α-CAs from Photosynthetic Organisms
Abstract
1. Introduction
2. Higher Plants
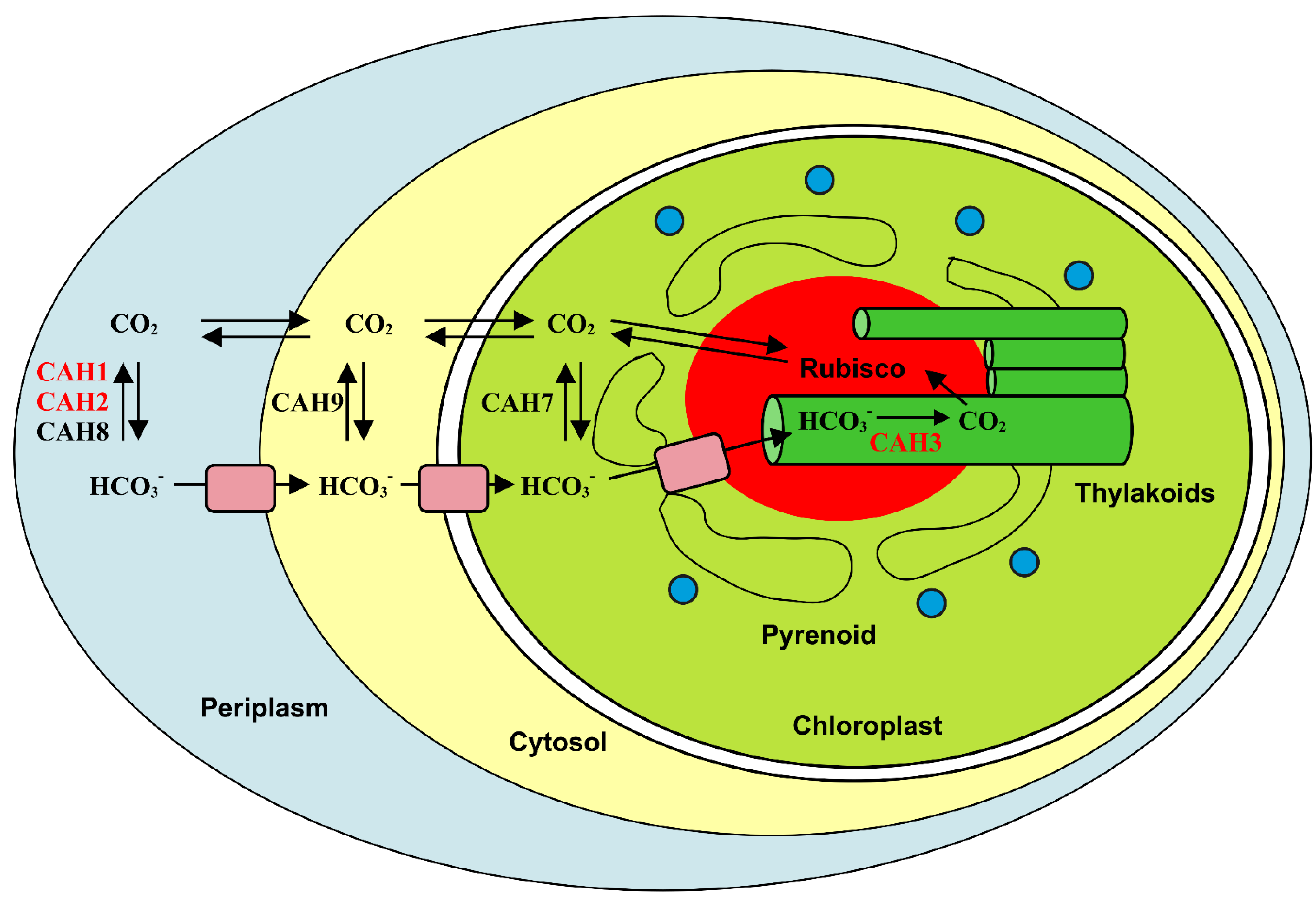
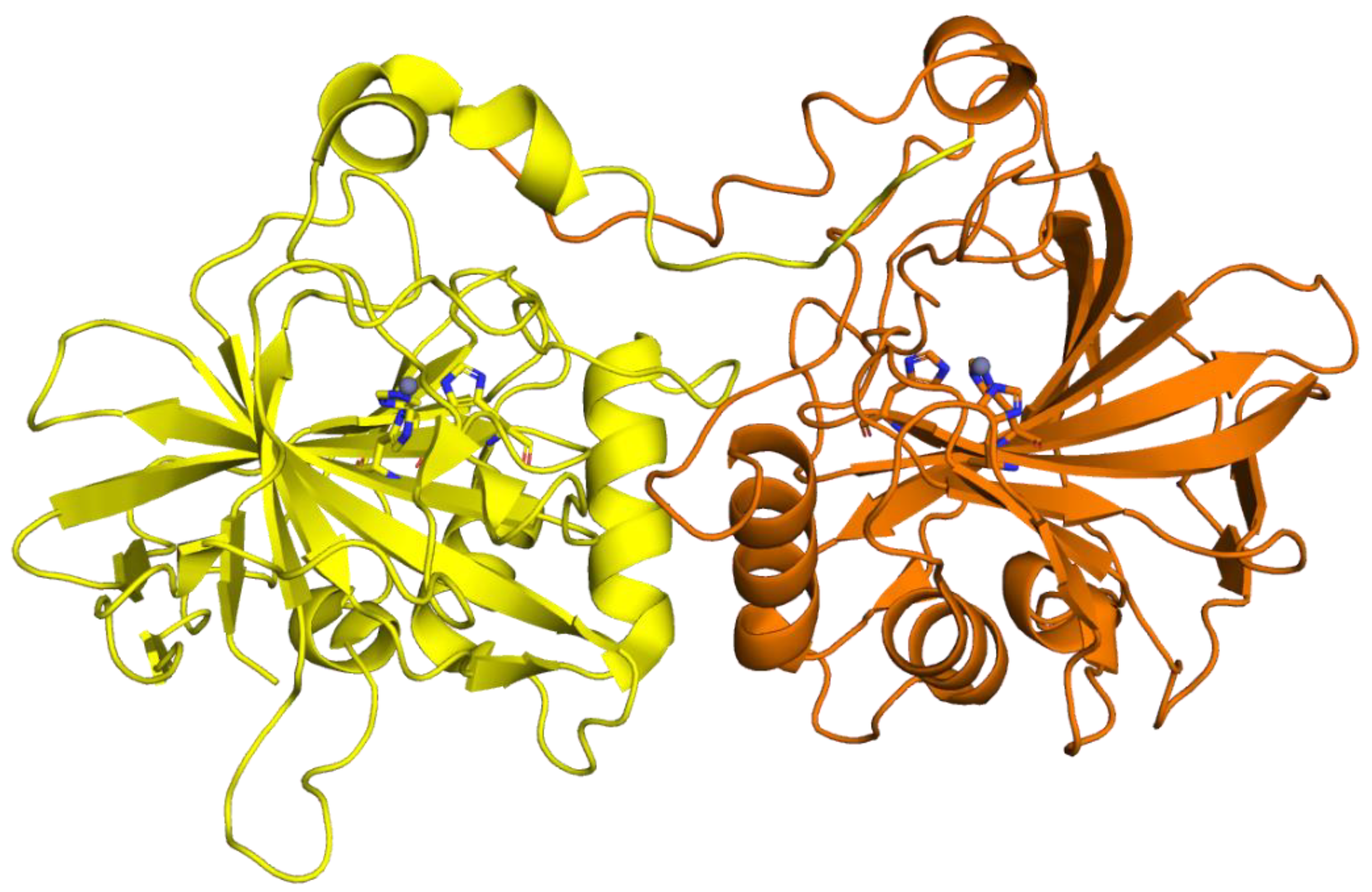
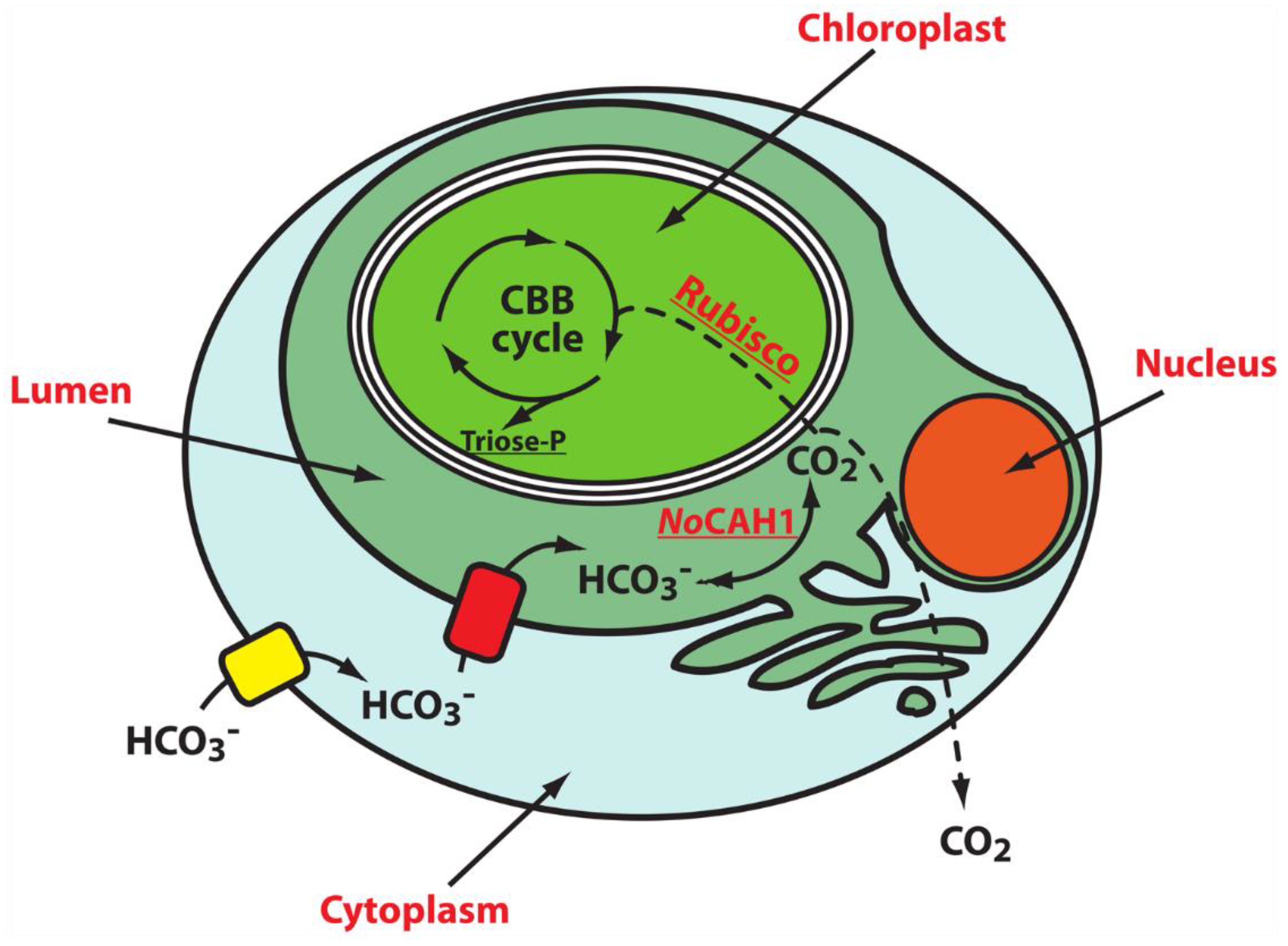
3. Algae
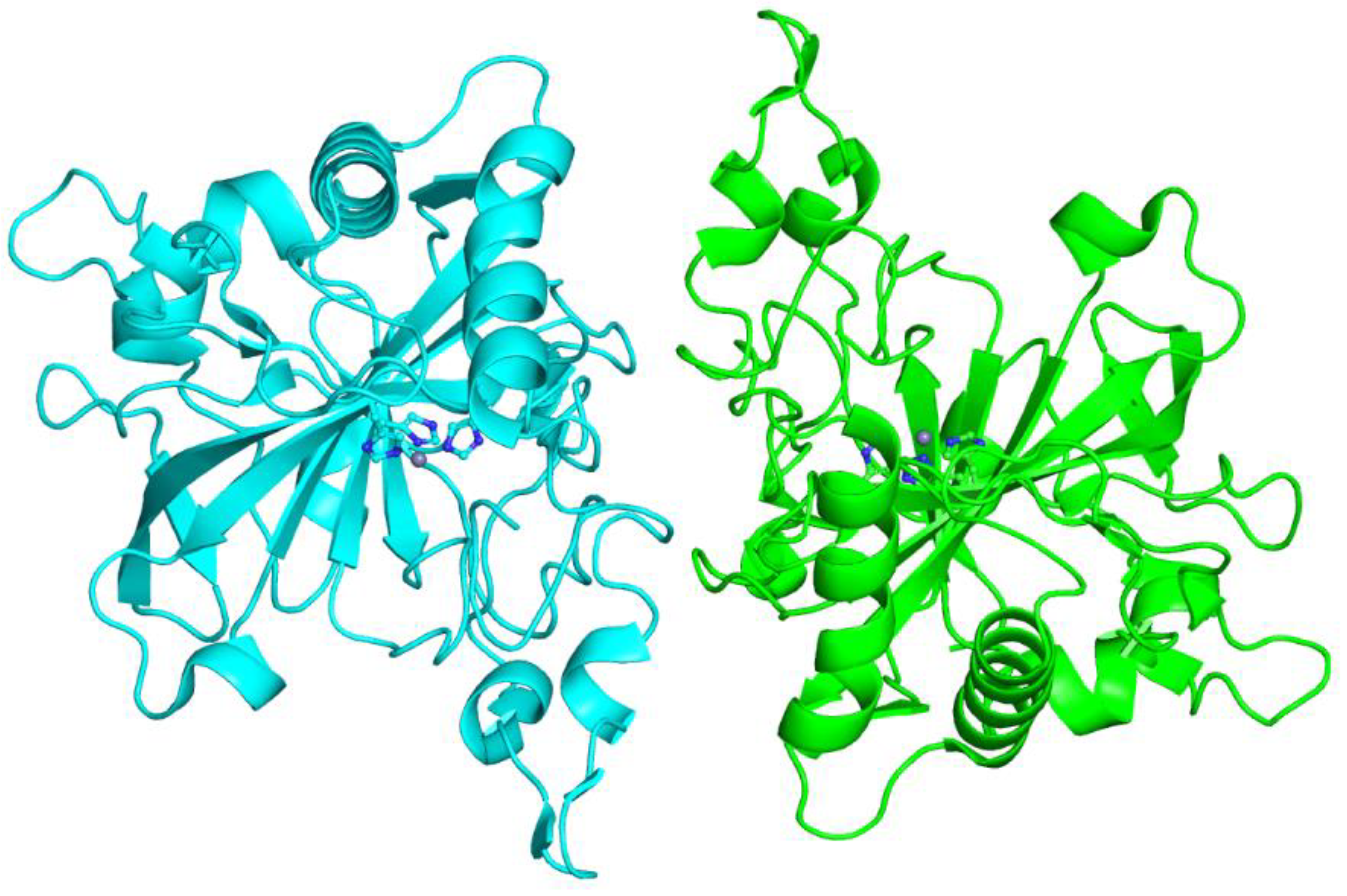
4. Cyanobacteria
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Monti, D.M.; De Simone, G.; Langella, E.; Supuran, C.T.; Di Fiore, A.; Monti, S.M. Insights into the Role of Reactive Sulfhydryl Groups of Carbonic Anhydrase III and VII during Oxidative Damage. J. Enzym. Inhib. Med. Chem. 2017, 32, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Monti, D.M.; Scaloni, A.; De Simone, G.; Monti, S.M. Protective Role of Carbonic Anhydrases III and VII in Cellular Defense Mechanisms upon Redox Unbalance. Oxid. Med. Cell. Longev. 2018, 2018, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Buonanno, M.; Langella, E.; Zambrano, N.; Succoio, M.; Sasso, E.; Alterio, V.; Di Fiore, A.; Sandomenico, A.; Supuran, C.T.; Scaloni, A.; et al. Disclosing the Interaction of Carbonic Anhydrase IX with Cullin-Associated NEDD8-Dissociated Protein 1 by Molecular Modeling and Integrated Binding Measurements. ACS Chem. Biol. 2017, 12, 1460–1465. [Google Scholar] [CrossRef] [PubMed]
- Buanne, P.; Renzone, G.; Monteleone, F.; Vitale, M.; Monti, S.M.; Sandomenico, A.M.; Garbi, C.; Montanaro, D.; Accardo, M.; Troncone, G.; et al. Characterization of Carbonic Anhydrase IX Interactome Reveals Proteins Assisting Its Nuclear Localization in Hypoxic Cells. J. Proteome Res. 2013, 12, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Lionetto, M.G.; Caricato, R.; Giordano, M.E.; Schettino, T. The Complex Relationship between Metals and Carbonic Anhydrase: New Insights and Perspectives. Int. J. Mol. Sci. 2016, 17, 127. [Google Scholar] [CrossRef]
- DiMario, R.J.; Machingura, M.C.; Waldrop, G.L.; Moroney, J.V. The Many Types of Carbonic Anhydrases in Photosynthetic Organisms. Plant Sci. 2018, 268, 11–17. [Google Scholar] [CrossRef]
- Alterio, V.; Di Fiore, A.; D’Ambrosio, K.; Supuran, C.T.; De Simone, G. Multiple Binding Modes of Inhibitors to Carbonic Anhydrases: How to Design Specific Drugs Targeting 15 Different Isoforms? Chem. Rev. 2012, 112, 4421–4468. [Google Scholar] [CrossRef]
- DiMario, R.J.; Clayton, H.; Mukherjee, A.; Ludwig, M.; Moroney, J.V. Plant Carbonic Anhydrases: Structures, Locations, Evolution, and Physiological Roles. Mol. Plant 2017, 10, 30–46. [Google Scholar] [CrossRef]
- Monti, S.M.; Supuran, C.T.; De Simone, G. Carbonic Anhydrase IX as a Target for Designing Novel Anticancer Drugs. Curr. Med. Chem. 2012, 19, 821–830. [Google Scholar] [CrossRef]
- Esbaugh, A.J.; Tufts, B.L. The Structure and Function of Carbonic Anhydrase Isozymes in the Respiratory System of Vertebrates. Respir. Physiol. Neurobiol. 2006, 154, 185–198. [Google Scholar] [CrossRef]
- Rowlett, R.S. Structure and Catalytic Mechanism of the Beta-Carbonic Anhydrases. Biochim. Biophys. Acta 2010, 1804, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Dathan, N.A.; Alterio, V.; Troiano, E.; Vullo, D.; Ludwig, M.; De Simone, G.; Supuran, C.T.; Monti, S.M. Biochemical Characterization of the Chloroplastic β-Carbonic Anhydrase from Flaveria Bidentis (L.) “Kuntze”. J. Enzym. Inhib. Med. Chem. 2014, 29, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Kimber, M.; Pai, E. The Active Site Architecture of Pisum Sativum Beta-Carbonic Anhydrase Is a Mirror Image of That of Alpha-Carbonic Anhydrases. EMBO J. 2000, 19, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.M.; De Simone, G.; Dathan, N.A.; Ludwig, M.; Vullo, D.; Scozzafava, A.; Capasso, C.; Supuran, C.T. Kinetic and Anion Inhibition Studies of a β-Carbonic Anhydrase (FbiCA 1) from the C4 Plant Flaveria Bidentis. Bioorg. Med. Chem. Lett. 2013, 23, 1626–1630. [Google Scholar] [CrossRef]
