Characteristics of an In Vitro Mesenteric Lymph Node Cell Suspension Model and Its Possible Association with In Vivo Functional Evaluation
Abstract
:1. Introduction
2. Results
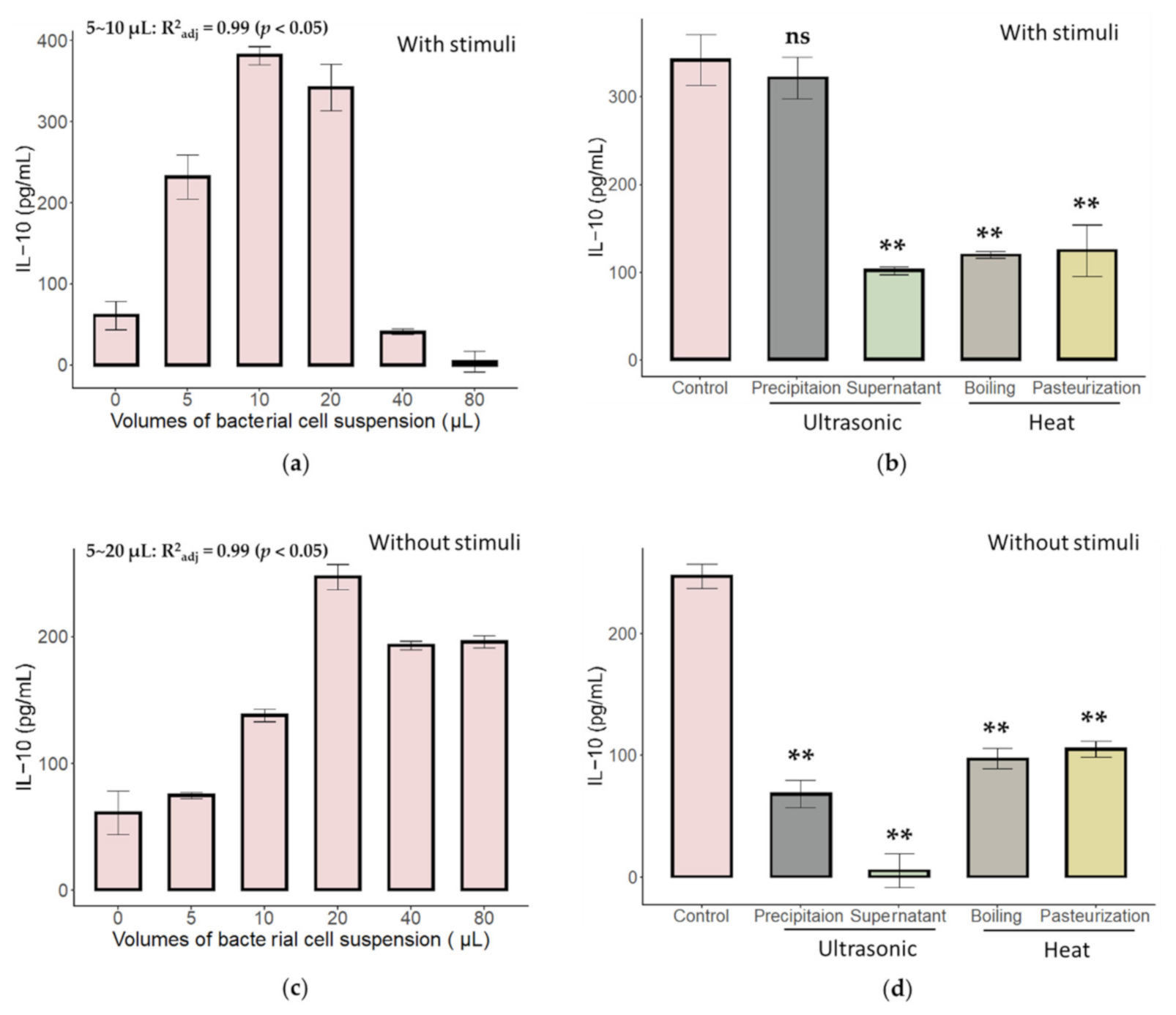
2.1. Characteristics of Immune Activation of C. butyricum and B. bifidum in an In Vitro MLN Model
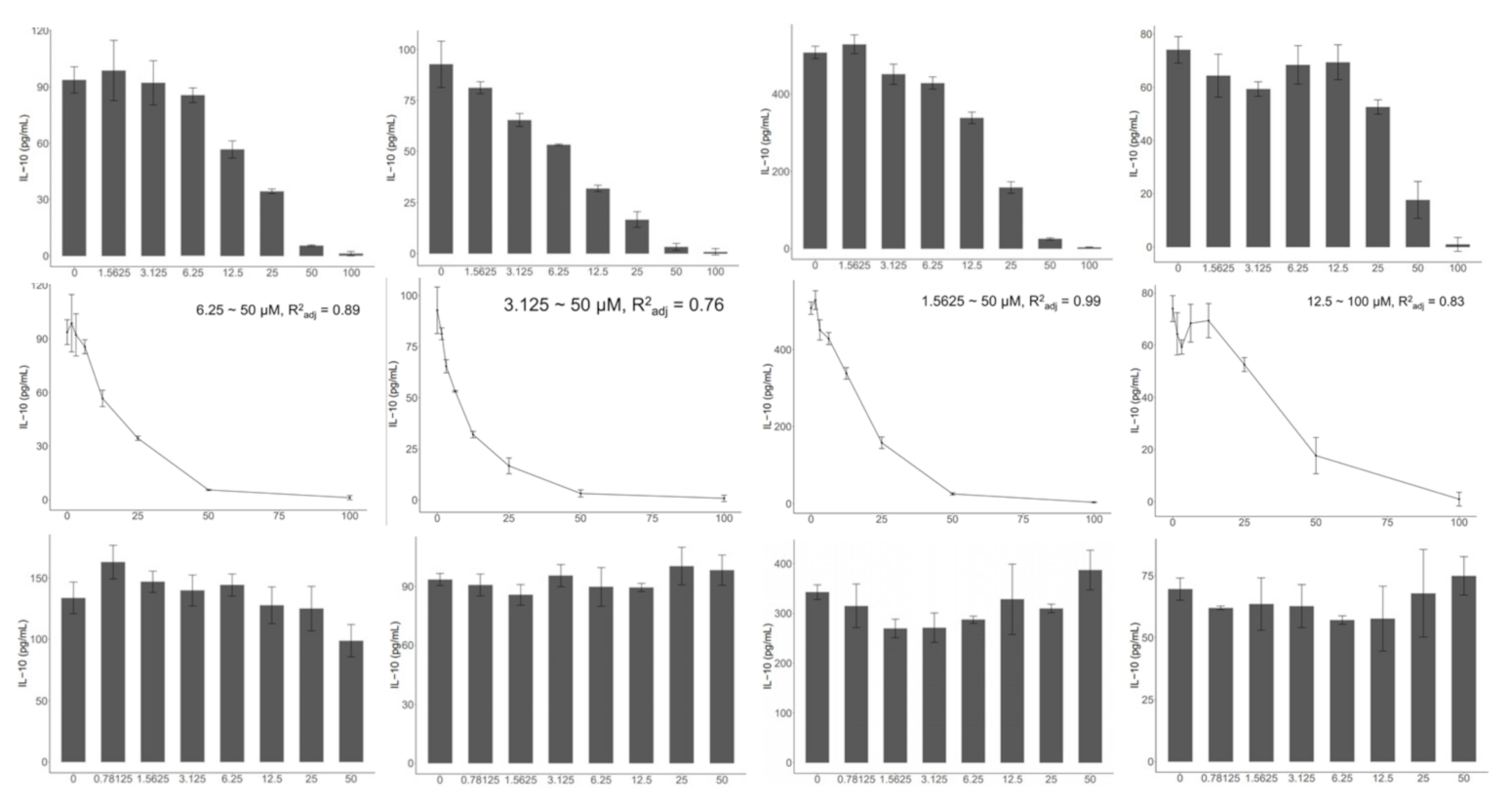
2.2. TLR2, but Not TLR4, Is Indispensable to the Activation of the In Vitro MLN Model
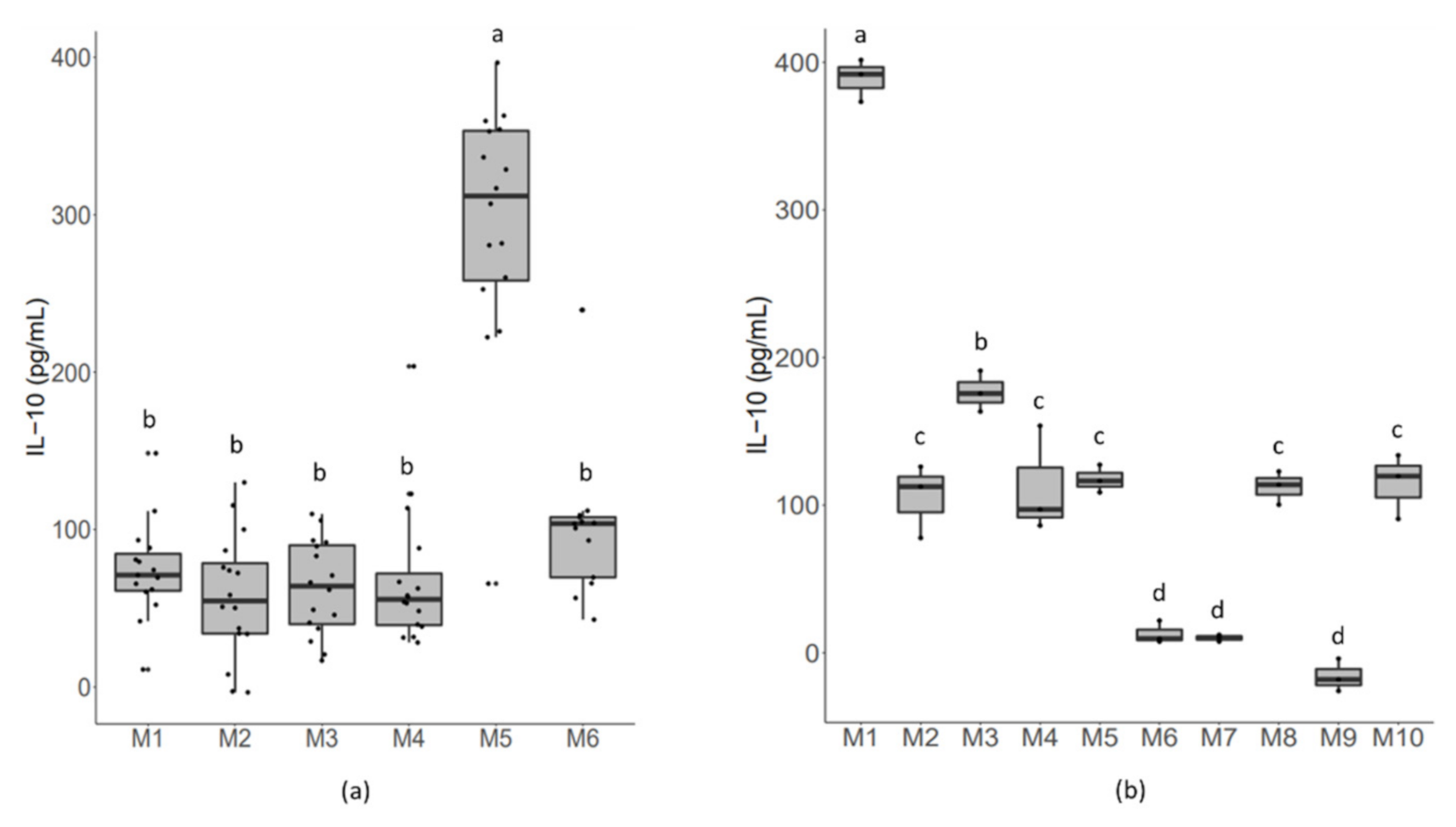
2.3. Cell Resources Exert an Effect on Bacterial-Induced Responsive Strength
2.4. Administering a Mixture of A. muciniphila and C. butyricum Significantly Increased the Mortality of TNBS-Challenged Mice
3. Discussion
4. Materials and Methods
4.1. Strains, Media, and Growth Conditions
4.2. In Vitro MLN Co-Culture Model
4.3. Animal Experiment
4.4. Statistical Analysis and Data Visualization
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Luccia, B.D.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8aa+ T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Qie, Y.; Park, J.; Kim, C.H. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016, 20, 202–214. [Google Scholar] [CrossRef] [Green Version]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef]
- Verma, R.; Lee, C.; Jeun, E.-J.; Yi, J.; Kim, K.S.; Ghosh, A.; Byun, S.; Lee, C.-G.; Kang, H.-J.; Kim, G.-C.; et al. Cell surface polysaccharides of Bifidobacterium bifidum induce the generation of Foxp3+ regulatory T cells. Sci. Immunol. 2018, 3, eaat6975. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, C.; Kujawski, M.; Chu, H.; Li, L.; Mazmanian, S.K.; Cantin, E.M. Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat. Commun. 2019, 10, 2153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuczma, M.P.; Szurek, E.A.; Cebula, A.; Chassaing, B.; Jung, Y.