Generating a Small Shuttle Vector for Effective Genetic Engineering of Methanosarcina mazei Allowed First Insights in Plasmid Replication Mechanism in the Methanoarchaeon
Abstract
1. Introduction
2. Results
2.1. Several Shortened Derivatives of the Shuttle Vector Are Stable in M. mazei
2.1.1. Elucidating Crucial Regions for Stable Replication in M. mazei
2.1.2. Smaller Shuttle Vectors Increase the Transformation Success
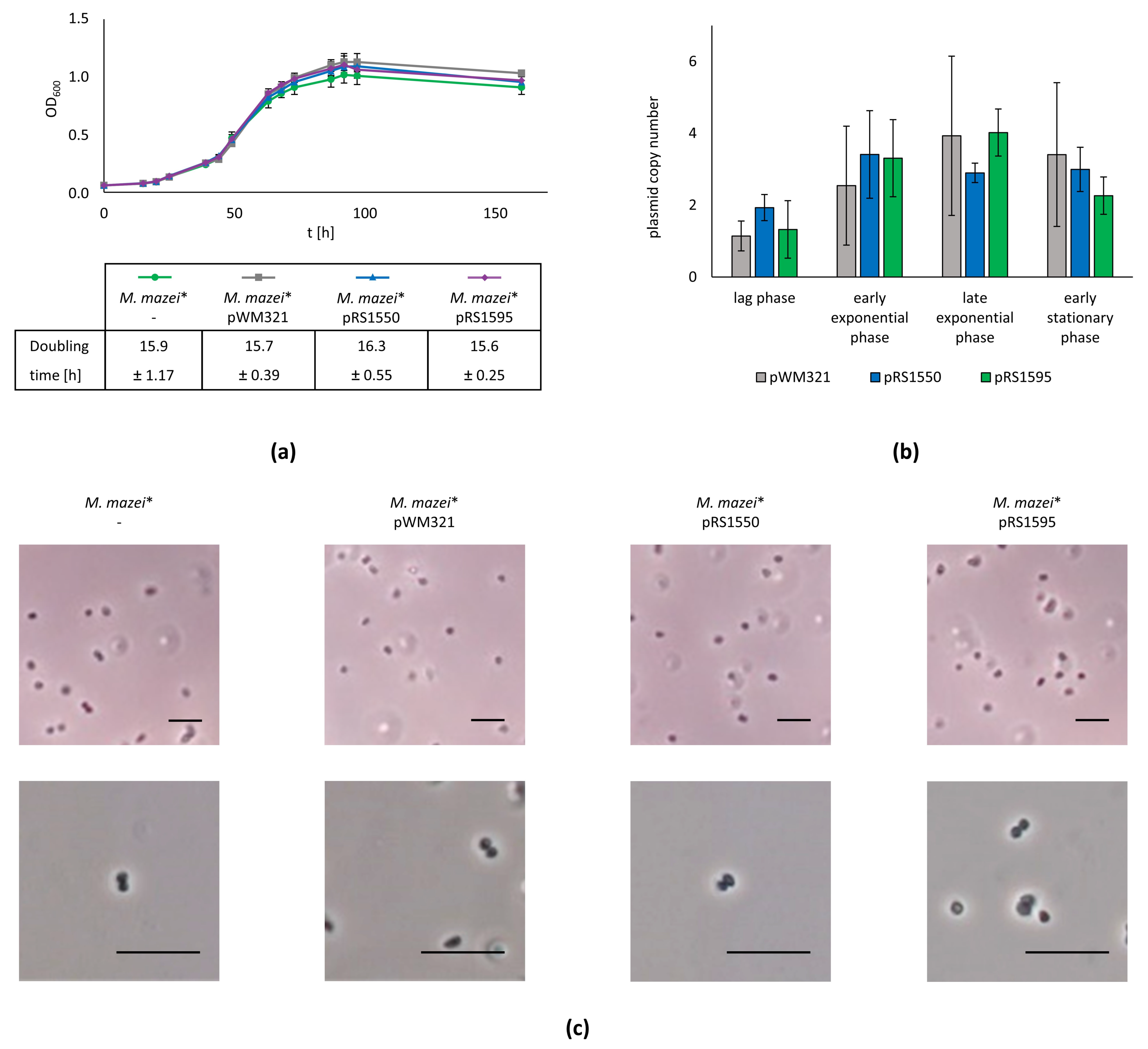
2.2. Smaller Shuttle Vectors in M. mazei Show the Same Phenotype and Copy Number as the Original pWM321
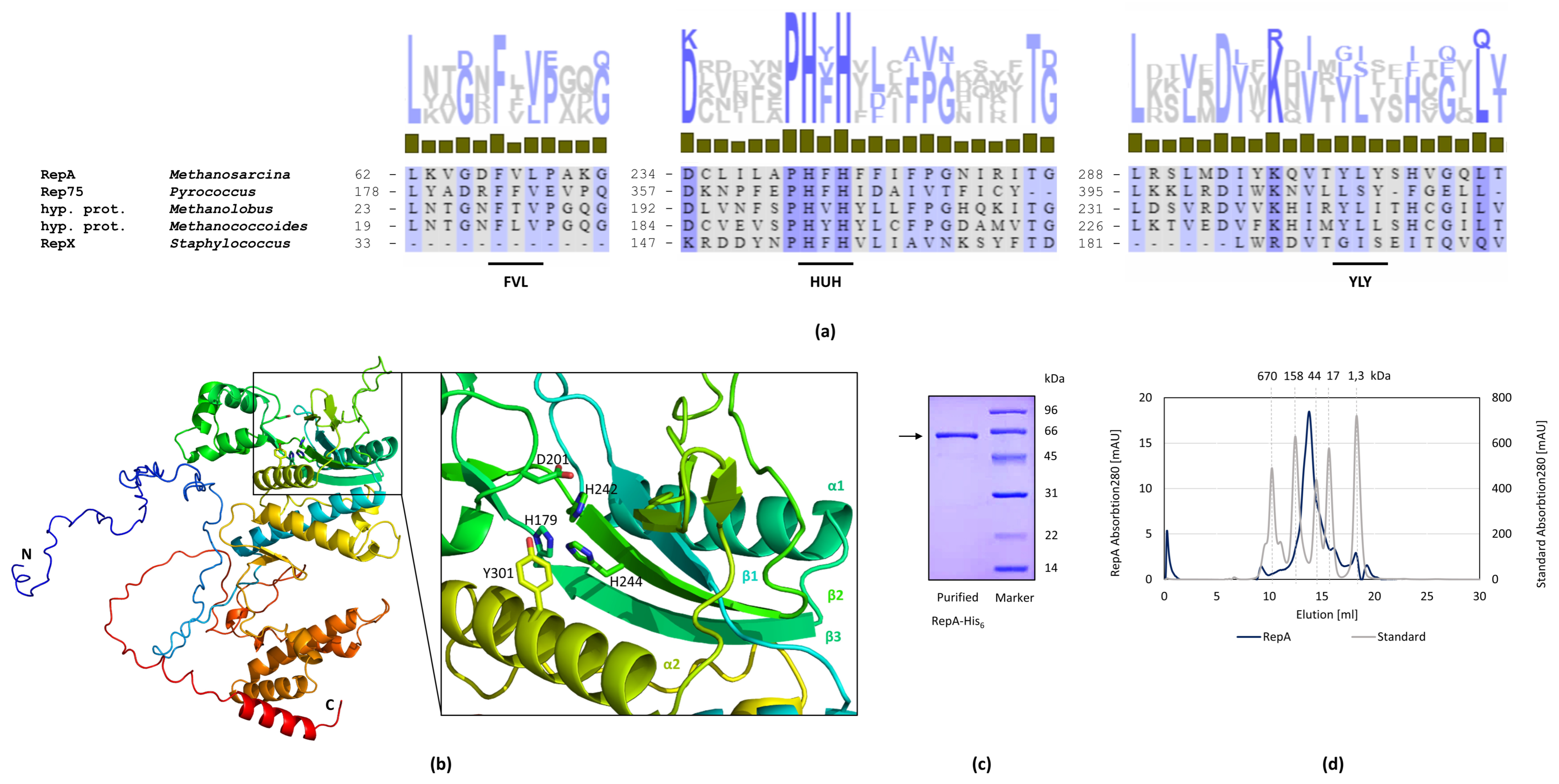
2.3. RepA Is the Essential Replication Protein
2.4. Interaction of RepA and DNA
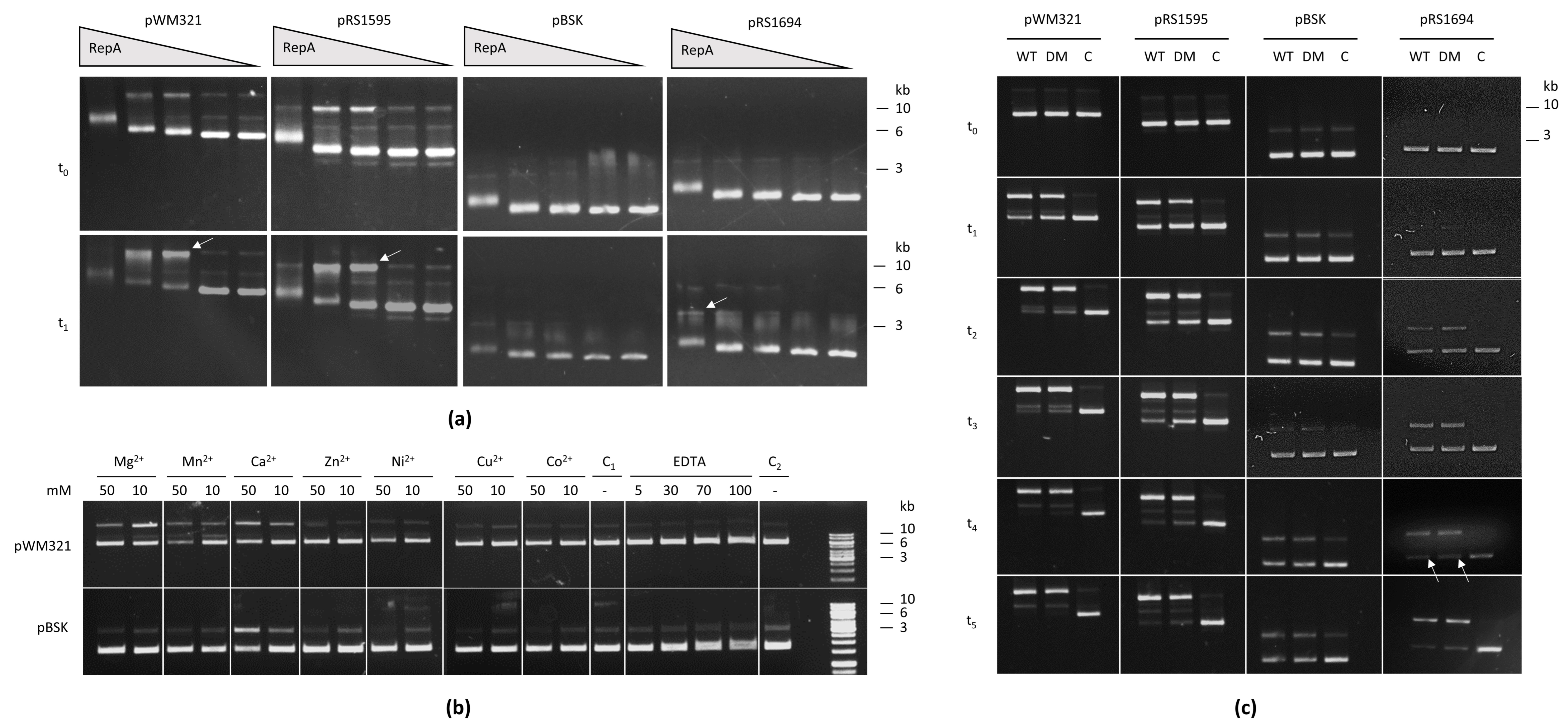
2.5. RepA Binds and Nicks Archaeal Plasmids
3. Discussion
3.1. Optimisation of Archaeal Shuttle Vectors for Biotechnological Applications
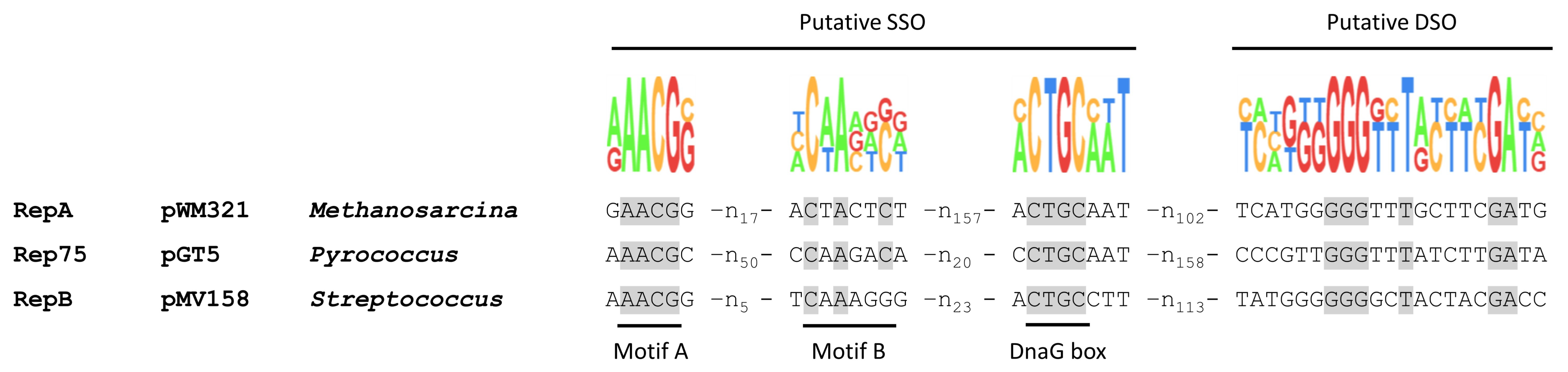
3.2. RepA Shows Conserved Motifs of Rolling Circle Initiator Proteins
3.3. The Double-Strand and the Single-Strand Origin
4. Materials and Methods
4.1. Strains and Plasmids
4.2. Generation of Plasmids and Construction of Mutant Strains
4.3. Growth of M. mazei
4.4. Copy Number Determination
4.5. Purification of RepA-His6
4.6. Size-Exclusion Chromatography
4.7. Nickase Assay and Electrophoretic Mobility Shift Assay
4.8. Folding Transition Analysis Using Tycho NT.6
4.9. Pull-Down Analysis
4.10. Bioinformatic Alignment and Protein Structure Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Thauer, R.K.; Kaster, A.-K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical background and biotechnological applications. AMB Express 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, K.; Ergal, İ.; Koller, M.; Basen, M.; Schuster, B.; Rittmann, S.K.