- Parisi, G.; Perales, M.; Fornasari, M.S.; Colaneri, A.; González-Schain, N.; Gómez-Casati, D.; Zimmermann, S.; Brennicke, A.; Araya, A.; Ferry, J.G.; et al. Gamma Carbonic Anhydrases in Plant Mitochondria. Plant Mol. Biol. 2004, 55, 193–207. [Google Scholar] [CrossRef]
- Cox, E.H.; McLendon, G.L.; Morel, F.M.M.; Lane, T.W.; Prince, R.C.; Pickering, I.J.; George, G.N. The Active Site Structure of Thalassiosira Weissflogii Carbonic Anhydrase 1. Biochemistry 2000, 39, 12128–12130. [Google Scholar] [CrossRef]
- Lane, T.W.; Morel, F.M.M. Regulation of Carbonic Anhydrase Expression by Zinc, Cobalt, and Carbon Dioxide in the Marine Diatom Thalassiosira Weissflogii. Plant Physiol. 2000, 123, 345–352. [Google Scholar] [CrossRef]
- Alterio, V.; Langella, E.; De Simone, G.; Monti, S.M. Cadmium-Containing Carbonic Anhydrase CDCA1 in Marine Diatom Thalassiosira Weissflogii. Mar. Drugs 2015, 13, 1688–1697. [Google Scholar] [CrossRef]
- Angeli, A.; Buonanno, M.; Donald, W.A.; Monti, S.M.; Supuran, C.T. The Zinc—but Not Cadmium—Containing ζ-Carbonic from the Diatom Thalassiosira Weissflogii Is Potently Activated by Amines and Amino Acids. Bioorg. Chem. 2018, 80, 261–265. [Google Scholar] [CrossRef]
- Viparelli, F.; Monti, S.M.; Simone, G.D..; Innocenti, A.; Scozzafava, A.; Xu, Y.; Morel, F.M.M.; Supuran, C.T. Inhibition of the R1 Fragment of the Cadmium-Containing ζ-Class Carbonic Anhydrase from the Diatom Thalassiosira Weissflogii with Anions. Bioorg. Med. Chem. Lett. 2010, 20, 4745–4748. [Google Scholar] [CrossRef]
- Alterio, V.; Langella, E.; Buonanno, M.; Esposito, D.; Nocentini, A.; Berrino, E.; Bua, S.; Polentarutti, M.; Supuran, C.T.; Monti, S.M.; et al. Zeta-Carbonic Anhydrases Show CS2 Hydrolase Activity: A New Metabolic Carbon Acquisition Pathway in Diatoms? Comput. Struct. Biotechnol. J. 2021, 19, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Fisher, G.M.; Andrews, K.T.; Poulsen, S.A.; Capasso, C.; Supuran, C.T. Discovery of a New Family of Carbonic Anhydrases in the Malaria Pathogen Plasmodium Falciparum--the η-Carbonic Anhydrases. Bioorg. Med. Chem. Lett. 2014, 24, 4389–4396. [Google Scholar] [CrossRef] [PubMed]
- Kikutani, S.; Nakajima, K.; Nagasato, C.; Tsuji, Y.; Miyatake, A.; Matsuda, Y. Thylakoid Luminal Θ-Carbonic Anhydrase Critical for Growth and Photosynthesis in the Marine Diatom Phaeodactylum Tricornutum. Proc. Natl. Acad. Sci. USA 2016, 113, 9828–9833. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E.L.; Clement, R.; Kosta, A.; Maberly, S.C.; Gontero, B. A New Widespread Subclass of Carbonic Anhydrase in Marine Phytoplankton. ISME J. 2019, 13, 2094–2106. [Google Scholar] [CrossRef] [PubMed]
- Nocentini, A.; Supuran, C.T.; Capasso, C. An Overview on the Recently Discovered Iota-Carbonic Anhydrases. J. Enzym. Inhib. Med. Chem. 2021, 36, 1988–1995. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; D’Ambrosio, K.; Ayoub, J.; Alterio, V.; De Simone, G. Chapter 2—α-Carbonic Anhydrases. In Carbonic Anhydrases; Supuran, C.T., Nocentini, A., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 19–54. ISBN 978-0-12-816476-1. [Google Scholar]