-J.; Kang, S.-M.; Fox, J.G.; Stecher, B.; Ignatowicz, L. Commensal epitopes drive differentiation of colonic Tregs. Sci. Adv. 2020, 6, eaaz3186. [Google Scholar] [CrossRef] [Green Version]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Backhed, F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S.; et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2020, 581, 475–479. [Google Scholar] [CrossRef]
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019, 12, 851–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geuking, M.B.; Cahenzli, J.; Lawson, M.; Ng, D.C.; Slack, E.; Hapfelmeier, S.; McCoy, K.D.; Macpherson, A.J. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 2011, 34, 794–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, S.-Y.; Park, W.-B. Production of cytokine and NO by RAW 264.7 macrophages and PBMC in vitro incubation with Flavonoids. Arch. Pharmacal Res. 2005, 28, 573–581. [Google Scholar] [CrossRef]
- Rinaldi, E.; Consonni, A.; Cordiglieri, C.; Sacco, G.; Crasa, C.; Fontana, A.; Morelli, L.; Elli, M.; Mantegazza, R.; Baggi, F. Therapeutic effect of Bifidobacterium administration on experimental autoimmune myasthenia gravis in lewis rats. Front. Immunol. 2019, 10, 2949. [Google Scholar] [CrossRef] [PubMed]
- Neff, C.P.; Rhodes, M.E.; Arnolds, K.L.; Collins, C.B.; Donnelly, J.; Nusbacher, N.; Jedlicka, P.; Schneider, J.M.; McCarter, M.D.; Shaffer, M.; et al. Diverse intestinal bacteria contain putative zwitterionic capsular polysaccharides with anti-inflammatory properties. Cell Host Microbe 2016, 20, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Kwon, H.K.; Lee, C.G.; So, J.S.; Chae, C.S.; Hwang, J.S.; Sahoo, A.; Nam, J.H.; Rhee, J.H.; Hwang, K.C.; Im, S.H. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. USA 2010, 107, 2159–2164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srutkova, D.; Schwarzer, M.; Hudcovic, T.; Zakostelska, Z.; Drab, V.; Spanova, A.; Rittich, B.; Kozakova, H.; Schabussova, I. Bifidobacterium longum CCM 7952 promotes epithelial barrier function and prevents acute DSS-induced colitis in strictly strain-specific manner. PLoS ONE 2015, 10, e0134050. [Google Scholar] [CrossRef]
- Kim, J.E.; Sharma, A.; Sharma, G.; Lee, S.Y.; Shin, H.S.; Rudra, D.; Im, S.-H. Lactobacillus pentosus modulates immune response by inducing IL-10 producing Tr1 cells. Immune Netw. 2019, 19, e39. [Google Scholar] [CrossRef]
- Li, M.; Wang, B.; Zhang, M.; Rantalainen, M.; Wang, S.; Zhou, H.; Zhang, Y.; Shen, J.; Pang, X.; Zhang, M.; et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc. Natl. Acad. Sci. USA 2008, 105, 2117–2122. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Verma, R.; Byun, S.; Jeun, E.J.; Kim, G.C.; Lee, S.; Kang, H.J.; Kim, C.J.; Sharma, G.; Lahiri, A.; et al. Structural specificities of cell surface beta-glucan polysaccharides determine commensal yeast mediated immuno-modulatory activities. Nat. Commun. 2021, 12, 3611. [Google Scholar] [CrossRef]
- Hofmann, T.; Klenow, S.; Borowicki, A.; Gill, C.I.; Pool-Zobel, B.L.; Glei, M. Gene expression profiles in human peripheral blood mononuclear cells as biomarkers for nutritional in vitro and in vivo investigations. Genes Nutr. 2010, 5, 309–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dillon, S.; Agrawal, S.; Banerjee, K.; Letterio, J.; Denning, T.L.; Oswald-Richter, K.; Kasprowicz, D.J.; Kellar, K.; Pare, J.; van Dyke, T.; et al. Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J. Clin. Investig. 