-M.R. Archaea Biotechnology. Biotechnol. Adv. 2020, 47, 107668. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.A.; Albers, S.-V.; Atomi, H.; Allers, T. Model organisms for genetics in the domain Archaea: Methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 2011, 35, 577–608. [Google Scholar] [CrossRef]
- Rother, M.; Metcalf, W.W. Genetic technologies for Archaea. Curr. Opin. Microbiol. 2005, 8, 745–751. [Google Scholar] [CrossRef]
- de Vrieze, J.; Hennebel, T.; Boon, N.; Verstraete, W. Methanosarcina: The rediscovered methanogen for heavy duty biomethanation. Bioresour. Technol. 2012, 112, 1–9. [Google Scholar] [CrossRef]
- Weidenbach, K.; Gutt, M.; Cassidy, L.; Chibani, C.; Schmitz, R.A. Small Proteins in Archaea, a Mainly Unexplored World. J. Bacteriol. 2022, 204, 128. [Google Scholar] [CrossRef]
- Deppenmeier, U.; Johann, A.; Hartsch, T.; Merkl, R.; Schmitz, R.A.; Martinez-Arias, R.; Henne, A.; Wiezer, A.; Bäumer, S.; Jacobi, C.; et al. The genome of Methanosarcina mazei: Evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 2002, 4, 453–461. [Google Scholar]
- Kohler, P.R.A.; Metcalf, W.W. Genetic manipulation of Methanosarcina spp. Front. Microbiol. 2012, 3, 259. [Google Scholar] [CrossRef]
- Metcalf, W.W.; Zhang, J.K.; Apolinario, E.; Wolfe, R.S. A genetic system for Archaea of the genus Methanosarcina: Liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 1997, 94, 2626–2631. [Google Scholar] [CrossRef] [PubMed]
- Sowers, K.R.; Gunsalus, R.P. Plasmid DNA from the acetotrophic methanogen Methanosarcina acetivorans. J. Bacteriol. 1988, 170, 4979–4982. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Masó, J.A.; Machón, C.; Bordanaba-Ruiseco, L.; Espinosa, M.; Coll, M.; del Solar, G. Plasmid Rolling-Circle Replication. Microbiol. Spectr. 2015, 3, PLAS-0035-2014. [Google Scholar] [CrossRef]