- D’Ambrosio, K.; Smaine, F.Z.; Carta, F.; De Simone, G.; Winum, J.Y.; Supuran, C.T. Development of Potent Carbonic Anhydrase Inhibitors Incorporating Both Sulfonamide and Sulfamide Groups. J. Med. Chem. 2012, 55, 6776–6783. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Truppo, E.; Supuran, C.T.; Alterio, V.; Dathan, N.; Bootorabi, F.; Parkkila, S.; Monti, S.M.; Simone, G. De Crystal Structure of the C183S/C217S Mutant of Human CA VII in Complex with Acetazolamide. Bioorg. Med. Chem. Lett. 2010, 20, 5023–5026. [Google Scholar] [CrossRef]
- Alterio, V.; De Simone, G.; Monti, S.M.; Scozzafava, A.; Supuran, C.T. Carbonic Anhydrase Inhibitors: Inhibition of Human, Bacterial, and Archaeal Isozymes with Benzene-1,3-Disulfonamides--Solution and Crystallographic Studies. Bioorg. Med. Chem. Lett. 2007, 17, 4201–4207. [Google Scholar] [CrossRef]
- Langella, E.; Buonanno, M.; Vullo, D.; Dathan, N.; Leone, M.; Supuran, C.T.; De Simone, G.; Monti, S.M. Biochemical, Biophysical and Molecular Dynamics Studies on the Proteoglycan-like Domain of Carbonic Anhydrase IX. Cell. Mol. Life Sci. 2018 7517 2018, 75, 3283–3296. [Google Scholar] [CrossRef]
- Moroney, J.V.; Bartlett, S.G.; Samuelsson, G. Carbonic Anhydrases in Plants and Algae. Plant. Cell Environ. 2001, 24, 141–153. [Google Scholar] [CrossRef]
- Wang, Y.; Stessman, D.J.; Spalding, M.H. The CO2 Concentrating Mechanism and Photosynthetic Carbon Assimilation in Limiting CO2: How Chlamydomonas Works against the Gradient. Plant J. 2015, 82, 429–448. [Google Scholar] [CrossRef] [PubMed]
- Tomar, V.; Sidhu, G.K.; Nogia, P.; Mehrotra, R.; Mehrotra, S. Regulatory Components of Carbon Concentrating Mechanisms in Aquatic Unicellular Photosynthetic Organisms. Plant Cell Rep. 2017, 36, 1671–1688. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, Y.; Nakajima, K.; Matsuda, Y. Molecular Aspects of the Biophysical CO2-Concentrating Mechanism and Its Regulation in Marine Diatoms. J. Exp. Bot. 2017, 68, 3763–3772. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Price, G.D. CO2 Concentrating Mechanisms in Cyanobacteria: Molecular Components, Their Diversity and Evolution. J. Exp. Bot. 2003, 54, 609–622. [Google Scholar] [CrossRef]
- Gee, C.W.; Niyogi, K.K. The Carbonic Anhydrase CAH1 Is an Essential Component of the Carbon-Concentrating Mechanism in Nannochloropsis Oceanica. Proc. Natl. Acad. Sci. USA 2017, 114, 4537–4542. [Google Scholar] [CrossRef]
- Ogren, W.L. Photorespiration: Pathways, Regulation, and Modification. Annu. Rev. Plant Physiol. 1984, 35, 415–442. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Fernie, A.R. Photorespiration: Players, Partners and Origin. Trends Plant Sci. 2010, 15, 330–336. [Google Scholar] [CrossRef]
- Badger, M. The Roles of Carbonic Anhydrases in Photosynthetic CO2 Concentrating Mechanisms. Photosynth. Res. 2003, 77, 83–94. [Google Scholar] [CrossRef]
- So, A.K.C.; Van Spall, H.G.C.; Coleman, J.R.; Espie, G.S. Catalytic Exchange of 18O from 13C18O-Labelled CO2 by Wild-Type Cells and EcaA, EcaB, and CcaA Mutants of the Cyanobacteria Synechococcus PCC7942 and Synechocystis PCC6803. Can. J. Bot. 1998, 76, 1153–1160. [Google Scholar] [CrossRef]
- Soltes-Rak, E.; Mulligan, M.E.; Coleman, J.R. Identification and Characterization of a Gene Encoding a Vertebrate-Type Carbonic Anhydrase in Cyanobacteria. J. Bacteriol. 1997, 179, 769. [Google Scholar] [CrossRef]
- Smith, K.S.; Ferry, J.G. Prokaryotic Carbonic Anhydrases. FEMS Microbiol. Rev. 2000, 24, 335–366. [Google Scholar] [CrossRef] [PubMed]
