Identification and Characterization of an Exonic Duplication in PALB2 in a Man with Synchronous Breast and Prostate Cancer
Abstract
1. Introduction
2. Results
2.1. Case Presentation and NGS Analysis
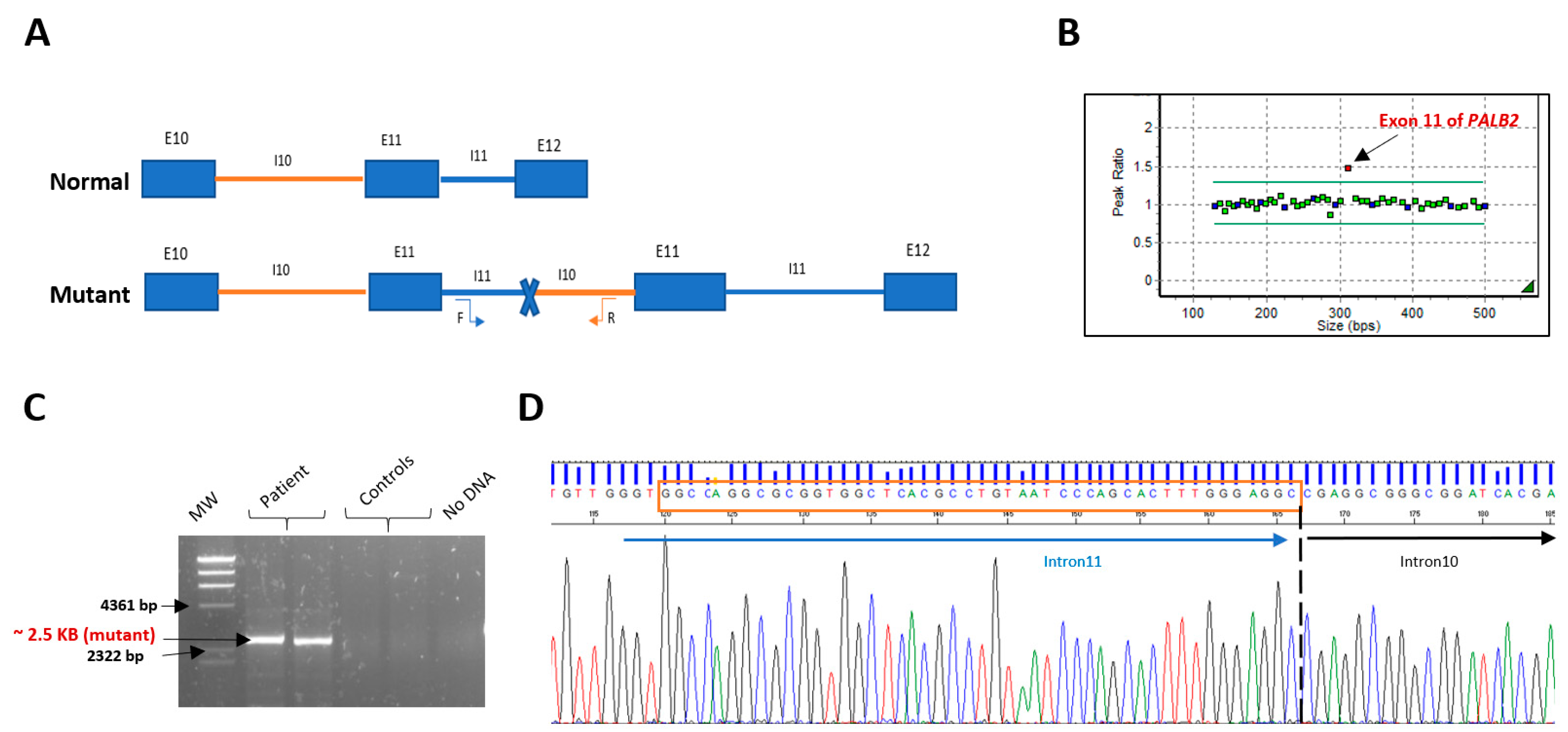
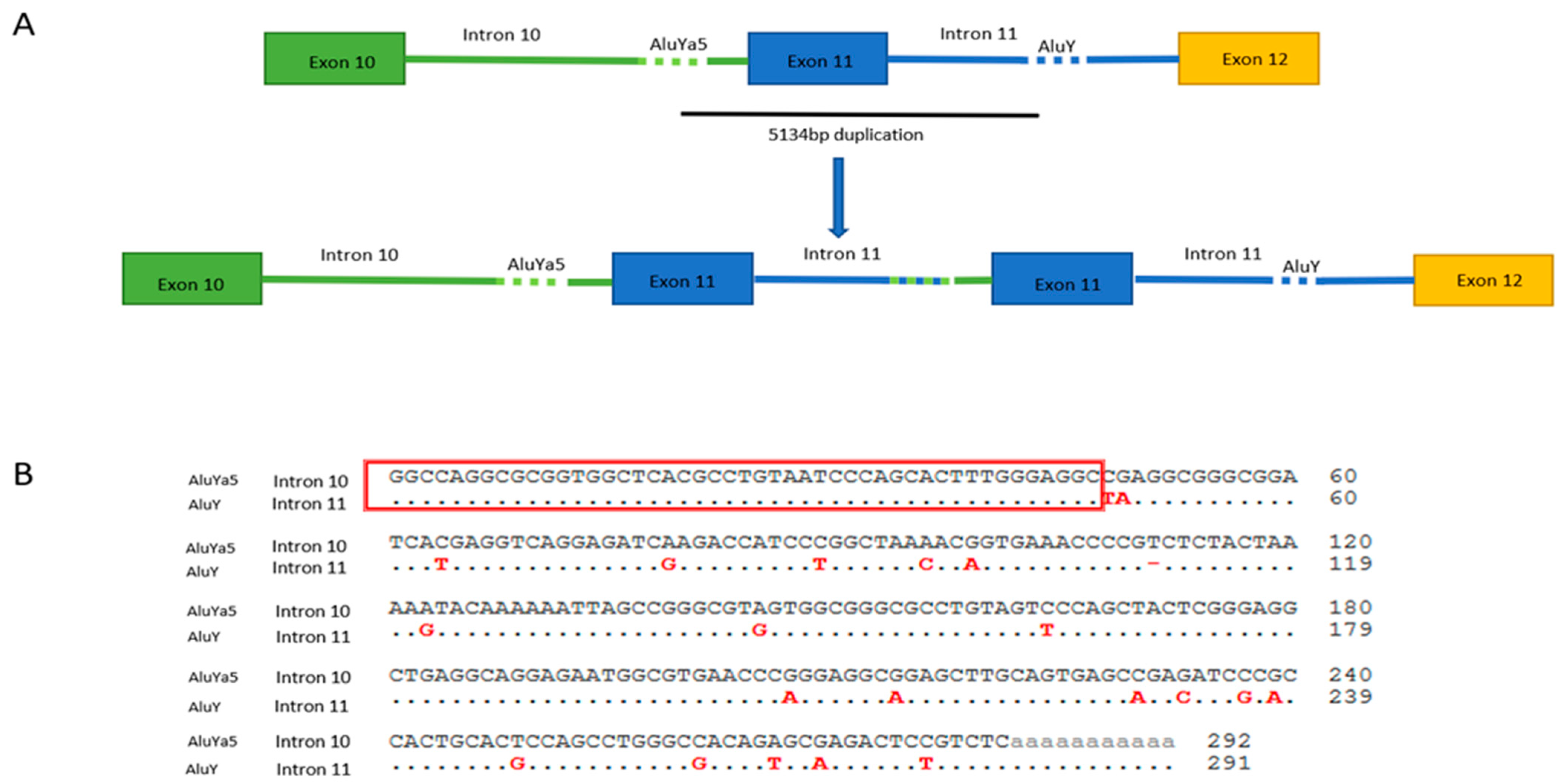
2.2. MLPA Analysis and Breakpoint Characterization
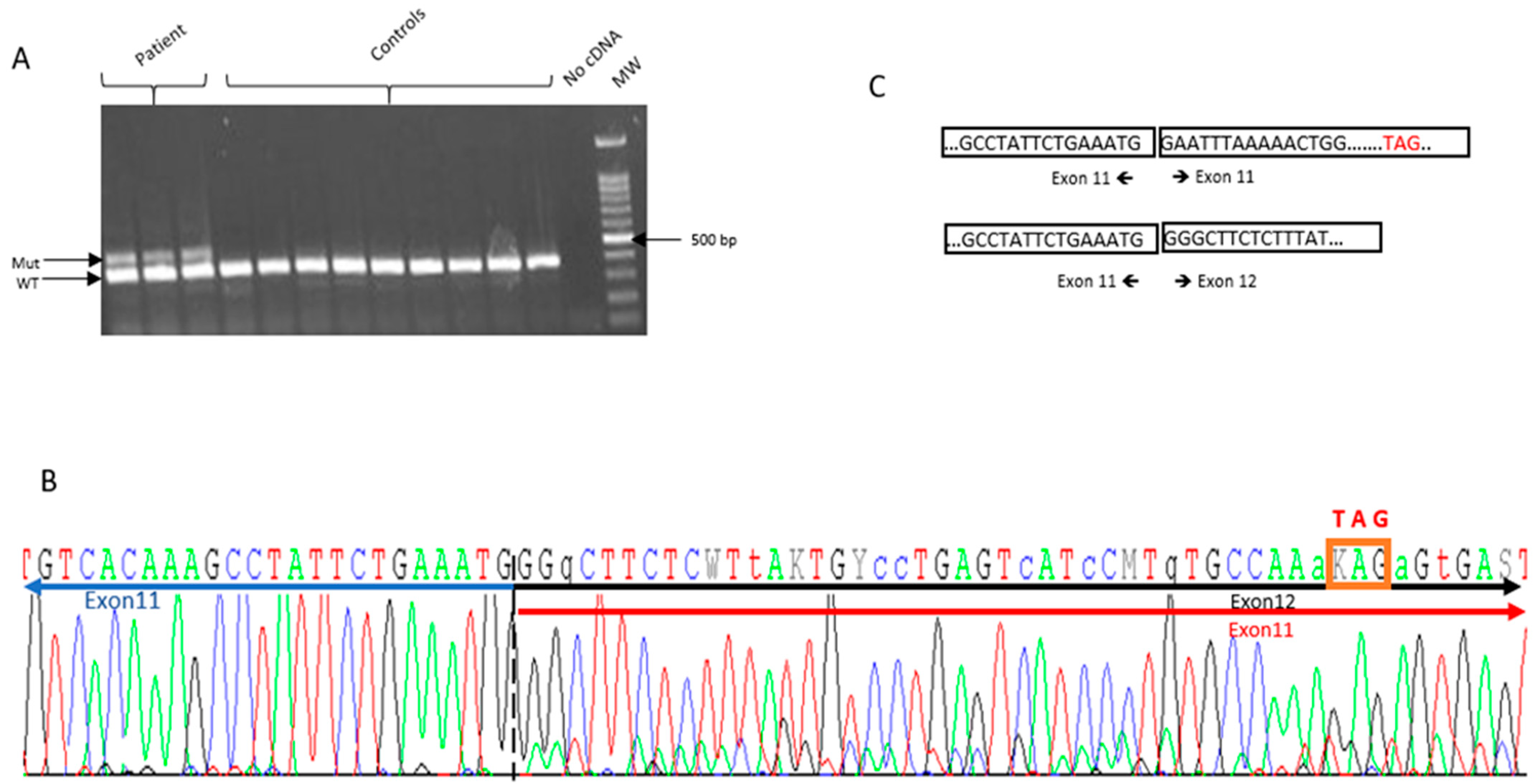
2.3. RNA Analysis
3. Discussion
4. Materials and Methods
4.1. Subjects and DNA Extraction
4.2. NGS Analysis and MLPA Confirmation
4.3. Long-Range PCR and Breakpoint Determination
4.4. RNA Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harbeck, N.; Penault-Llorca, F.; Cortes, J.; Gnant, M.; Houssami, N.; Poortmans, P.; Ruddy, K.; Tsang, J.; Cardoso, F. Breast Cancer. Nat. Rev. Dis. Primer 2019, 5, 66. [Google Scholar] [CrossRef]
- Kobayashi, H.; Ohno, S.; Sasaki, Y.; Matsuura, M. Hereditary Breast and Ovarian Cancer Susceptibility Genes (Review). Oncol. Rep. 2013, 30, 1019–1029. [Google Scholar] [CrossRef]
- Moretta, J.; Berthet, P.; Bonadona, V.; Caron, O.; Cohen-Haguenauer, O.; Colas, C.; Corsini, C.; Cusin, V.; De Pauw, A.; Delnatte, C.; et al. The French Genetic and Cancer Consortium guidelines for multigene panel analysis in hereditary breast and ovarian cancer predisposition. Bull. Cancer 2018, 105, 907–917. [Google Scholar] [CrossRef]
- Cobain, E.F.; Milliron, K.J.; Merajver, S.D. Updates on Breast Cancer Genetics: Clinical Implications of Detecting Syndromes of Inherited Increased Susceptibility to Breast Cancer. Semin. Oncol. 2016, 43, 528–535. [Google Scholar] [CrossRef]