2006, 116, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Steimle, A.; Michaelis, L.; Di Lorenzo, F.; Kliem, T.; Munzner, T.; Maerz, J.K.; Schafer, A.; Lange, A.; Parusel, R.; Gronbach, K.; et al. Weak agonistic LPS restores intestinal immune homeostasis. Mol. Ther. 2019, 27, 1974–1991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottman, N.; Reunanen, J.; Meijerink, M.; Pietila, T.E.; Kainulainen, V.; Klievink, J.; Huuskonen, L.; Aalvink, S.; Skurnik, M.; Boeren, S.; et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS ONE 2017, 12, e0173004. [Google Scholar] [CrossRef]
- Yamashita, M.; Katakura, Y.; Shim, S.-Y.; Matsumoto, S.-E.; Tamura, T.; Morihara, K.; Aiba, Y.; Teruya, K.; Tsuchiya, T.; Shirahata, S. Different individual immune responses elicited by in vitro immunization. Cytotechnology 2002, 40, 161–165. [Google Scholar] [CrossRef]
- Antoniou, E.; Margonis, G.A.; Angelou, A.; Pikouli, A.; Argiri, P.; Karavokyros, I.; Papalois, A.; Pikoulis, E. The TNBS-induced colitis animal model: An overview. Ann. Med. Surg. 2016, 11, 9–15. [Google Scholar] [CrossRef]
- Peran, L.; Camuesco, D.; Comalada, M.; Bailon, E.; Henriksson, A.; Xaus, J.; Zarzuelo, A.; Galvez, J. A comparative study of the preventative effects exerted by three probiotics, Bifidobacterium lactis, Lactobacillus casei and Lactobacillus acidophilus, in the TNBS model of rat colitis. J. Appl. Microbiol. 2007, 103, 836–844. [Google Scholar] [CrossRef]
- Uronis, J.M.; Arthur, J.C.; Keku, T.; Fodor, A.; Carroll, I.M.; Cruz, M.L.; Appleyard, C.B.; Jobin, C. Gut microbial diversity is reduced by the probiotic VSL#3 and correlates with decreased TNBS-induced colitis. Inflamm. Bowel Dis. 2011, 17, 289–297. [Google Scholar]
- Chandhni, P.R.; Pradhan, D.; Sowmya, K.; Gupta, S.; Kadyan, S.; Choudhary, R.; Gupta, A.; Gulati, G.; Mallappa, R.H.; Kaushik, J.K.; et al. Ameliorative effect of surface proteins of probiotic lactobacilli in colitis mouse models. Front. Microbiol. 2021, 12, 679773. [Google Scholar] [CrossRef]