- Langella, E.; Alterio, V.; D’Ambrosio, K.; Cadoni, R.; Winum, J.Y.; Supuran, C.T.; Monti, S.M.; De Simone, G.; Di Fiore, A. Exploring Benzoxaborole Derivatives as Carbonic Anhydrase Inhibitors: A Structural and Computational Analysis Reveals Their Conformational Variability as a Tool to Increase Enzyme Selectivity. J. Enzym. Inhib. Med. Chem. 2019, 34, 1498–1505. [Google Scholar] [CrossRef]
- De Simone, G.; Langella, E.; Esposito, D.; Supuran, C.T.; Monti, S.M.; Winum, J.Y.; Alterio, V. Insights into the Binding Mode of Sulphamates and Sulphamides to HCA II: Crystallographic Studies and Binding Free Energy Calculations. J. Enzym. Inhib. Med. Chem. 2017, 32, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- Momayyezi, M.; McKown, A.D.; Bell, S.C.S.; Guy, R.D. Emerging Roles for Carbonic Anhydrase in Mesophyll Conductance and Photosynthesis. Plant J. 2020, 101, 831–844. [Google Scholar] [CrossRef]
- Villarejo, A.; Burén, S.; Larsson, S.; Déjardin, A.; Monné, M.; Rudhe, C.; Karlsson, J.; Jansson, S.; Lerouge, P.; Rolland, N.; et al. Evidence for a Protein Transported through the Secretory Pathway En Route to the Higher Plant Chloroplast. Nat. Cell Biol. 2005, 7, 1224–1231. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Rivero, A.; Shutova, T.; Román, M.J.; Villarejo, A.; Martinez, F. Phosphorylation Controls the Localization and Activation of the Lumenal Carbonic Anhydrase in Chlamydomonas Reinhardtii. PLoS ONE 2012, 7, e49063. [Google Scholar] [CrossRef]
- Rudenko, N.N.; Ignatova, L.K.; Fedorchuk, T.P.; Ivanov, B.N. Carbonic Anhydrases in Photosynthetic Cells of Higher Plants. Biochemistry. (Mosc). 2015, 80, 674–687. [Google Scholar] [CrossRef]
- Burén, S.; Ortega-Villasante, C.; Blanco-Rivero, A.; Martínez-Bernardini, A.; Shutova, T.; Shevela, D.; Messinger, J.; Bako, L.; Villarejo, A.; Samuelsson, G. Importance of Post-Translational Modifications for Functionality of a Chloroplast-Localized Carbonic Anhydrase (CAH1) in Arabidopsis Thaliana. PLoS ONE 2011, 6, e21021. [Google Scholar] [CrossRef]
- Friso, G.; Giacomelli, L.; Ytterberg, A.J.; Peltier, J.B.; Rudella, A.; Sun, Q.; Van Wijk, K.J. In-Depth Analysis of the Thylakoid Membrane Proteome of Arabidopsis Thaliana Chloroplasts: New Proteins, New Functions, and a Plastid Proteome Database. Plant Cell 2004, 16, 478–499. [Google Scholar] [CrossRef]
- Fedorchuk, T.P.; Opanasenko, V.K.; Rudenko, N.N.; Ivanov, B.N. Bicarbonate-Induced Stimulation of Photophosphorylation in Isolated Thylakoids: Effects of Carbonic Anhydrase Inhibitors. Biol. Membr. 2018, 35, 34–41. [Google Scholar] [CrossRef]
- Fedorchuk, T.P.; Kireeva, I.A.; Opanasenko, V.K.; Terentyev, V.V.; Rudenko, N.N.; Borisova-Mubarakshina, M.M.; Ivanov, B.N. Alpha Carbonic Anhydrase 5 Mediates Stimulation of ATP Synthesis by Bicarbonate in Isolated Arabidopsis Thylakoids. Front. Plant Sci. 2021, 12, 891. [Google Scholar] [CrossRef]
- Zhurikova, E.M.; Ignatova, L.K.; Rudenko, N.N.; Mudrik, V.A.; Vetoshkina, D.V.; Ivanov, B.N. Participation of Two Carbonic Anhydrases of the Alpha Family in Photosynthetic Reactions in Arabidopsis Thaliana. Biochemistry. (Mosc). 2016, 81, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.N.; Ignatova, L.K.; Nadeeva-Zhurikova, E.M.; Fedorchuk, T.P.; Ivanov, B.N.; Borisova-Mubarakshina, M.M. Advances in Understanding the Physiological Role and Locations of Carbonic Anhydrases in C3 Plant Cells. Protoplasma 2021, 258, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, N.N.; Ivanov, B.N. Unsolved Problems of Carbonic Anhydrases Functioning in Photosynthetic Cells of Higher C3 Plants. Biochemistry. (Mosc). 2021, 86, 1243–1255. [Google Scholar] [CrossRef] [PubMed]
