Selecting for CRISPR-Edited Knock-In Cells
Abstract
1. Introduction
2. Strategies to Increase HDR-Dependent CRISPR-Cas9 Mediated Genome Editing
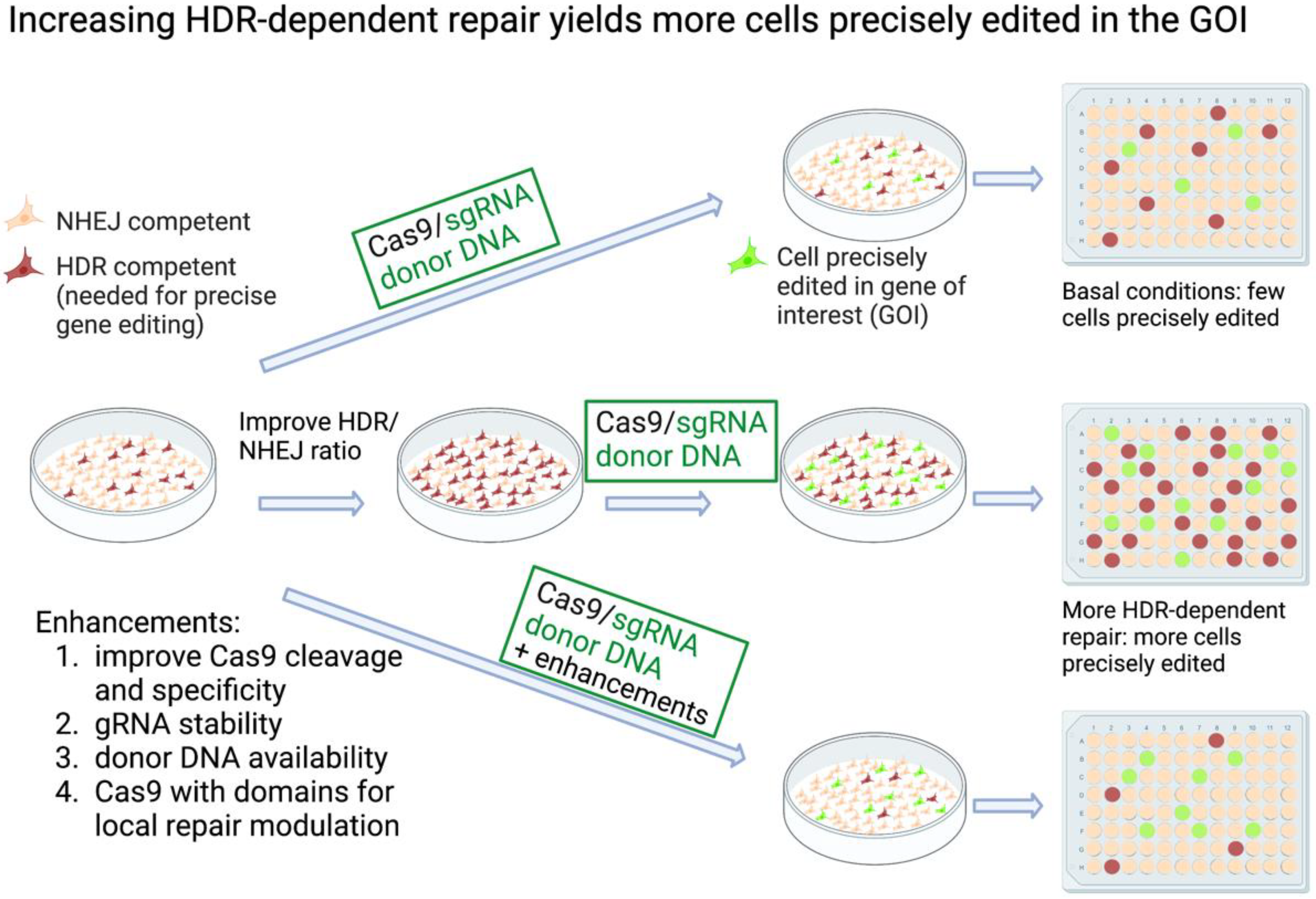
2.1. Inhibiting NHEJ/Promoting HDR Globally to Increase CRISPR Knock-In Editing
2.2. Improving Cas9 Cleavage and Specificity
2.3. Modifications of the Guide RNA and Donor DNA to Improve Editing Efficiency
2.4. Promoting HDR Specifically at the Break Site
2.5. Editing without a DSB—Circumventing the HDR Requirement
3. Selecting Edited Cells
3.1. Identifying and Selecting for CRISPR-Edited Cells
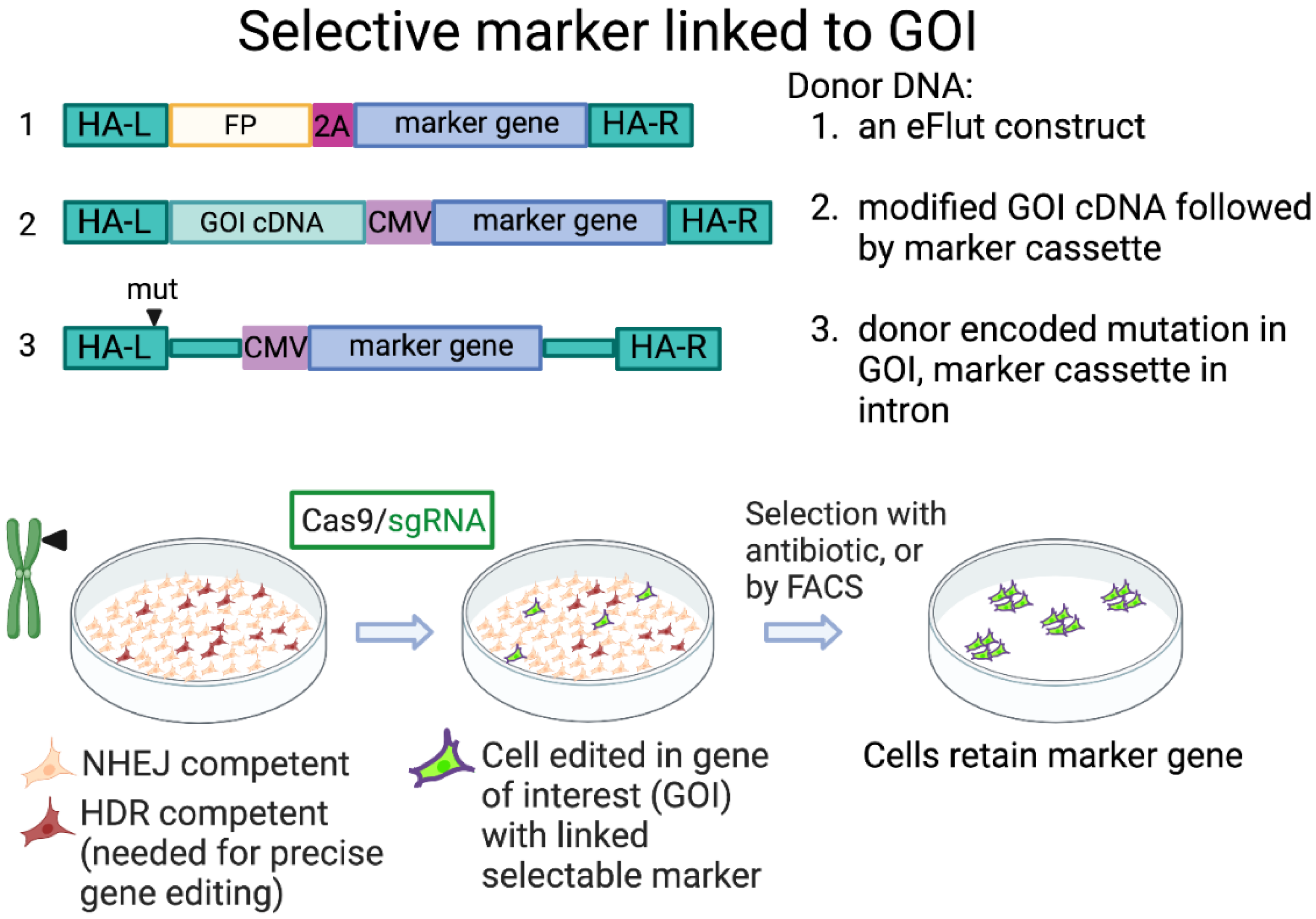
3.2. Selection Marker Expressed with Gene of Interest
3.3. Selection Involving Co-Editing
4. Scarless and Semi-Scarless Selection
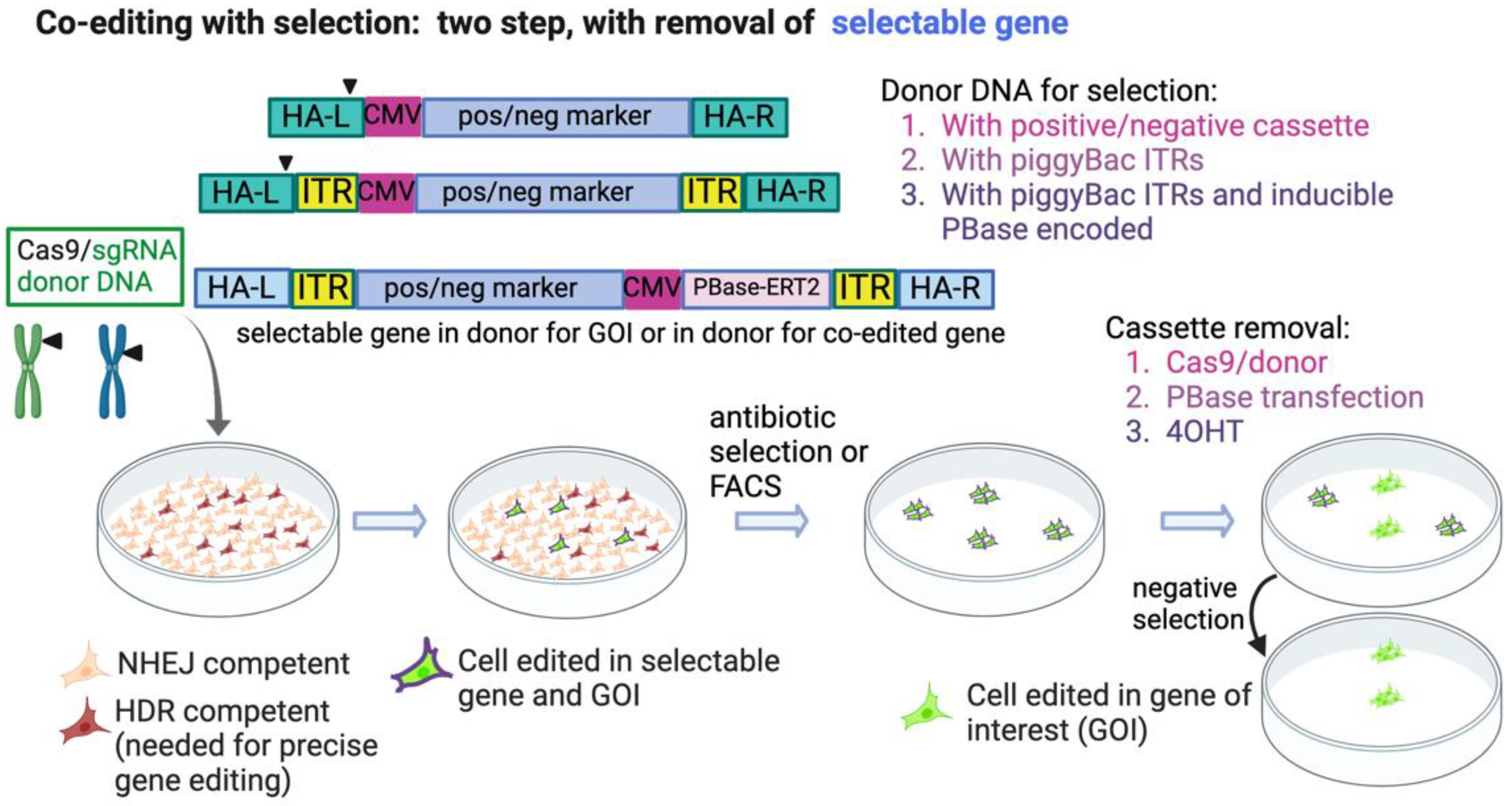
4.1. Two Step Editing—Insert, then Remove, Selectable Marker
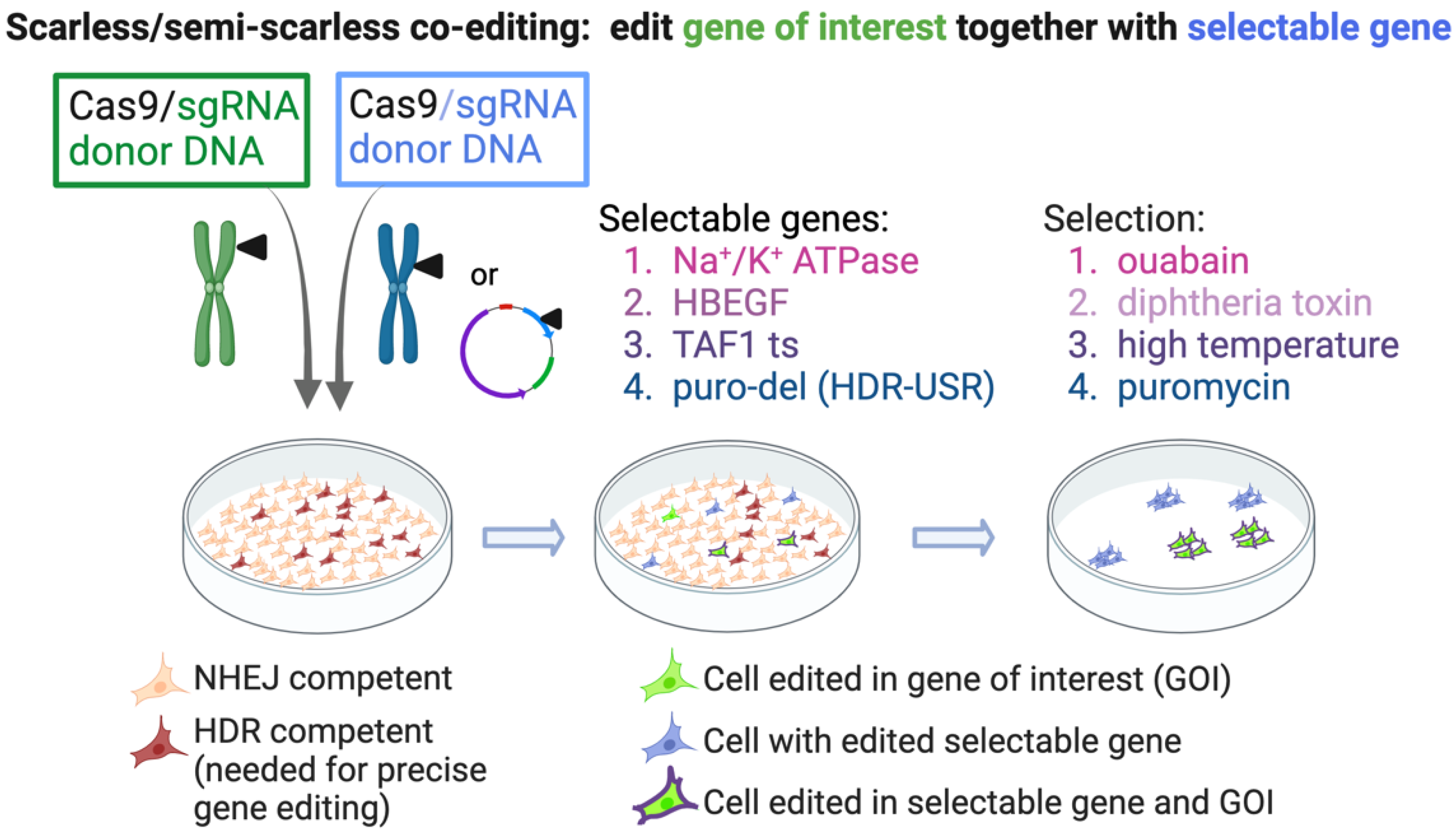
4.2. Co-Editing of an Endogenous Gene for Selection
4.3. Scarless Selection—By Editing a Plasmid-Based Marker
5. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Takata, M.; Sasaki, M.S.; Sonoda, E.; Morrison, C.; Hashimoto, M.; Utsumi, H.; Yamaguchi-Iwai, Y.; Shinohara, A.; Takeda, S. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998, 17, 5497–5508. [Google Scholar] [CrossRef]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drablos, F.; Saetrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef]
- Branzei, D.; Foiani, M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008, 9, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, Z.; Li, A.; Xie, X.; Liu, J.; Li, S.; Li, Y.; Wang, B.; Hu, L.; Yang, L.; et al. Strategies for High-Efficiency Mutation Using the CRISPR/Cas System. Front. Cell Dev. Biol. 2021, 9, 803252. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Ren, S.; Yu, S.; Pan, H.; Li, T.; Ge, S.; Zhang, J.; Xia, N. Methods Favoring Homology-Directed Repair Choice in Response to CRISPR/Cas9 Induced-Double Strand Breaks. Int J. Mol. Sci 2020, 21, 6461. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.D.; Richardson, C.D.; Corn, J.E. Advances in genome editing through control of DNA repair pathways. Nat. Cell Biol. 2019, 21, 1468–1478. [Google Scholar] [CrossRef]