- Del Solar, G.; Giraldo, R.; Ruiz-Echevarría, M.J.; Espinosa, M.; Díaz-Orejas, R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998, 62, 434–464. [Google Scholar] [CrossRef]
- Lilly, J.; Camps, M. Mechanisms of Theta Plasmid Replication. Microbiol. Spectr. 2015, 3, PLAS-0029-2014. [Google Scholar] [CrossRef]
- Erauso, G.; Marsin, S.; Benbouzid-Rollet, N.; Baucher, M.F.; Barbeyron, T.; Zivanovic, Y.; Prieur, D.; Forterre, P. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: Evidence for rolling-circle replication in a hyperthermophile. J. Bacteriol. 1996, 178, 3232–3237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Soler, N.; Justome, A.; Quevillon-Cheruel, S.; Lorieux, F.; le Cam, E.; Marguet, E.; Forterre, P. The rolling-circle plasmid pTN1 from the hyperthermophilic archaeon Thermococcus nautilus. Mol. Microbiol. 2007, 66, 357–370. [Google Scholar] [CrossRef]
- Zhou, L.; Zhou, M.; Sun, C.; Han, J.; Lu, Q.; Zhou, J.; Xiang, H. Precise Determination, Cross-Recognition, and Functional Analysis of the Double-Strand Origins of the Rolling-Circle Replication Plasmids in Haloarchaea. J. Bacteriol. 2008, 190, 5710–5719. [Google Scholar] [CrossRef] [PubMed]
- Fink, C.; Beblawy, S.; Enkerlin, A.M.; Mühling, L.; Angenent, L.T.; Molitor, B. A Shuttle-Vector System Allows Heterologous Gene Expression in the Thermophilic Methanogen Methanothermobacter thermautotrophicus ΔH. mBio 2021, 12, e02766-21. [Google Scholar] [CrossRef]
- Ehlers, C.; Weidenbach, K.; Veit, K.; Deppenmeier, U.; Metcalf, W.W.; Schmitz, R.A. Development of genetic methods and construction of a chromosomal glnK1 mutant in Methanosarcina mazei strain Gö1. Mol. Genet. Genom. 2005, 273, 290–298. [Google Scholar] [CrossRef]