- Burén, S. Targeting and Function of CAH1-Characterization of a Novel Protein Pathway to the Plant Cell Chloroplast; Umeå University: Umeå, Sweden, 2010. [Google Scholar]
- Rudenko, N.N.; Vetoshkina, D.V.; Fedorchuk, T.P.; Ivanov, B.N. Effect of Light Intensity under Different Photoperiods on Expression Level of Carbonic Anhydrase Genes of the α- and β-Families in Arabidopsis Thaliana Leaves. Biochemistry. (Mosc). 2017, 82, 1025–1035. [Google Scholar] [CrossRef]
- Rudenko, N.N.; Fedorchuk, T.P.; Vetoshkina, D.V.; Zhurikova, E.M.; Ignatova, L.K.; Ivanov, B.N. Influence of Knockout of At4g20990 Gene Encoding α-CA4 on Photosystem II Light-Harvesting Antenna in Plants Grown under Different Light Intensities and Day Lengths. Protoplasma 2018, 255, 69–78. [Google Scholar] [CrossRef]
- Ignatova, L.; Zhurikova, E.; Ivanov, B. The Presence of the Low Molecular Mass Carbonic Anhydrase in Photosystem II of C3 Higher Plants. J. Plant Physiol. 2019, 232, 94–99. [Google Scholar] [CrossRef]
- Makita, Y.; Shimada, S.; Kawashima, M.; Kondou-Kuriyama, T.; Toyoda, T.; Matsui, M. MOROKOSHI: Transcriptome Database in Sorghum Bicolor. Plant Cell Physiol. 2015, 56, e6. [Google Scholar] [CrossRef]
- Tang, H.; Krishnakumar, V.; Bidwell, S.; Rosen, B.; Chan, A.; Zhou, S.; Gentzbittel, L.; Childs, K.L.; Yandell, M.; Gundlach, H.; et al. An Improved Genome Release (Version Mt4.0) for the Model Legume Medicago Truncatula. BMC Genom. 2014, 15, 1–14. [Google Scholar] [CrossRef]
- Moroney, J.V.; Ma, Y.; Frey, W.D.; Fusilier, K.A.; Pham, T.T.; Simms, T.A.; DiMario, R.J.; Yang, J.; Mukherjee, B. The Carbonic Anhydrase Isoforms of Chlamydomonas Reinhardtii: Intracellular Location, Expression, and Physiological Roles. Photosynth. Res. 2011, 109, 133–149. [Google Scholar] [CrossRef]
- Aspatwar, A.; Haapanen, S.; Parkkila, S. An Update on the Metabolic Roles of Carbonic Anhydrases in the Model Alga Chlamydomonas Reinhardtii. Metabolites 2018, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Allen, A.E.; Kikutani, S.; Endo, Y.; Bowler, C.; Matsuda, Y. Localization of Putative Carbonic Anhydrases in Two Marine Diatoms, Phaeodactylum Tricornutum and Thalassiosira Pseudonana. Photosynth. Res. 2011, 109, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Samukawa, M.; Shen, C.; Hopkinson, B.M.; Matsuda, Y. Localization of Putative Carbonic Anhydrases in the Marine Diatom, Thalassiosira Pseudonana. Photosynth. Res. 2014, 121, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Coleman, J.R.; Berry, J.A.; Togasaki, R.K.; Grossman, A.R. Identification of Extracellular Carbonic Anhydrase of Chlamydomonas Reinhardtii. Plant Physiol. 1984, 76, 472–477. [Google Scholar] [CrossRef]
- Yang, S.Y.; Tsuzuki, M.; Miyachi, S. Carbonic Anhydrase of Chlamydomonas: Purification and Studies on Its Induction Using Antiserum against Chlamydomonas Carbonic Anhydrase. Plant Cell Physiol. 1985, 26, 25–34. [Google Scholar] [CrossRef]
- Fukuzawa, H.; Fujiwara, S.; Yamamoto, Y.; Dionisio-Sese, M.L.; Miyachi, S. CDNA Cloning, Sequence, and Expression of Carbonic Anhydrase in Chlamydomonas Reinhardtii: Regulation by Environmental CO2 Concentration. Proc. Natl. Acad. Sci. USA 1990, 87, 4383–4387. [Google Scholar] [CrossRef]
- Fujiwara, S.; Fukuzawa, H.; Tachiki, A.; Miyachi, S. Structure and Differential Expression of Two Genes Encoding Carbonic Anhydrase in Chlamydomonas Reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 9779–9783. [Google Scholar] [CrossRef]
- Ishida, S.; Muto, S.; Miyachi, S. Structural Analysis of Periplasmic Carbonic Anhydrase 1 of Chlamydomonas Reinhardtii. Eur. J. Biochem. 1993, 214, 9–16. [Google Scholar] [CrossRef]