- Campos, F.A.B.; Rouleau, E.; Torrezan, G.T.; Carraro, D.M.; Casali da Rocha, J.C.; Mantovani, H.K.; da Silva, L.R.; de Toledo Osório, C.A.B.; Moraes Sanches, S.; Caputo, S.M.; et al. Genetic Landscape of Male Breast Cancer. Cancers 2021, 13, 3535. [Google Scholar] [CrossRef] [PubMed]
- Sy, S.M.H.; Huen, M.S.Y.; Chen, J. PALB2 Is an Integral Component of the BRCA Complex Required for Homologous Recombination Repair. Proc. Natl. Acad. Sci. USA 2009, 106, 7155–7160. [Google Scholar] [CrossRef]
- Reid, S.; Schindler, D.; Hanenberg, H.; Barker, K.; Hanks, S.; Kalb, R.; Neveling, K.; Kelly, P.; Seal, S.; Freund, M.; et al. Biallelic Mutations in PALB2 Cause Fanconi Anemia Subtype FA-N and Predispose to Childhood Cancer. Nat. Genet. 2007, 39, 162–164. [Google Scholar] [CrossRef]
- Xia, B.; Dorsman, J.C.; Ameziane, N.; de Vries, Y.; Rooimans, M.A.; Sheng, Q.; Pals, G.; Errami, A.; Gluckman, E.; Llera, J.; et al. Fanconi Anemia Is Associated with a Defect in the BRCA2 Partner PALB2. Nat. Genet. 2007, 39, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Hruban, R.H.; Kamiyama, M.; Borges, M.; Zhang, X.; Parsons, D.W.; Lin, J.C.-H.; Palmisano, E.; Brune, K.; Jaffee, E.M.; et al. Exomic Sequencing Identifies PALB2 as a Pancreatic Cancer Susceptibility Gene. Science 2009, 324, 217. [Google Scholar] [CrossRef]
- Antoniou, A.C.; Casadei, S.; Heikkinen, T.; Barrowdale, D.; Pylkäs, K.; Roberts, J.; Lee, A.; Subramanian, D.; De Leeneer, K.; Fostira, F.; et al. Breast-Cancer Risk in Families with Mutations in PALB2. N. Engl. J. Med. 2014, 371, 497–506. [Google Scholar] [CrossRef]
