying this particular variant of ACTG1 and the 28th patient worldwide documented with an array of symptoms resulting from the expression of defective γ-actin molecules providing a clear diagnosis of B-WS.
Currently, the ACTG1 variants listed in HGMD (69 including the variant presented here) fall into three non-overlapping groups associated with different symptomology: sensorineural deafness DFNA20/26, B-WS and symptomology apparently unrelated to either deafness or B-WS (Table 1, white background, grey background and green background, respectively). This leads to distinct molecular and clinical diagnoses, prognoses, therapies and genetic counseling for each group.
However, we note that, for all 69 (confirmed pathogenic and possibly pathogenic) ACTG1 variants recorded in the HGMD to date, the associated pathologies are fully consistent with the hypervariable penetrance of ACTG1-related B-WS traits. These include the apparently non-syndromic deafness-associated variants (37/69), variants carrying a confirmed B-WS diagnosis (19/69) and variants reporting pathologies other than deafness or B-WS (13/69; variants 2, 12, 13, 14, 23, 27, 31, 37, 40, 46, 47, 49 and 69 in Table 1). Without exception, the specific disparate phenotypes reported for these (microcephaly, polymicrogyria, pachygyria, microlissencephaly, intellectual disability, coloboma of the eye, congenital diaphragmatic hernia, autism, multiple congenital anomalies) are consistent with hypervariable penetrance of the phenotypic spectrum of B-WS.
There are also nine positions in the amino acid sequence of ACTG1 (Table 1, inverted arrows; variants # 15/16, 18/19, 24/25, 33/34, 38/39, 41/42, 49/50, 56/57, 62/63) where two distinct substitutions arise from separate point mutations within the same codon. This leads to two different variants with an associated pathology listed in the HGMD. In most cases (7/9), the associated pathology is the same for both variants (deafness in 5/9 positions; B-WS in 2/9 positions); in two further cases (Table 1, positions p.181 and p.256), we see B-WS for one variant (#34, p.Ala181Val; #50 p.Arg256Trp), with either deafness (#33) or pachygyria (#49), respectively, for the alternative variant. Variant #34, (p.Ala181Val) represents our patient; different substitutions at the same position in ACTG1 result in B-WS in our patient rather than isolated deafness, an apparently different phenotype. Deafness and pachygyria, however, are present in 76% and 89% respectively, of formally diagnosed ACTG1-related B-WS cases where an appropriate examination was performed (Table 1). We note that while a highly specific intragenic correlation of ACTG1 mutation with a particular trait cannot at present be formally excluded without rigorous molecular mapping, the phenotype is identical for both variants in 7/9 of positions dispersed between positions p.82 and p.332 (77%) of the ACTG1 protein sequence (375 residues). Further, in 9/9 positions, the phenotype is entirely consistent with hypervariable penetrance of traits within the spectrum for B-WS.
In addition, familial studies for DFNA20/26 NSHL involving a single segregating variant (p.Ala58Val) for ACTG1-associated sensorineural deafness reflect both full-blown B-WS (with early onset, progressive deafness, ptosis, coloboma, seizures, developmental delay), a manifestation of isolated classic B-WS traits (seizures, early-onset progressive deafness) and apparently isolated non-syndromic deafness with anomalous optic nerves in successive generations of a family [10] (Table 2: #3–5).
In short, all mutations in ACTG1 listed in the HGMD are associated with phenotypes consistent with hypervariable expression of B-WS traits. An extensive cellular network of first-level interaction partners exists for γ-actin molecules [1]. Together with normal variation in the patient’s genetic landscape, this implies considerable variability inherent in the mutation load impacting actin physiology and, therefore, actinopathies arising from a single pathogenic variant in different patient backgrounds. A simple interpretation is, therefore, that all these differing phenotypes, including deafness, reflect variable trait manifestation of undiagnosed B-WS. We therefore contend the phenotypes reported in 37/69 deafness-associated ACTG1 variants, as well as the 18/69 confirmed variants with a pathology consistent with isolated B-WS traits (Table 1), though currently associated with neither B-WS nor deafness, result from variable penetrance of molecular variants underlying actinopathies capable of causing B-WS.