- Boer, D.R.; Ruíz-Masó, J.A.; López-Blanco, J.R.; Blanco, A.G.; Vives-Llàcer, M.; Chacón, P.; Usón, I.; Gomis-Rüth, F.X.; Espinosa, M.; Llorca, O.; et al. Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains. EMBO J. 2009, 28, 1666–1678. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ngugi, D.K.; Blom, J.; Ali, S.; Ferry, J.G.; Stingl, U. Draft Genome Sequence of an Obligately Methylotrophic Methanogen, Methanococcoides methylutens, Isolated from Marine Sediment. Genome Announc. 2014, 2, e01184-14. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S.; Weisblum, B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 1982, 150, 815–825. [Google Scholar] [CrossRef] [PubMed]
- van der Lelie, D.; Bron, S.; Venema, G.; Oskam, L. Similarity of minus origins of replication and flanking open reading frames of plasmids pUB110, pTB913 and pMV158. Nucleic Acids Res. 1989, 17, 7283–7294. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dupureur, C.M. Roles of metal ions in nucleases. Curr. Opin. Chem. Biol. 2008, 12, 250–255. [Google Scholar] [CrossRef]
- Bowen, L.M.; Dupureur, C.M. Investigation of restriction enzyme cofactor requirements: A relationship between metal ion properties and sequence specificity. Biochemistry 2003, 42, 12643–12653. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Lu, Y. Putative Extracellular Electron Transfer in Methanogenic Archaea. Front. Microbiol. 2021, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Blasco-Gómez, R.; Batlle-Vilanova, P.; Villano, M.; Balaguer, M.D.; Colprim, J.; Puig, S. On the Edge of Research and Technological Application: A Critical Review of Electromethanogenesis. Int. J. Mol. Sci. 2017, 18, 874. [Google Scholar] [CrossRef]