- Shutova, T.; Kenneweg, H.; Buchta, J.; Nikitina, J.; Terentyev, V.; Chernyshov, S.; Andersson, B.; Allakhverdiev, S.I.; Klimov, V.V.; Dau, H.; et al. The Photosystem II-Associated Cah3 in Chlamydomonas Enhances the O2 Evolution Rate by Proton Removal. EMBO J. 2008, 27, 782–791. [Google Scholar] [CrossRef]
- Küken, A.; Sommer, F.; Yaneva-Roder, L.; Mackinder, L.C.M.; Höhne, M.; Geimer, S.; Jonikas, M.C.; Schroda, M.; Stitt, M.; Nikoloski, Z.; et al. Effects of Microcompartmentation on Flux Distribution and Metabolic Pools in Chlamydomonas Reinhardtii Chloroplasts. Elife 2018, 7, e37960. [Google Scholar] [CrossRef]
- Yagawa, Y.; Aizawa, K.; Yang, S.Y.; Miyachi, S. Release of Carbonic Anhydrase from the Cell Surface of Chlamydomonas Reinhardtii by Trypsin. Plant Cell Physiol. 1986, 27, 215–221. [Google Scholar] [CrossRef]
- Kimpel, D.L.; Togasaki, R.K.; Miyachi, S. Carbonic Anhydrase in Chlamydomonas Reinhardtii. I. Localization. Plant Cell Physiol. 1983, 24, 255–259. [Google Scholar] [CrossRef]
- Karlsson, J.; Ciarke, A.K.; Chen, Z.Y.; Hugghins, S.Y.; Park, Y.I.; Husic, H.D.; Moroney, J.V.; Samuelsson, G. A Novel Alpha-Type Carbonic Anhydrase Associated with the Thylakoid Membrane in Chlamydomonas Reinhardtii Is Required for Growth at Ambient CO2. EMBO J. 1998, 17, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Hiltonen, T.; Husic, H.D.; Ramazanov, Z.; Samuelsson, G. Intracellular Carbonic Anhydrase of Chlamydomonas Reinhardtii. Plant Physiol. 1995, 109, 533–539. [Google Scholar] [CrossRef]
- Badger, M.R.; Kaplan, A.; Berry, J.A. Internal Inorganic Carbon Pool of Chlamydomonas Reinhardtii. Plant Physiol. 1980, 66, 407–413. [Google Scholar] [CrossRef]
- Moroney, J.V.; Husic, H.D.; Tolbert, N.E. Effect of Carbonic Anhydrase Inhibitors on Inorganic Carbon Accumulation by Chlamydomonas Reinhardtii. Plant Physiol. 1985, 79, 177–183. [Google Scholar] [CrossRef]
- Moroney, J.V.; Ynalvez, R.A. Proposed Carbon Dioxide Concentrating Mechanism in Chlamydomonas Reinhardtii. Eukaryot. Cell 2007, 6, 1251. [Google Scholar] [CrossRef]
- Markelova, A.G.; Sinetova, M.P.; Kupriyanova, E.V.; Pronina, N.A. Distribution and Functional Role of Carbonic Anhydrase Cah3 Associated with Thylakoid Membranes in the Chloroplast and Pyrenoid of Chlamydomonas Reinhardtii. Russ. J. Plant Physiol. 2009, 56, 761–768. [Google Scholar] [CrossRef]
- Terentyev, V.V.; Shukshina, A.K.; Shitov, A.V. Carbonic Anhydrase CAH3 Supports the Activity of Photosystem II under Increased PH. Biochim. Biophys. acta. Bioenerg. 2019, 1860, 582–590. [Google Scholar] [CrossRef]
- Suzuki, K.; Yang, S.Y.; Shimizu, S.; Morishita, E.C.; Jiang, J.; Zhang, F.; Hoque, M.M.; Sato, Y.; Tsunoda, M.; Sekiguchi, T.; et al. The Unique Structure of Carbonic Anhydrase ACA1 from Chlamydomonas Reinhardtii. Acta Crystallogr. D. Biol. Crystallogr. 2011, 67, 894–901. [Google Scholar] [CrossRef]
- Benlloch, R.; Shevela, D.; Hainzl, T.; Grundström, C.; Shutova, T.; Messinger, J.; Samuelsson, G.; Elisabeth Sauer-Eriksson, A. Crystal Structure and Functional Characterization of Photosystem II-Associated Carbonic Anhydrase CAH3 in Chlamydomonas Reinhardtii. Plant Physiol. 2015, 167, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Touw, W.G.; Baakman, C.; Black, J.; Te Beek, T.A.H.; Krieger, E.; Joosten, R.P.; Vriend, G. A Series of PDB-Related Databanks for Everyday Needs. Nucleic Acids Res. 2015, 43, D364–D368. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of Protein Secondary Structure: Pattern Recognition of Hydrogen-bonded and Geometrical Features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.; Gokhman, I.; Pick, U.; Zamir, A. A Salt-Resistant Plasma Membrane Carbonic Anhydrase Is Induced by Salt in Dunaliella Salina. J. Biol. Chem. 1996, 271, 17718–17723. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.; Pick, U.; Zamir, A. A Salt-Induced 60-Kilodalton Plasma Membrane Protein Plays a Potential Role in the Extreme Halotolerance of the Alga Dunaliella. Plant Physiol. 1994, 106, 1359–1365. [Google Scholar] [CrossRef] [PubMed]
- Bageshwar, U.K.; Premkumar, L.; Gokhman, I.; Savchenko, T.; Sussman, J.L.; Zamir, A. Natural Protein Engineering: A Uniquely Salt-Tolerant, but Not Halophilic, Alpha-Type Carbonic Anhydrase from Algae Proliferating in Low- to Hyper-Saline Environments. Protein Eng. Des. Sel. 2004, 17, 191–200. [Google Scholar] [CrossRef]
- Premkumar, L.; Greenblatt, H.M.; Bageshwar, U.K.; Savchenko, T.; Gokhman, I.; Sussman, J.L.; Zamir, A. Three-Dimensional Structure of a Halotolerant Algal Carbonic Anhydrase Predicts Halotolerance of a Mammalian Homolog. Proc. Natl. Acad. Sci. USA 2005, 102, 7493–7498. [Google Scholar] [CrossRef]
- Bieger, B.; Essen, L.O.; Oesterhelt, D. Crystal Structure of Halophilic Dodecin: A Novel, Dodecameric Flavin Binding Protein from Halobacterium Salinarum. Structure 2003, 11, 375–385. [Google Scholar] [CrossRef]
- Moog, D.; Stork, S.; Reislöhner, S.; Grosche, C.; Maier, U.G. In Vivo Localization Studies in the Stramenopile Alga Nannochloropsis Oceanica. Protist 2015, 166, 161–171. [Google Scholar] [CrossRef]
- Murakami, R.; Hashimoto, H. Unusual Nuclear Division in Nannochloropsis Oculata (Eustigmatophyceae, Heterokonta) Which May Ensure Faithful Transmission of Secondary Plastids. Protist 2009, 160, 41–49. [Google Scholar] [CrossRef]
- Cavalier-Smith, T.; Van Der Giezen, M.; Martin, W.; Wilkins, A.; Doolittle, W.F.; Allen, J.F.; Leaver, C.J.; Whatley, F.R.; Howe, C.J. Genomic Reduction and Evolution of Novel Genetic Membranes and Protein-Targeting Machinery in Eukaryote-Eukaryote Chimaeras (Meta-Algae). Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2003, 358, 109–134. [Google Scholar] [CrossRef] [PubMed]
- Razzak, M.A.; Lee, J.M.; Lee, D.W.; Kim, J.H.; Yoon, H.S.; Hwang, I. Expression of Seven Carbonic Anhydrases in Red Alga Gracilariopsis Chorda and Their Subcellular Localization in a Heterologous System, Arabidopsis Thaliana. Plant Cell Rep. 2019, 38, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Whitton, B.A.; Potts, M. Introduction to the Cyanobacteria. In Ecology of Cyanobacteria II; Whitton, B., Ed.; Springer: Dordrecht, The Netherlands, 2012; pp. 1–13. ISBN 9789400738553. [Google Scholar]
- Schopf, J.W.; Packer, B.M. Early Archean (3.3-Billion to 3.5-Billion-Year-Old) Microfossils from Warrawoona Group, Australia. Science 1987, 237, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Cannon, G.C.; Bradburne, C.E.; Aldrich, H.C.; Baker, S.H.; Heinhorst, S.; Shively, J.M. Microcompartments in Prokaryotes: Carboxysomes and Related Polyhedra. Appl. Environ. Microbiol. 2001, 67, 5351. [Google Scholar] [CrossRef] [PubMed]
- So, A.K.C.; Espie, G.S. Cyanobacterial Carbonic Anhydrases. Can. J. Bot. 2005, 83, 721–734. [Google Scholar] [CrossRef]
- Kupriyanova, E.V.; Sinetova, M.A.; Cho, S.M.; Park, Y.I.; Markelova, A.G.; Los, D.A.; Pronina, N.A. Specific Features of the System of Carbonic Anhydrases of Alkaliphilic Cyanobacteria. Russ. J. Plant Physiol. 2013, 60, 465–471. [Google Scholar] [CrossRef]