Given the progressive nature and hypervariable trait penetrance of ACTG1-related B-WS and DFNA20/26 deafness, which are particularly well showcased in the familial context of segregating ACTG1 pathogenic alleles, we hope that this perspective of the overlapping etiology may be clinically useful and, crucially, inform specific molecular diagnoses and genetic counseling, in addition to therapeutic handling.
4. Materials and Methods
Array Comparative Genomic Hybridization (aCGH)
We used the CGX 3x720K Whole-Genome array (Roche NimbleGen, Madison, WI, USA) as per the manufacturer’s instructions and scanned slides into image files using an MS 200 Microarray Scanner (Roche NimbleGen, Madison, WI, USA). Data were analyzed using DEVA (Roche NimbleGen, Madison, WI, USA) and Genoglyphix Analysis Software (Perkin Elmer, Waltham, MA, USA).
Exome sequencing (ES)
Genomic DNA was extracted from peripheral blood leukocytes of the patient and his parents using an automatic magnetic bead-based method using MagNA Pure 96 system, Roche, Germany. Whole-exome sequencing libraries were prepared using Agilent SureSelect XT Human All Exon V6 sample preparation kits and the Illumina NovaSeq 6000 sequencer (Illumina, CA, USA), via 2 × 100 bp reads. Genomic data processing was based on an in-house pipeline, and the reads were aligned with Burrows–Wheeler Aligner (BWA) v0.7.16 software to the GRCh38 reference genome. The Genome Analysis Toolkit (GATK) HaplotypeCaller v4.0b4 was used for variant calling and the Ensembl Variant Effect Predictor (VEP) v96 was used to annotate the variants.
For variant prioritization, the ES data were first analyzed for the known pathogenic or likely pathogenic variants reported in the ClinVar database. The in silico gene panel was used for rare variants (under 0.01% in the gnomAD v2 database) in genes associated with microcephaly consisting of over 800 genes. Those genes were manually selected from several gene panels customized for patients with microcephaly, and additional genes were selected from various databases such as OMIM or DECIPHER. The detailed gene list is included in Supplementary Table S1. Variant pathogenicity prediction and ACMG classification were carried out with the use of the VarSome website [55] and the dnNSFP v4.2 database [56].
Polymerase chain reaction was performed by use of the FastStart Taq DNA Polymerase, dNTPack kit (Roche) as per the manufacturer’s instructions and the following primers: forward: TCCAGGTTTCTCATTTGGTTTCT; reverse: CCCGACAGCACCGTGTT (100 ng template DNA; annealing temperature, 58 °C; polymerase activity time, 1 min; 35 cycles; product length, 759 nt).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms23020692/s1.
Author Contributions
Conceptualization: P.G.; data curation: M.D., A.K.-K., E.B.-O., M.J., E.K., D.L.G., M.I.F., M.B.-F., J.B. and P.G.; formal analysis: M.D., A.K.-K. and P.G.; funding acquisition: P.G.; investigation: M.D., A.K.-K. and P.G.; methodology: P.G.; project administration: P.G.; resources: A.K.-K. and P.G.; software: M.D.; supervision: P.G.; validation: M.D. and P.G.; visualization: P.G.; writing—original draft preparation: P.G. and D.L.G.; writing—review and editing: M.D., A.K.-K., E.B.-O., D.L.G., M.I.F., M.B.-F. and P.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Centre, Poland 2015/19/B/NZ2/01824.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of Institute of Mother and Child, Warsaw, Poland (Project identification code: 29/2016 from 23 June 2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank the patient and his family members for their participation in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pollard, T.D.; Cooper, J.A. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 1986, 55, 987–1035. [Google Scholar] [CrossRef]