- Wood, A.G.; Whitman, W.B.; Konisky, J. A newly-isolated marine methanogen harbors a small cryptic plasmid. Arch. Microbiol. 1985, 142, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Meile, L.; Kiener, A.; Leisinger, T. A plasmid in the archaebacterium Methanobacterium thermoautotrophicum. Mol. Genet. Genom. 1983, 191, 480–484. [Google Scholar] [CrossRef]
- Tumbula, D.L.; Bowen, T.L.; Whitman, W.B. Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J. Bacteriol. 1997, 179, 2976–2986. [Google Scholar] [CrossRef]
- Feinbaum, R. Introduction to Plasmid Biology. Curr. Protoc. Mol. Biol. 1998, 41, 19. [Google Scholar] [CrossRef]
- Smyshlyaev, G.; Bateman, A.; Barabas, O. Sequence analysis of tyrosine recombinases allows annotation of mobile genetic elements in prokaryotic genomes. Mol. Syst. Biol. 2021, 17, e9880. [Google Scholar] [CrossRef] [PubMed]
- Badel, C.; Da Cunha, V.; Oberto, J. Archaeal tyrosine recombinases. FEMS Microbiol. Rev. 2021, 45, 9658. [Google Scholar] [CrossRef] [PubMed]
- Ilyina, T.V.; Koonin, E.V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992, 20, 3279–3285. [Google Scholar] [CrossRef] [PubMed]
- Chandler, M.; de Dyda, F.L.C.F.; Hickman, A.B.; Moncalian, G.; Ton-Hoang, B. Breaking and joining single-stranded DNA: The HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013, 11, 525–538. [Google Scholar] [CrossRef]
- Lorenzo-Díaz, F.; Fernández-López, C.; Garcillán-Barcia, M.P.; Espinosa, M. Bringing them together: Plasmid pMV158 rolling circle replication and conjugation under an evolutionary perspective. Plasmid 2014, 74 (Suppl. 2), 15–31. [Google Scholar] [CrossRef] [PubMed]
- Ekundayo, B.; Bleichert, F. Origins of DNA replication. PLoS Genet. 2019, 15, e1008320. [Google Scholar] [CrossRef] [PubMed]