- Sledzinski, P.; Dabrowska, M.; Nowaczyk, M.; Olejniczak, M. Paving the way towards precise and safe CRISPR genome editing. Biotechnol. Adv. 2021, 49, 107737. [Google Scholar] [CrossRef]
- Chu, V.T.; Weber, T.; Wefers, B.; Wurst, W.; Sander, S.; Rajewsky, K.; Kuhn, R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015, 33, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef]
- Paulsen, B.S.; Mandal, P.K.; Frock, R.L.; Boyraz, B.; Yadav, R.; Upadhyayula, S.; Gutierrez-Martinez, P.; Ebina, W.; Fasth, A.; Kirchhausen, T.; et al. Ectopic expression of RAD52 and dn53BP1 improves homology-directed repair during CRISPR-Cas9 genome editing. Nat. Biomed. Eng. 2017, 1, 878–888. [Google Scholar] [CrossRef] [PubMed]
- Canny, M.D.; Moatti, N.; Wan, L.C.K.; Fradet-Turcotte, A.; Krasner, D.; Mateos-Gomez, P.A.; Zimmermann, M.; Orthwein, A.; Juang, Y.C.; Zhang, W.; et al. Inhibition of 53BP1 favors homology-dependent DNA repair and increases CRISPR-Cas9 genome-editing efficiency. Nat. Biotechnol. 2018, 36, 95–102. [Google Scholar] [CrossRef]
- Shao, S.; Ren, C.; Liu, Z.; Bai, Y.; Chen, Z.; Wei, Z.; Wang, X.; Zhang, Z.; Xu, K. Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52. Int. J. Biochem. Cell Biol. 2017, 92, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Smirnikhina, S.A.; Zaynitdinova, M.I.; Sergeeva, V.A.; Lavrov, A.V. Improving Homology-Directed Repair in Genome Editing Experiments by Influencing the Cell Cycle. Int J. Mol. Sci. 2022, 23, 5992. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Staahl, B.T.; Alla, R.K.; Doudna, J.A. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 2014, 3, e04766. [Google Scholar] [CrossRef]
- Rahman, S.H.; Bobis-Wozowicz, S.; Chatterjee, D.; Gellhaus, K.; Pars, K.; Heilbronn, R.; Jacobs, R.; Cathomen, T. The nontoxic cell cycle modulator indirubin augments transduction of adeno-associated viral vectors and zinc-finger nuclease-mediated gene targeting. Hum. Gene Ther. 2013, 24, 67–77. [Google Scholar] [CrossRef]