- Southey, M.C.; Goldgar, D.E.; Winqvist, R.; Pylkäs, K.; Couch, F.; Tischkowitz, M.; Foulkes, W.D.; Dennis, J.; Michailidou, K.; van Rensburg, E.J.; et al. PALB2, CHEK2 and ATM Rare Variants and Cancer Risk: Data from COGS. J. Med. Genet. 2016, 53, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Pakkanen, S.; Wahlfors, T.; Siltanen, S.; Patrikainen, M.; Matikainen, M.P.; Tammela, T.L.J.; Schleutker, J. PALB2 Variants in Hereditary and Unselected Finnish Prostate Cancer Cases. J. Negat. Results Biomed. 2009, 8, 12. [Google Scholar] [CrossRef]
- Eeles, R.A.; Olama, A.A.A.; Benlloch, S.; Saunders, E.J.; Leongamornlert, D.A.; Tymrakiewicz, M.; Ghoussaini, M.; Luccarini, C.; Dennis, J.; Jugurnauth-Little, S.; et al. Identification of 23 New Prostate Cancer Susceptibility Loci Using the ICOGS Custom Genotyping Array. Nat. Genet. 2013, 45, 385–391. [Google Scholar] [CrossRef]
- Yang, X.; Leslie, G.; Doroszuk, A.; Schneider, S.; Allen, J.; Decker, B.; Dunning, A.M.; Redman, J.; Scarth, J.; Plaskocinska, I.; et al. Cancer Risks Associated With Germline PALB2 Pathogenic Variants: An International Study of 524 Families. J. Clin. Oncol. 2020, 38, 674–685. [Google Scholar] [CrossRef]
- Wokołorczyk, D.; Kluźniak, W.; Stempa, K.; Rusak, B.; Huzarski, T.; Gronwald, J.; Gliniewicz, K.; Kashyap, A.; Morawska, S.; Dębniak, T.; et al. PALB2 Mutations and Prostate Cancer Risk and Survival. Br. J. Cancer 2021, 125, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Erkko, H.; Xia, B.; Nikkilä, J.; Schleutker, J.; Syrjäkoski, K.; Mannermaa, A.; Kallioniemi, A.; Pylkäs, K.; Karppinen, S.-M.; Rapakko, K.; et al. A Recurrent Mutation in PALB2 in Finnish Cancer Families. Nature 2007, 446, 316–319. [Google Scholar] [CrossRef]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef]