- Berks, B.C.; Sargent, F.; Palmer, T. The Tat Protein Export Pathway. Mol. Microbiol. 2000, 35, 260–274. [Google Scholar] [CrossRef]
- Wang, D.; Li, Q.; Li, W.; Xing, J.; Su, Z. Improvement of Succinate Production by Overexpression of a Cyanobacterial Carbonic Anhydrase in Escherichia Coli. Enzym. Microb. Technol. 2009, 45, 491–497. [Google Scholar] [CrossRef]
- Kupriyanova, E.V.; Sinetova, M.A.; Bedbenov, V.S.; Pronina, N.A.; Los, D.A. Putative Extracellular α-Class Carbonic Anhydrase, EcaA, of Synechococcus Elongatus PCC 7942 Is an Active Enzyme: A Sequel to an Old Story. Microbiology 2018, 164, 576–586. [Google Scholar] [CrossRef]
- So, A.K.C.; Espie, G.S. Cloning, Characterization and Expression of Carbonic Anhydrase from the Cyanobacterium Synechocystis PCC6803. Plant Mol. Biol. 1998, 37, 205–215. [Google Scholar] [CrossRef]
- Kupriyanova, E.V.; Lebedeva, N.V.; Dudoladova, M.V.; Gerasimenko, L.M.; Alekseeva, S.G.; Pronina, N.A.; Zavarzin, G.A. Carbonic Anhydrase Activity of Alkalophilic Cyanobacteria from Soda Lakes. Russ. J. Plant Physiol. 2003, 50, 532–539. [Google Scholar] [CrossRef]
- Kupriyanova, E.V.; Markelova, A.G.; Lebedeva, N.V.; Gerasimenko, L.M.; Zavarzin, G.A.; Pronina, N.A. Carbonic Anhydrase of the Alkaliphilic Cyanobacterium Microcoleus Chthonoplastes. Microbiology 2004, 73, 255–259. [Google Scholar] [CrossRef]
- Kupriyanova, E.V.; Sinetova, M.A.; Mironov, K.S.; Novikova, G.V.; Dykman, L.A.; Rodionova, M.V.; Gabrielyan, D.A.; Los, D.A. Highly Active Extracellular α-Class Carbonic Anhydrase of Cyanothece Sp. ATCC 51142. Biochimie 2019, 160, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Kupriyanova, E.V.; Sinetova, M.A.; Leusenko, A.V.; Voronkov, A.S.; Los, D.A. A Leader Peptide of the Extracellular Cyanobacterial Carbonic Anhydrase Ensures the Efficient Secretion of Recombinant Proteins in Escherichia Coli. J. Biotechnol. 2022, 344, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Dudoladova, M.V.; Markelova, A.G.; Lebedeva, N.V.; Pronina, N.A. Compartmentation of α- and β-Carbonic Anhydrases in Cells of Halo- and Alkalophilic Cyanobacteria Rhabdoderma Lineare. Russ. J. Plant Physiol. 2004, 51, 806–814. [Google Scholar] [CrossRef]
- Dudoladova, M.V.; Kupriyanova, E.V.; Markelova, A.G.; Sinetova, M.P.; Allakhverdiev, S.I.; Pronina, N.A. The Thylakoid Carbonic Anhydrase Associated with Photosystem II Is the Component of Inorganic Carbon Accumulating System in Cells of Halo- and Alkaliphilic Cyanobacterium Rhabdoderma Lineare. Biochim. Biophys. Acta-Bioenerg. 2007, 1767, 616–623. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Langella, E.; Di Fiore, A.; Alterio, V.; Monti, S.M.; De Simone, G.; D’Ambrosio, K. α-CAs from Photosynthetic Organisms. Int. J. Mol. Sci. 2022, 23, 12045. https://doi.org/10.3390/ijms231912045
Langella E, Di Fiore A, Alterio V, Monti SM, De Simone G, D’Ambrosio K. α-CAs from Photosynthetic Organisms. International Journal of Molecular Sciences. 2022; 23(19):12045. https://doi.org/10.3390/ijms231912045
Chicago/Turabian StyleLangella, Emma, Anna Di Fiore, Vincenzo Alterio, Simona Maria Monti, Giuseppina De Simone, and Katia D’Ambrosio. 2022. "α-CAs from Photosynthetic Organisms" International Journal of Molecular Sciences 23, no. 19: 12045. https://doi.org/10.3390/ijms231912045
APA StyleLangella, E., Di Fiore, A., Alterio, V., Monti, S. M., De Simone, G., & D’Ambrosio, K. (2022). α-CAs from Photosynthetic Organisms. International Journal of Molecular Sciences, 23(19), 12045. https://doi.org/10.3390/ijms231912045