- Gutschner, T.; Haemmerle, M.; Genovese, G.; Draetta, G.F.; Chin, L. Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep. 2016, 14, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Maresca, M.; Lin, V.G.; Guo, N.; Yang, Y. Obligate ligation-gated recombination (ObLiGaRe): Custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013, 23, 539–546. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Wyvekens, N.; Khayter, C.; Foden, J.A.; Thapar, V.; Reyon, D.; Goodwin, M.J.; Aryee, M.J.; Joung, J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014, 32, 569–576. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577–582. [Google Scholar] [CrossRef]
- Allen, D.; Rosenberg, M.; Hendel, A. Using Synthetically Engineered Guide RNAs to Enhance CRISPR Genome Editing Systems in Mammalian Cells. Front. Genome Ed. 2020, 2, 617910. [Google Scholar] [CrossRef] [PubMed]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Rahdar, M.; McMahon, M.A.; Prakash, T.P.; Swayze, E.E.; Bennett, C.F.; Cleveland, D.W. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. USA 2015, 112, E7110–E7117. [Google Scholar] [CrossRef]
- Lee, K.; Mackley, V.A.; Rao, A.; Chong, A.T.; Dewitt, M.A.; Corn, J.E.; Murthy, N. Synthetically modified guide RNA and donor DNA are a versatile platform for CRISPR-Cas9 engineering. Elife 2017, 6, e25312. [Google Scholar] [CrossRef] [PubMed]
- Roth, T.L.; Puig-Saus, C.; Yu, R.; Shifrut, E.; Carnevale, J.; Li, P.J.; Hiatt, J.; Saco, J.; Krystofinski, P.; Li, H.; et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature 2018, 559, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Shy, B.R.; Vykunta, V.S.; Ha, A.; Talbot, A.; Roth, T.L.; Nguyen, D.N.; Pfeifer, W.G.; Chen, Y.Y.; Blaeschke, F.; Shifrut, E.; et al. High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Tran, N.T.; Bashir, S.; Li, X.; Rossius, J.; Chu, V.T.; Rajewsky, K.; Kuhn, R. Enhancement of Precise Gene Editing by the Association of Cas9 With Homologous Recombination Factors. Front. Genet. 2019, 10, 365. [Google Scholar] [CrossRef]
- Charpentier, M.; Khedher, A.H.Y.; Menoret, S.; Brion, A.; Lamribet, K.; Dardillac, E.; Boix, C.; Perrouault, L.; Tesson, L.; Geny, S.; et al. CtIP fusion to Cas9 enhances transgene integration by homology-dependent repair. Nat. Commun. 2018, 9, 1133. [Google Scholar] [CrossRef]
- Reuven, N.; Adler, J.; Broennimann, K.; Myers, N.; Shaul, Y. Recruitment of DNA Repair MRN Complex by Intrinsically Disordered Protein Domain Fused to Cas9 Improves Efficiency of CRISPR-Mediated Genome Editing. Biomolecules 2019, 9, 584. [Google Scholar] [CrossRef]