- Ozaki, H.; Kinoshita, K.; Hori, M. Mechanism of the intestinal dysmotility in the inflammatory bowel diseases: Possible involvement of muscularis resident macrophages. Nihon Yakurigaku Zasshi. Folia Pharmacol. Jpn. 2003, 122, 46–50. [Google Scholar]
- Han, K.; Singh, K.; Rodman, M.J.; Hassanzadeh, S.; Wu, K.; Nguyen, A.; Huffstutler, R.D.; Seifuddin, F.; Dagur, P.K.; Saxena, A.; et al. Fasting-induced FOXO4 blunts human CD4(+) T helper cell responsiveness. Nat. Metab. 2021, 3, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Ansaldo, E.; Slayden, L.C.; Ching, K.L.; Koch, M.A.; Wolf, N.K.; Plichta, D.R.; Brown, E.M.; Graham, D.B.; Xavier, R.J.; Moon, J.J.; et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 2019, 364, 1179–1184. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Chatelier, E.L.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meroni, E.; Stakenborg, N.; Gomez-Pinilla, P.J.; De Hertogh, G.; Goverse, G.; Matteoli, G.; Verheijden, S.; Boeckxstaens, G.E. Functional characterization of oxazolone-induced colitis and survival improvement by vagus nerve stimulation. PLoS ONE 2018, 13, e0197487. [Google Scholar] [CrossRef]
- Skelly, A.N.; Sato, Y.; Kearney, S.; Honda, K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 2019, 19, 305–323. [Google Scholar] [CrossRef]
- Mistry, P.; Laird, M.H.; Schwarz, R.S.; Greene, S.; Dyson, T.; Snyder, G.A.; Xiao, T.S.; Chauhan, J.; Fletcher, S.; Toshchakov, V.Y.; et al. Inhibition of TLR2 signaling by small molecule inhibitors targeting a pocket within the TLR2 TIR domain. Proc. Natl. Acad. Sci. USA 2015, 112, 5455–5460. [Google Scholar] [CrossRef] [Green Version]