- Horak, P.; Weischenfeldt, J.; von Amsberg, G.; Beyer, B.; Schütte, A.; Uhrig, S.; Gieldon, L.; Klink, B.; Feuerbach, L.; Hübschmann, D.; et al. Response to Olaparib in a PALB2 Germline Mutated Prostate Cancer and Genetic Events Associated with Resistance. Cold Spring Harb. Mol. Case Stud. 2019, 5, a003657. [Google Scholar] [CrossRef]
- Farmer, H.; McCabe, N.; Lord, C.J.; Tutt, A.N.J.; Johnson, D.A.; Richardson, T.B.; Santarosa, M.; Dillon, K.J.; Hickson, I.; Knights, C.; et al. Targeting the DNA Repair Defect in BRCA Mutant Cells as a Therapeutic Strategy. Nature 2005, 434, 917–921. [Google Scholar] [CrossRef]
- Schaeffer, E.; Srinivas, S.; Antonarakis, E.S.; Armstrong, A.J.; Bekelman, J.E.; Cheng, H.; D’Amico, A.V.; Davis, B.J.; Desai, N.; Dorff, T.; et al. NCCN Guidelines Insights: Prostate Cancer, Version 1.2021. J. Natl. Compr. Cancer Netw. JNCCN 2021, 19, 134–143. [Google Scholar] [CrossRef]
- Seong, M.-W.; Cho, S.I.; Kim, K.H.; Chung, I.Y.; Kang, E.; Lee, J.W.; Park, S.K.; Lee, M.H.; Choi, D.H.; Yom, C.K.; et al. A Multi-Institutional Study of the Prevalence of BRCA1 and BRCA2 Large Genomic Rearrangements in Familial Breast Cancer Patients. BMC Cancer 2014, 14, 645. [Google Scholar] [CrossRef][Green Version]
- Janatova, M.; Kleibl, Z.; Stribrna, J.; Panczak, A.; Vesela, K.; Zimovjanova, M.; Kleiblova, P.; Dundr, P.; Soukupova, J.; Pohlreich, P. The PALB2 Gene Is a Strong Candidate for Clinical Testing in BRCA1- and BRCA2-Negative Hereditary Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2013, 22, 2323–2332. [Google Scholar] [CrossRef]