- Jayavaradhan, R.; Pillis, D.M.; Goodman, M.; Zhang, F.; Zhang, Y.; Andreassen, P.R.; Malik, P. CRISPR-Cas9 fusion to dominant-negative 53BP1 enhances HDR and inhibits NHEJ specifically at Cas9 target sites. Nat. Commun. 2019, 10, 2866. [Google Scholar] [CrossRef]
- Benitez, E.K.; Lomova Kaufman, A.; Cervantes, L.; Clark, D.N.; Ayoub, P.G.; Senadheera, S.; Osborne, K.; Sanchez, J.M.; Crisostomo, R.V.; Wang, X.; et al. Global and Local Manipulation of DNA Repair Mechanisms to Alter Site-Specific Gene Editing Outcomes in Hematopoietic Stem Cells. Front. Genome Ed. 2020, 2, 601541. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Wolff, J.H.; Haldrup, J.; Thomsen, E.A.; Andersen, S.; Mikkelsen, J.G. piggyPrime: High-Efficacy Prime Editing in Human Cells Using piggyBac-Based DNA Transposition. Front. Genome Ed. 2021, 3, 786893. [Google Scholar] [CrossRef]
- Eggenschwiler, R.; Gschwendtberger, T.; Felski, C.; Jahn, C.; Langer, F.; Sterneckert, J.; Hermann, A.; Luhmann, J.; Steinemann, D.; Haase, A.; et al. A selectable all-in-one CRISPR prime editing piggyBac transposon allows for highly efficient gene editing in human cell lines. Sci. Rep. 2021, 11, 22154. [Google Scholar] [CrossRef]
- Miyaoka, Y.; Chan, A.H.; Judge, L.M.; Yoo, J.; Huang, M.; Nguyen, T.D.; Lizarraga, P.P.; So, P.L.; Conklin, B.R. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat. Methods 2014, 11, 291–293. [Google Scholar] [CrossRef]
- McCormick, M. Sib selection. Methods Enzymol. 1987, 151, 445–449. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Steyer, B.; Bu, Q.; Cory, E.; Jiang, K.; Duong, S.; Sinha, D.; Steltzer, S.; Gamm, D.; Chang, Q.; Saha, K. Scarless Genome Editing of Human Pluripotent Stem Cells via Transient Puromycin Selection. Stem Cell Rep. 2018, 10, 642–654. [Google Scholar] [CrossRef]
- Stewart-Ornstein, J.; Lahav, G. Dynamics of CDKN1A in Single Cells Defined by an Endogenous Fluorescent Tagging Toolkit. Cell Rep. 2016, 14, 1800–1811. [Google Scholar] [CrossRef]
- Bak, R.O.; Dever, D.P.; Porteus, M.H. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nat. Protoc. 2018, 13, 358–376. [Google Scholar] [CrossRef] [PubMed]