- Wright, L.D.; Johnson, C.M.; Grossman, A.D. Identification of a Single Strand Origin of Replication in the Integrative and Conjugative Element ICEBs1 of Bacillus subtilis. PLoS Genet. 2015, 11, e1005556. [Google Scholar] [CrossRef]
- Thomas, C.D.; Balson, D.F.; Shaw, W.V. In vitro studies of the initiation of staphylococcal plasmid replication. Specificity of RepD for its origin (oriD) and characterization of the Rep-ori tyrosyl ester intermediate. J. Biol. Chem. 1990, 265, 5519–5530. [Google Scholar] [CrossRef]
- Marsin, S.; Forterre, P. The active site of the rolling circle replication protein Rep75 is involved in site-specific nuclease, ligase and nucleotidyl transferase activities. Mol. Microbiol. 1999, 33, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Miller, V.L.; Mekalanos, J.J. A novel suicide vector and its use in construction of insertion mutations: Osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 1988, 170, 2575–2583. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Gutt, M.; Jordan, B.; Weidenbach, K.; Gudzuhn, M.; Kiessling, C.; Cassidy, L.; Helbig, A.; Tholey, A.; Pyper, D.J.; Kubatova, N.; et al. High complexity of Glutamine synthetase regulation in Methanosarcina mazei: Small protein 26 interacts and enhances glutamine synthetase activity. FEBS J. 2021, 288, 5350–5373. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
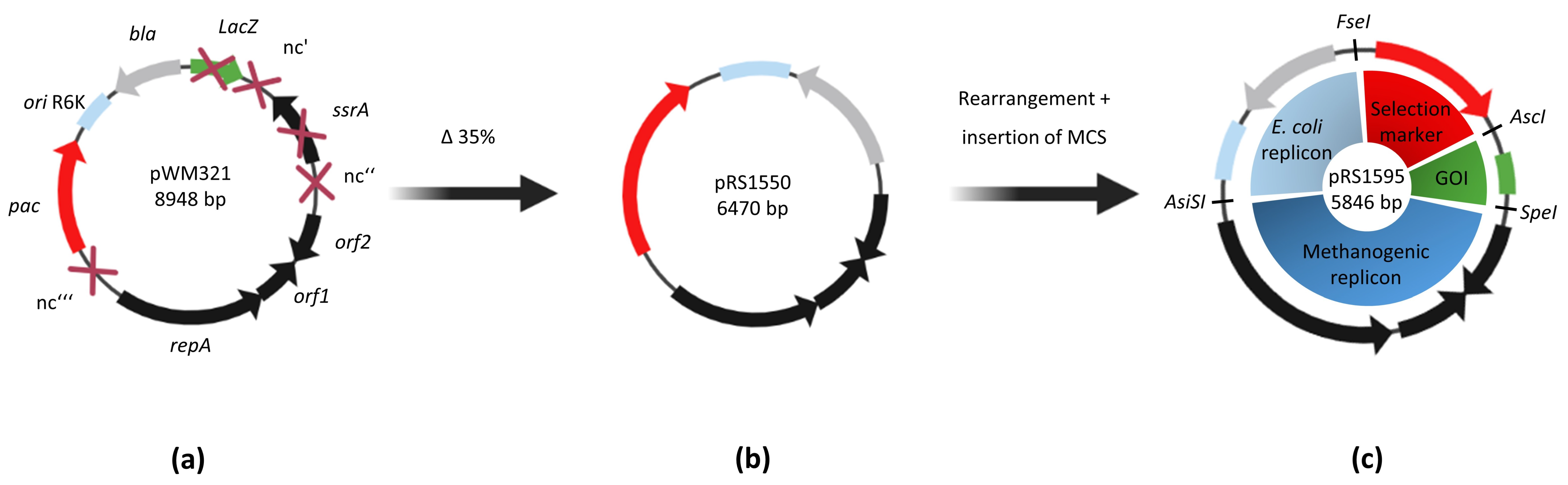

| Name | Size (kbp) | Deleted Region | Area in pWM321 | Stability in M. mazei |
|---|---|---|---|---|
| pRS1532 | ~8.2 | ∆nc‘1 | ∆526–1287 | + |
| pRS1515 | ~8.0 | ∆ssrA | ∆1316–2287 | + |
| pRS1531 | ~8.4 | ∆nc‘‘1 | ∆2325–2778 | – |
| pRS1518 | ~7.9 | ∆orf1+2 | ∆2811–3845 | – |
| pRS1522 | ~7.2 | ∆repA | ∆3908–5688 | – |
| pRS1523 | ~8.5 | ∆nc‘‘‘1 | ∆5712–6149 | + |