- Yang, C.; Arnold, A.G.; Trottier, M.; Sonoda, Y.; Abu-Rustum, N.R.; Zivanovic, O.; Robson, M.E.; Stadler, Z.K.; Walsh, M.F.; Hyman, D.M.; et al. Characterization of a Novel Germline PALB2 Duplication in a Hereditary Breast and Ovarian Cancer Family. Breast Cancer Res. Treat. 2016, 160, 447–456. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Richardson, M.E.; Chong, H.; Mu, W.; Conner, B.R.; Hsuan, V.; Willett, S.; Lam, S.; Tsai, P.; Pesaran, T.; Chamberlin, A.C.; et al. DNA Breakpoint Assay Reveals a Majority of Gross Duplications Occur in Tandem Reducing VUS Classifications in Breast Cancer Predisposition Genes. Genet. Med. 2019, 21, 683–693. [Google Scholar] [CrossRef]
- Tischkowitz, M.; Balmaña, J.; Foulkes, W.D.; James, P.; Ngeow, J.; Schmutzler, R.; Voian, N.; Wick, M.J.; Stewart, D.R.; Pal, T. Management of Individuals with Germline Variants in PALB2: A Clinical Practice Resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1416–1423. [Google Scholar] [CrossRef]
- Parenti, S.; Rabacchi, C.; Marino, M.; Tenedini, E.; Artuso, L.; Castellano, S.; Carretta, C.; Mallia, S.; Cortesi, L.; Toss, A.; et al. Characterization of New ATM Deletion Associated with Hereditary Breast Cancer. Genes 2021, 12, 136. [Google Scholar] [CrossRef]