- Erba, H.P.; Eddy, R.; Shows, T.; Kedes, L.; Gunning, P. Structure, chromosome location, and expression of the human gamma-actin gene: Differential evolution, location, and expression of the cytoskeletal beta- and gamma-actin genes. Mol. Cell Biol. 1988, 8, 1775–1789. [Google Scholar] [CrossRef]
- Höfer, D.; Ness, W.; Drenckhahn, D. Sorting of actin isoforms in chicken auditory hair cells. J. Cell Sci. 1997, 110, 765–770. [Google Scholar] [CrossRef]
- Hudspeth, A.J. How the ear’s works work. Nature 1989, 341, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Blanchoin, L.; Mullins, R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000, 29, 545–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilney, L.G.; Egelman, E.H.; DeRosier, D.J.; Saunder, J.C. Actin filaments, stereocilia, and hair cells of the bird cochlea. II. Packing of actin filaments in the stereocilia and in the cuticular plate and what happens to the organization when the stereocilia are bent. J. Cell Biol. 1983, 96, 822–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirokawa, N.; Tilney, L.G. Interactions between actin filaments and between actin filaments and membranes in quick-frozen and deeply etched hair cells of the chick ear. J. Cell Biol. 1982, 95, 249–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verloes, A.; Di Donato, N.; Masliah-Planchon, J.; Jongmans, M.; Abdul-Raman, O.A.; Albrecht, B.; Allanson, J.; Brunner, H.; Bertola, D.; Chassaing, N.; et al. Baraitser-Winter cerebrofrontofacial syndrome: Delineation of the spectrum in 42 cases. Eur. J. Hum. Genet. 2015, 23, 292–301. [Google Scholar] [CrossRef]
- Di Donato, N.; Kuechler, A.; Vergano, S.; Heinritz, W.; Bodurtha, J.; Merchant, S.R.; Breningstall, G.; Ladda, R.; Sell, S.; Altmüller, J.; et al. Update on the ACTG1-associated Baraitser-Winter cerebrofrontofacial syndrome. Am. J. Med. Genet. A 2016, 170, 2644–2651. [Google Scholar] [CrossRef]
- Kemerley, A.; Sloan, C.; Pfeifer, W.; Smith, R.; Drack, A. A novel mutation in ACTG1 causing Baraitser-Winter syndrome with extremely variable expressivity in three generations. Ophthalmic Genet. 2017, 38, 152–156. [Google Scholar] [CrossRef]
- Accogli, A.; Severino, M.; Riva, A.; Madia, F.; Balagura, G.; Iacomino, M.; Carlini, B.; Baldassari, S.; Giacomini, T.; Croci, C.; et al. Targeted re-sequencing in malformations of cortical development: Genotype-phenotype correlations. Seizure 2020, 80, 145–152. [Google Scholar] [CrossRef]
- Wang, H.; Guan, J.; Lan, L.; Yu, L.; Xie, L.; Liu, X.; Yang, J.; Zhao, C.; Wang, D.; Wang, Q. A novel de novo mutation of ACTG1 in two sporadic non-syndromic hearing loss cases. Sci. China Life Sci. 2018, 61, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Miyajima, H.; Moteki, H.; Day, T.; Nishio, S.Y.; Murata, T.; Ikezono, T.; Takeda, H.; Abe, S.; Iwasaki, S.; Takahashi, M.; et al. Novel, ACTG1 mutations in patients identified by massively parallel DNA sequencing cause progressive hearing loss. Sci. Rep. 2020, 10, 7056. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Miyagawa, M.; Nishio, S.Y.; Ichinose, A.; Iwasaki, S.; Murata, T.; Kitajiri, S.; Usami, S. Mutational spectrum and clinical features of patients with ACTG1 mutations identified by massively parallel DNA sequencing. Ann. Otol. Rhinol. Laryngol. 2015, 124, 84S–93S. [Google Scholar] [CrossRef] [PubMed]
- De Heer, A.M.; Huygen, P.L.; Collin, R.W.; Oostrik, J.; Kremer, H.; Cremers, C.W. Audiometric and vestibular features in a second Dutch, DFNA20/26 family with a novel mutation in ACTG1. Ann. Otol. Rhinol. Laryngol. 2009, 118, 382–390. [Google Scholar] [CrossRef]