- Ono, Y.; Maejima, Y.; Saito, M.; Sakamoto, K.; Horita, S.; Shimomura, K.; Inoue, S.; Kotani, J. TAK-242, a specific inhibitor of Toll-like receptor 4 signalling, prevents endotoxemia-induced skeletal muscle wasting in mice. Sci. Rep. 2020, 10, 694. [Google Scholar] [CrossRef] [Green Version]
- Kassambara, A.; Kosinski, M.; Biecek, P.; Fabian, S.; Survminer: Drawing Survival Curves Using ‘ggplot2’. R Packages. Available online: http://CRAN.R-project.org/package=survminer (accessed on 17 October 2021).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, S.; Li, J.; Qu, D.; Tian, F.; Yu, L.; Zhang, H.; Chen, W.; Zhao, J.; Zhai, Q. Characteristics of an In Vitro Mesenteric Lymph Node Cell Suspension Model and Its Possible Association with In Vivo Functional Evaluation. Int. J. Mol. Sci. 2022, 23, 1003. https://doi.org/10.3390/ijms23021003
Feng S, Li J, Qu D, Tian F, Yu L, Zhang H, Chen W, Zhao J, Zhai Q. Characteristics of an In Vitro Mesenteric Lymph Node Cell Suspension Model and Its Possible Association with In Vivo Functional Evaluation. International Journal of Molecular Sciences. 2022; 23(2):1003. https://doi.org/10.3390/ijms23021003
Chicago/Turabian StyleFeng, Saisai, Jing Li, Dingwu Qu, Fengwei Tian, Leilei Yu, Hao Zhang, Wei Chen, Jianxin Zhao, and Qixiao Zhai. 2022. "Characteristics of an In Vitro Mesenteric Lymph Node Cell Suspension Model and Its Possible Association with In Vivo Functional Evaluation" International Journal of Molecular Sciences 23, no. 2: 1003. https://doi.org/10.3390/ijms23021003
APA StyleFeng, S., Li, J., Qu, D., Tian, F., Yu, L., Zhang, H., Chen, W., Zhao, J., & Zhai, Q. (2022). Characteristics of an In Vitro Mesenteric Lymph Node Cell Suspension Model and Its Possible Association with In Vivo Functional Evaluation. International Journal of Molecular Sciences, 23(2), 1003. https://doi.org/10.3390/ijms23021003