- Dever, D.P.; Bak, R.O.; Reinisch, A.; Camarena, J.; Washington, G.; Nicolas, C.E.; Pavel-Dinu, M.; Saxena, N.; Wilkens, A.B.; Mantri, S.; et al. CRISPR/Cas9 beta-globin gene targeting in human haematopoietic stem cells. Nature 2016, 539, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Scheller, S.H.; Rashad, Y.; Saleh, F.M.; Willingham, K.A.; Reilich, A.; Lin, D.; Izadpanah, R.; Alt, E.U.; Braun, S.E. Biallelic, Selectable, Knock-in Targeting of CCR5 via CRISPR-Cas9 Mediated Homology Directed Repair Inhibits HIV-1 Replication. Front. Immunol. 2022, 13, 821190. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, Y.; Zhong, L.; Wu, K.; Zhang, R.; Kang, T.; Wu, S.; Wu, Y. Efficient gene editing through an intronic selection marker in cells. Cell Mol. Life Sci. 2022, 79, 111. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Sun, Y.; Fang, Y.; Deng, J.; Mu, L.; Xu, K.; Mymryk, J.S.; Zhang, Z. A Universal Surrogate Reporter for Efficient Enrichment of CRISPR/Cas9-Mediated Homology-Directed Repair in Mammalian Cells. Mol. Ther.-Nucleic Acids 2019, 19, 775–789. [Google Scholar] [CrossRef]
- Shy, B.R.; MacDougall, M.S.; Clarke, R.; Merrill, B.J. Co-incident insertion enables high efficiency genome engineering in mouse embryonic stem cells. Nucleic Acids Res. 2016, 44, 7997–8010. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Li, G.H.; Fu, J.; Fu, Y.W.; Zhang, L.; Chen, W.; Arakaki, C.; Zhang, J.P.; Wen, W.; Zhao, M.; et al. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018, 46, 10195–10215. [Google Scholar] [CrossRef] [PubMed]
- Mitzelfelt, K.A.; McDermott-Roe, C.; Grzybowski, M.N.; Marquez, M.; Kuo, C.T.; Riedel, M.; Lai, S.; Choi, M.J.; Kolander, K.D.; Helbling, D.; et al. Efficient Precision Genome Editing in iPSCs via Genetic Co-targeting with Selection. Stem Cell Rep. 2017, 8, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.; Jasin, M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature 2000, 405, 697–700. [Google Scholar] [CrossRef]
- Ikeda, K.; Uchida, N.; Nishimura, T.; White, J.; Martin, R.M.; Nakauchi, H.; Sebastiano, V.; Weinberg, K.I.; Porteus, M.H. Efficient scarless genome editing in human pluripotent stem cells. Nat. Methods 2018, 15, 1045–1047. [Google Scholar] [CrossRef]