- Chacon-Camacho, O.F.; Barragán-Arévalo, T.; Villarroel, C.E.; Almanza-Monterrubio, M.; Zenteno, J.C. Previously undescribed phenotypic findings and novel ACTG1 gene pathogenic variants in Baraitser-Winter cerebrofrontofacial syndrome. Eur. J. Med. Genet. 2020, 63, 103877. [Google Scholar] [CrossRef] [PubMed]
- Morgan, A.; Lenarduzzi, S.; Cappellani, S.; Pecile, V.; Morgutti, M.; Orzan, E.; Ghiselli, S.; Ambrosetti, U.; Brumat, M.; Gajendrarao, P.; et al. Genomic studies in a large cohort of hearing impaired haract patients revealed several new alleles, a rare case of uniparental disomy (UPD) and the importance to search for copy number variations. Front Genet. 2018, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Rainger, J.; Williamson, K.A.; Soares, D.C.; Truch, J.; Kurian, D.; Gillessen-Kaesbach, G.; Seawright, A.; Prendergast, J.; Halachev, M.; Wheeler, A.; et al. A recurrent de novo mutation in ACTG1 causes isolated ocular coloboma. Hum. Mutat. 2017, 38, 942–946. [Google Scholar] [CrossRef] [Green Version]
- Gieldon, L.; Mackenroth, L.; Kahlert, A.K.; Lemke, J.R.; Porrmann, J.; Schallner, J.; von der Hagen, M.; Markus, S.; Weidensee, S.; Novotna, B.; et al. Diagnostic value of partial exome sequencing in developmental disorders. PLoS ONE 2018, 13, e0201041. [Google Scholar] [CrossRef]
- Poirier, K.; Martinovic, J.; Laquerrière, A.; Cavallin, M.; Fallet-Bianco, C.; Desguerre, I.; Valence, S.; Grande-Goburghun, J.; Francannet, C.; Deleuze, J.F.; et al. Rare, ACTG1 variants in fetal microlissencephaly. Eur. J. Med. Genet. 2015, 58, 416–418. [Google Scholar] [CrossRef] [PubMed]
- Sloan-Heggen, C.M.; Bierer, A.O.; Shearer, A.E.; Kolbe, D.L.; Nishimura, C.J.; Frees, K.L.; Ephraim, S.S.; Shibata, S.B.; Booth, K.T.; Campbell, C.A.; et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum. Genet. 2016, 135, 441–450. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Yang, T.; Wei, S.; DeWan, A.T.; Morell, R.J.; Elfenbein, J.L.; Fisher, R.A.; Leal, S.M.; Smith, R.J.; Friderici, K.H. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26). Am. J. Hum. Genet. 2003, 73, 1082–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morín, M.; Bryan, K.E.; Mayo-Merino, F.; Goodyear, R.; Mencía, A.; Modamio-Høybjør, S.; del Castillo, I.; Cabalka, J.M.; Richardson, G.; Moreno, F.; et al. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 2009, 18, 3075–3089. [Google Scholar] [CrossRef] [Green Version]
- Rivière, J.B.; van Bon, B.W.; Hoischen, A.; Kholmanskikh, S.S.; O’Roak, B.J.; Gilissen, C.; Gijsen, S.; Sullivan, C.T.; Christian, S.L.; Abdul-Rahman, O.A.; et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012, 44, 440–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Li, H.; Ren, X.; Mao, H.; Zhu, Q.; Zhu, Z.; Yang, R.; Yuan, W.; Liu, J.; Wang, Q.; et al. Novel, ACTG1 mutation causing autosomal dominant non-syndromic hearing impairment in a Chinese family. J. Genet. Genom. 2008, 35, 553–558. [Google Scholar] [CrossRef]
- Turner, T.N.; Wilfert, A.B.; Bakken, T.E.; Bernier, R.A.; Pepper, M.R.; Zhang, Z.; Torene, R.I.; Retterer, K.; Eichler, E.E. Sex-based analysis of de novo variants in neurodevelopmental disorders. Am. J. Hum. Genet. 2019, 105, 1274–1285. [Google Scholar] [CrossRef] [PubMed]
- Cabanillas, R.; Diñeiro, M.; Cifuentes, G.A.; Castillo, D.; Pruneda, P.C.; Álvarez, R.; Sánchez-Durán, N.; Capín, R.; Plasencia, A.; Viejo-Díaz, M.; et al. Comprehensive genomic diagnosis of non-syndromic and syndromic hereditary hearing loss in Spanish patients. BMC Med. Genom. 2018, 11, 58. [Google Scholar] [CrossRef] [Green Version]