- Houdayer, C.; Caux-Moncoutier, V.; Krieger, S.; Barrois, M.; Bonnet, F.; Bourdon, V.; Bronner, M.; Buisson, M.; Coulet, F.; Gaildrat, P.; et al. Guidelines for Splicing Analysis in Molecular Diagnosis Derived from a Set of 327 Combined in Silico/in Vitro Studies on BRCA1 and BRCA2 Variants. Hum. Mutat. 2012, 33, 1228–1238. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouras, A.; Lafaye, C.; Leone, M.; Kherraf, Z.-E.; Martin-Denavit, T.; Fert-Ferrer, S.; Calender, A.; Boutry-Kryza, N. Identification and Characterization of an Exonic Duplication in PALB2 in a Man with Synchronous Breast and Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 667. https://doi.org/10.3390/ijms23020667
Bouras A, Lafaye C, Leone M, Kherraf Z-E, Martin-Denavit T, Fert-Ferrer S, Calender A, Boutry-Kryza N. Identification and Characterization of an Exonic Duplication in PALB2 in a Man with Synchronous Breast and Prostate Cancer. International Journal of Molecular Sciences. 2022; 23(2):667. https://doi.org/10.3390/ijms23020667
Chicago/Turabian StyleBouras, Ahmed, Cyril Lafaye, Melanie Leone, Zine-Eddine Kherraf, Tanguy Martin-Denavit, Sandra Fert-Ferrer, Alain Calender, and Nadia Boutry-Kryza. 2022. "Identification and Characterization of an Exonic Duplication in PALB2 in a Man with Synchronous Breast and Prostate Cancer" International Journal of Molecular Sciences 23, no. 2: 667. https://doi.org/10.3390/ijms23020667
APA StyleBouras, A., Lafaye, C., Leone, M., Kherraf, Z.-E., Martin-Denavit, T., Fert-Ferrer, S., Calender, A., & Boutry-Kryza, N. (2022). Identification and Characterization of an Exonic Duplication in PALB2 in a Man with Synchronous Breast and Prostate Cancer. International Journal of Molecular Sciences, 23(2), 667. https://doi.org/10.3390/ijms23020667