- Li, M.A.; Turner, D.J.; Ning, Z.; Yusa, K.; Liang, Q.; Eckert, S.; Rad, L.; Fitzgerald, T.W.; Craig, N.L.; Bradley, A. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011, 39, e148. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Wang, J.; Beyer, A.I.; Teque, F.; Cradick, T.J.; Qi, Z.; Chang, J.C.; Bao, G.; Muench, M.O.; Yu, J.; et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc. Natl. Acad. Sci. USA 2014, 111, 9591–9596. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Ye, L.; Chang, J.C.; Beyer, A.I.; Wang, J.; Muench, M.O.; Kan, Y.W. Seamless gene correction of beta-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014, 24, 1526–1533. [Google Scholar] [CrossRef]
- Kondrashov, A.; Duc Hoang, M.; Smith, J.G.W.; Bhagwan, J.R.; Duncan, G.; Mosqueira, D.; Munoz, M.B.; Vo, N.T.N.; Denning, C. Simplified Footprint-Free Cas9/CRISPR Editing of Cardiac-Associated Genes in Human Pluripotent Stem Cells. Stem Cells Dev. 2018, 27, 391–404. [Google Scholar] [CrossRef]
- Singh, A.M.; Perry, D.W.; Steffey, V.V.A.; Miller, K.; Allison, D.W. Decoding the Epigenetic Heterogeneity of Human Pluripotent Stem Cells with Seamless Gene Editing. Methods Mol. Biol. 2016, 1516, 153–169. [Google Scholar] [CrossRef]
- Li, X.; Burnight, E.R.; Cooney, A.L.; Malani, N.; Brady, T.; Sander, J.D.; Staber, J.; Wheelan, S.J.; Joung, J.K.; McCray, P.B., Jr.; et al. piggyBac transposase tools for genome engineering. Proc. Natl. Acad. Sci. USA 2013, 110, E2279–E2287. [Google Scholar] [CrossRef]
- Arias-Fuenzalida, J.; Jarazo, J.; Qing, X.; Walter, J.; Gomez-Giro, G.; Nickels, S.L.; Zaehres, H.; Scholer, H.R.; Schwamborn, J.C. FACS-Assisted CRISPR-Cas9 Genome Editing Facilitates Parkinson’s Disease Modeling. Stem Cell Rep. 2017, 9, 1423–1431. [Google Scholar] [CrossRef]
- Yusa, K.; Zhou, L.; Li, M.A.; Bradley, A.; Craig, N.L. A hyperactive piggyBac transposase for mammalian applications. Proc. Natl. Acad. Sci. USA 2011, 108, 1531–1536. [Google Scholar] [CrossRef]