- Baux, D.; Vaché, C.; Blanchet, C.; Willems, M.; Baudoin, C.; Moclyn, M.; Faugère, V.; Touraine, R.; Isidor, B.; Dupin-Deguine, D.; et al. Combined genetic approaches yield a 48% diagnostic rate in a large cohort of French hearing-impaired patients. Sci. Rep. 2017, 7, 16783. [Google Scholar] [CrossRef] [Green Version]
- Stranneheim, H.; Lagerstedt-Robinson, K.; Magnusson, M.; Kvarnung, M.; Nilsson, D.; Lesko, N.; Engvall, M.; Anderlid, B.M.; Arnell, H.; Johansson, C.B.; et al. Integration of whole genome sequencing into a healthcare setting: High diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med. 2021, 13, 40. [Google Scholar] [CrossRef]
- Miyagawa, M.; Naito, T.; Nishio, S.Y.; Kamatani, N.; Usami, S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of Japanese deafness patients. PLoS ONE 2013, 8, e71381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Longoni, M.; High, F.A.; Qi, H.; Joy, M.P.; Hila, R.; Coletti, C.M.; Wynn, J.; Loscertales, M.; Shan, L.; Bult, C.J.; et al. Genome-wide enrichment of damaging de novo variants in patients with isolated and complex congenital diaphragmatic hernia. Hum. Genet. 2017, 136, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.I.; Oh, S.K.; Kim, D.B.; Choi, S.Y.; Kim, U.K.; Lee, K.Y.; Lee, S.H. Targeted massive parallel sequencing: The effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J. Rare Dis. 2012, 7, 60. [Google Scholar] [CrossRef] [Green Version]
- Retterer, K.; Juusola, J.; Cho, M.T.; Vitazka, P.; Millan, F.; Gibellini, F.; Vertino-Bell, A.; Smaoui, N.; Neidich, J.; Monaghan, K.G.; et al. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016, 18, 696–704. [Google Scholar] [CrossRef] [Green Version]
- Vontell, R.; Supramaniam, V.G.; Davidson, A.; Thornton, C.; Marnerides, A.; Holder-Espinasse, M.; Lillis, S.; Yau, S.; Jansson, M.; Hagberg, H.E.; et al. Post-mortem characterization of a case with an ACTG1 variant, agenesis of the corpus callosum and neuronal heterotopia. Front Physiol. 2019, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Stutterd, C.A.; Brock, S.; Stouffs, K.; Fanjul-Fernandez, M.; Lockhart, P.J.; McGillivray, G.; Mandelstam, S.; Pope, K.; Delatycki, M.B.; Jansen, A.; et al. Genetic heterogeneity of polymicrogyria: Study of 123 patients using deep sequencing. Brain Commun. 2020, 3, fcaa221. [Google Scholar] [CrossRef]
- Thiffault, I.; Farrow, E.; Zellmer, L.; Berrios, C.; Miller, N.; Gibson, M.; Caylor, R.; Jenkins, J.; Faller, D.; Soden, S.; et al. Clinical genome sequencing in an unbiased pediatric cohort. Genet. Med. 2019, 21, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, T.; Imaizumi, T.; Yamamoto-Shimojima, K.; Lu, Y.; Yanagishita, T.; Shimada, S.; Chong, P.F.; Kira, R.; Ueda, R.; Ishiyama, A.; et al. Genomic backgrounds of Japanese patients with undiagnosed neurodevelopmental disorders. Brain Dev. 2019, 41, 776–782. [Google Scholar] [CrossRef]
- Yuan, Y.; Gao, X.; Huang, B.; Lu, J.; Wang, G.; Lin, X.; Qu, Y.; Dai, P. Phenotypic heterogeneity in a DFNA20/26 family segregating a novel ACTG1 mutation. BMC Genet. 2016, 17, 33. [Google Scholar] [CrossRef] [Green Version]
- Homma, T.K.; Freire, B.L.; Honjo Kawahira, R.S.; Dauber, A.; Funari, M.F.A.; Lerario, A.M.; Nishi, M.Y.; Albuquerque, E.V.; Vasques, G.A.; Collett-Solberg, P.F.; et al. Genetic disorders in prenatal onset syndromic short stature identified by exome sequencing. J. Pediatr. 2019, 215, 192–198. [Google Scholar] [CrossRef]
- Jin, S.C.; Homsy, J.; Zaidi, S.; Lu, Q.; Morton, S.; DePalma, S.R.; Zeng, X.; Qi, H.; Chang, W.; Sierant, M.C.; et al. Contribution of rare inherited and de novo variants in 2, 871 congenital heart disease probands. Nat. Genet. 2017, 49, 1593–1601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zazo Seco, C.; Wesdorp, M.; Feenstra, I.; Pfundt, R.; Hehir-Kwa, J.Y.; Lelieveld, S.H.; Castelein, S.; Gilissen, C.; de Wijs, I.J.; Admiraal, R.J.; et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur. J. Hum. Genet. 2017, 25, 308–314. [Google Scholar] [CrossRef] [Green Version]