| pRS1550 | ~6.5 | ∆nc‘ + ∆ssrA + parts of ∆nc‘‘1 ∆nc‘‘‘1 | ∆29–2556 ∆5712–6149 | + |
| pRS1595 | ~5.8 | ∆nc‘ + ∆ssrA + parts of ∆nc‘‘ ∆nc‘‘‘1 | ∆29–2556 ∆5712–6149 | + |
| Strain/Plasmid | Genotype/Relevant Characteristics | Source/Reference |
|---|---|---|
| M. mazei* | Potential cell wall mutant | [20] |
| E. coli DH5α | General cloning strain | [43] |
| E. coli JM109 λpir | General cloning strain | [43] |
| E. coli BL21-CodonPlus®®-RIL | Overexpression strain with broader codon usage | Stratagene, La Jolla, CA, USA |
| pWM321 | Shuttle vector E. coli–Methanosarcina (8.9 kbp) | [11] |
| pET28a(+) | expression vector, N-term. His-tag | Novagene®®, Merck Millipore, Darmstadt, Germany |
| pRS1452 | pWM321∆pac | This work |
| pEX-K248_pac_JT | Synthesised pac gene under the control of pmcrB | Eurofins Genomics Life Science Services, Ebersberg, Germany |
| pRS1515 | pRS1452∆ssrA+pac | This work |
| pRS1518 | pRS1452∆orf1+2+pac | This work |
| pRS1522 | pRS1452∆repA+pac | This work |
| pRS1523 | pRS1452∆nc‘‘‘+pac 1 | This work |
| pRS1531 | pRS1452∆nc‘‘+pac 1 | This work |
| pRS1532 | pRS1452∆nc‘+pac 1 | This work |
| pRS1550 | pRS1452∆29–2556, ∆5712–6149+pac | This work |
| pRS1559 | pET28a(+)repA | This work |
| pEX-K168_MCS_JT | Synthesised MCS | Eurofins Genomics Life Science Services, Ebersberg, Germany |
| pEX-K248_pac2_JT | Synthesised pac gene under the control of pmcrB | Eurofins Genomics Life Science Services, Ebersberg, Germany |
| pRS1595 | Modular shuttle vector | This work |
| pRS1625 | pET28a(+)repA_DM | This work |
| pBluescript II KS (+) | General cloning vector | Stratagene, La Jolla, CA, USA |
| pRS1694 | pBSK with 345 bp region of pWM321 (ncts. 5088–5432) | This work |
| Plasmid | Forward Primer | Reverse Primer | Purpose |
|---|---|---|---|