- Agudelo, D.; Duringer, A.; Bozoyan, L.; Huard, C.C.; Carter, S.; Loehr, J.; Synodinou, D.; Drouin, M.; Salsman, J.; Dellaire, G.; et al. Marker-free coselection for CRISPR-driven genome editing in human cells. Nat. Methods 2017, 14, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Akrap, N.; Cerboni, S.; Porritt, M.J.; Wimberger, S.; Lundin, A.; Moller, C.; Firth, M.; Gordon, E.; Lazovic, B.; et al. Universal toxin-based selection for precise genome engineering in human cells. Nat. Commun. 2021, 12, 497. [Google Scholar] [CrossRef]
- Reuven, N.; Adler, J.; Myers, N.; Shaul, Y. CRISPR Co-Editing Strategy for Scarless Homology-Directed Genome Editing. Int. J. Mol. Sci. 2021, 22, 3741. [Google Scholar] [CrossRef]
- Ma, L.; Xing, J.; Li, Q.; Zhang, Z.; Xu, K. Development of a universal antibiotic resistance screening reporter for improving efficiency of cytosine and adenine base editing. J. Biol. Chem. 2022, 298, 102103. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.A.; Talas, A.; Kulcsar, P.I.; Biczok, Z.; Krausz, S.L.; Varady, G.; Welker, E. PEAR, a flexible fluorescent reporter for the identification and enrichment of successfully prime edited cells. Elife 2022, 11, e69504. [Google Scholar] [CrossRef] [PubMed]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef] [PubMed]
- Nahmad, A.D.; Reuveni, E.; Goldschmidt, E.; Tenne, T.; Liberman, M.; Horovitz-Fried, M.; Khosravi, R.; Kobo, H.; Reinstein, E.; Madi, A.; et al. Frequent aneuploidy in primary human T cells after CRISPR-Cas9 cleavage. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]

| Selection Strategy | Description | Scarless? | Reference |
|---|---|---|---|
| Marker linked to GOI | eFlut plasmid sets for generating dsDNA donors with selectable genes by PCR- for tagging N- or C-termini of endogenous genes | No | [50] |
| Foreign selection cassettes are combined in a donor DNA for modifying a GOI | No | [51] | |
| Two donor DNAs with different fluorescent protein-expressing selection cassettes for selection of biallelically edited cells | No | [53] | |
| GEIS (Gene Editing through an Intronic Selection marker) | Yes, if cassette does not interfere with splicing or GOI expression | [54] | |
| Co-editing with independent locus | Co-editing of GOI alongside selectable gene requiring HDR | No | [57,58] |
| COIN (co-incidental insertion) | No | [56] | |
| Scarless selection | Gene edit is itself selectable: FACS sorting for TCR in primary human T cells | Yes | [35] |