- Mutai, H.; Suzuki, N.; Shimizu, A.; Torii, C.; Namba, K.; Morimoto, N.; Kudoh, J.; Kaga, K.; Kosaki, K.; Matsunaga, T. Diverse spectrum of rare deafness genes underlies early-childhood hearing loss in Japanese patients: A cross-sectional, multi-center next-generation sequencing study. Orphanet J. Rare Dis. 2013, 8, 172. [Google Scholar] [CrossRef] [Green Version]
- Van Wijk, E.; Krieger, E.; Kemperman, M.H.; de Leenheer, E.M.; Huygen, P.L.; Cremers, C.W.; Cremers, F.P.; Kremer, H. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26). J. Med. Genet. 2003, 40, 879–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyagawa, M.; Nishio, S.Y.; Ikeda, T.; Fukushima, K.; Usami, S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and EAS. PLoS ONE 2013, 8, e75793. [Google Scholar] [CrossRef] [PubMed]
- Park, G.; Gim, J.; Kim, A.R.; Han, K.H.; Kim, H.S.; Oh, S.H.; Park, T.; Park, W.Y.; Choi, B.Y. Multiphasic analysis of whole exome sequencing data identifies a novel mutation of ACTG1 in a nonsyndromic hearing loss family. BMC Genom. 2013, 14, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Q.; Zhu, H.; Qian, X.; Chen, Z.; Yao, J.; Lu, Y.; Cao, X.; Xing, G. Targeted genomic capture and massively parallel sequencing to identify novel variants causing Chinese hereditary hearing loss. J. Transl. Med. 2014, 12, 311. [Google Scholar] [CrossRef] [Green Version]
- Vona, B.; Müller, T.; Nanda, I.; Neuner, C.; Hofrichter, M.A.; Schröder, J.; Bartsch, O.; Läßig, A.; Keilmann, A.; Schraven, S.; et al. Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet. Med. 2014, 16, 945–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakuma, N.; Moteki, H.; Takahashi, M.; Nishio, S.Y.; Arai, Y.; Yamashita, Y.; Oridate, N.; Usami, S. An effective screening strategy for deafness in combination with a next-generation sequencing platform: A consecutive analysis. J. Hum. Genet. 2016, 61, 253–261. [Google Scholar] [CrossRef]
- Rendtorff, N.D.; Zhu, M.; Fagerheim, T.; Antal, T.L.; Jones, M.; Teslovich, T.M.; Gillanders, E.M.; Barmada, M.; Teig, E.; Trent, J.M.; et al. A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian, DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur. J. Hum. Genet. 2006, 14, 1097–1105. [Google Scholar] [CrossRef]
- Sommen, M.; Schrauwen, I.; Vandeweyer, G.; Boeckx, N.; Corneveaux, J.J.; van den Ende, J.; Boudewyns, A.; de Leenheer, E.; Janssens, S.; Claes, K.; et al. DNA diagnostics of hereditary hearing loss: A targeted resequencing approach combined with a mutation classification system. Hum. Mutat. 2016, 37, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Zarrei, M.; Burton, C.L.; Engchuan, W.; Young, E.J.; Higginbotham, E.J.; MacDonald, J.R.; Trost, B.; Chan, A.J.S.; Walker, S.; Lamoureux, S.; et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. NPJ Genom. Med. 2019, 4, 26. [Google Scholar] [CrossRef] [Green Version]
- Sorrentino, U.; Piccolo, C.; Rigon, C.; Brasson, V.; Trevisson, E.; Boaretto, F.; Martini, A.; Cassina, M. DFNA20/26 and other ACTG1-associated phenotypes: A case report and review of the literature. Audiol. Res. 2021, 11, 582–593. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, J.E.; Lee, C.; Kim, A.R.; Kim, B.J.; Park, W.Y.; Ki, C.S.; Lee, J. Genomic analysis of korean patient with microcephaly. Front. Genet. 2021, 11, 543528. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4, a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).