| Two-step scarless selection: insert, then remove selectable marker | Two CRISPR editing steps—first step CRISPR to introduce selectable marker and edit GOI, second CRISPR to remove selectable marker | Yes | [60] |
| Selectable cassette flanking mutation in GOI; removal using piggyBac transposase | Co-editing of selectable cassette followed by removal by transfected piggyBac transposase | Yes, if cassette not re-inserted by transposase | [62,63,64] |
| Co-editing with removable selection cassette expressing PBase-ERT2 for induction of cassette removal with 4OHT | Yes, if cassette not re-inserted by transposase | [56] | |
| Editing of GOI with donor containing removable selection cassette. Removal via transfection with excision-only PBase | Yes | [65] | |
| Two excisable fluorescent donor cassettes to permit sorting of biallelic clones, removal of cassettes with excision-only PBase | Yes | [67] | |
| Co-editing with selectable endogenous gene | Co-editing of sodium/potassium pump for resistance to ouabain | Semi | [69] |
| Co-editing of HBEGF for resistance to diphtheria toxin, “Xential” | Semi | [70] | |
| Co-editing of TAF1ts, selective gene restored to wt sequence | Semi | [71] | |
| Co-editing with plasmid-encoded selectable gene | “HDR-USR” (universal surrogate reporter) Truncated puro-resistance gene is corrected on plasmid | Yes | [55] |
| “ACBE-ARSR” Adenine and cytosine base editing antibiotic resistance screening reporter | Yes | [72] | |
| “PEAR” Prime editor activity reporter | Yes | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reuven, N.; Shaul, Y. Selecting for CRISPR-Edited Knock-In Cells. Int. J. Mol. Sci. 2022, 23, 11919. https://doi.org/10.3390/ijms231911919
Reuven N, Shaul Y. Selecting for CRISPR-Edited Knock-In Cells. International Journal of Molecular Sciences. 2022; 23(19):11919. https://doi.org/10.3390/ijms231911919
Chicago/Turabian StyleReuven, Nina, and Yosef Shaul. 2022. "Selecting for CRISPR-Edited Knock-In Cells" International Journal of Molecular Sciences 23, no. 19: 11919. https://doi.org/10.3390/ijms231911919
APA StyleReuven, N., & Shaul, Y. (2022). Selecting for CRISPR-Edited Knock-In Cells. International Journal of Molecular Sciences, 23(19), 11919. https://doi.org/10.3390/ijms231911919