| pRS1452 | 5′-CGCCCGCCCCACGAC-3′ | 5′-CCTGCAGGTTTTGATGTAGTTTCTTACTAC-3′ | ∆6190–7150 from pWM321 |
| pRS1532 | 5′-GATCCCGCAGATTATGGAAC-3′ | 5′-CAATTTCACACAGGAAACAGC-3′ | ∆526–1287 from pWM321 |
| pRS1515 | 5′-GTATGTAAATAAATACTTTGTGC-3′ | 5′-GAAATAATGTTCCATAATCTGC-3′ | ∆1316–2287 from pWM321 |
| pRS1531 | 5′-TTGTCGAAGAACTTCCAAAC-3′ | 5′-TAAATGACATCTATGCACAAAG-3′ | ∆2325–2778 from pWM321 |
| pRS1518 | 5′-CGTATCACTTTAGGCTTTAAG-3′ | 5′-GATCGGTCTACTGTTTGGAAG-3′ | ∆2811–3845 from pWM321 |
| pRS1522 | 5′-GAATAAGATTAACGCCTACC-3′ | 5′-CGTTCAACAAGGCTTTTG-3′ | ∆3908–5688 from pWM321 |
| pRS1523 | 5′-CACTATCAAATGACATTGTAGTAAG-3′ | 5′-TAAGGTAGGCGTTAATCTTATTC-3′ | ∆5712–6149 from pWM321 |
| pRS1550 | 5′-GTCACAACATTCACAAAAATAG-3′ 5′-CACTATCAAATGACATTGTAGTAAG-3′ | 5′-CCTGAATGGCGAATGGTTAAGG-3′ 5′-TAAGGTAGGCGTTAATCTTATTC-3′ | ∆29–2556 and ∆5712–6149 from pWM321 |
| pRS1571 | 5′-GCGATCGCAACCTGCAGGTTCACTG-3′ | 5′-GATGTAGTTTCTTACTACAATGTC-3′ | Introduce AsiSI site in pRS1550 |
| pRS1577 | 5′-GGCGCGCCTTAACTAGTCGCCATTCAG-3′ | 5′-GGTGGCACTGGCCGGCCAAATGTGCGCG-3′ | Introduce FseI/ SpeI site in pRS1550 |
| pRS1559 | 5′-CGACAGGAAATGTCATATGAGTTCTGATTTTAG-3′ | 5′-GCCCAAAAGCCTTCTCGAGCGTGGCATCTC-3′ | RepA overexpression |
| pRS1625 | 5′-GAGAGAAAAGATAAGTCACCTGC-3′ | 5′-CTTTTCTCTCACGTTGGGCAG-3′ | RepA _DM overexpression |
| pRS1694 | 5′-GTTTTGGGCCCGGTTCGC-3′ | 5′-CTCCCGGGCCCAAGTCCATCGAAGC-3′ | Introduce putative DSO/ SSO in pBSK |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomsen, J.; Schmitz, R.A. Generating a Small Shuttle Vector for Effective Genetic Engineering of Methanosarcina mazei Allowed First Insights in Plasmid Replication Mechanism in the Methanoarchaeon. Int. J. Mol. Sci. 2022, 23, 11910. https://doi.org/10.3390/ijms231911910
Thomsen J, Schmitz RA. Generating a Small Shuttle Vector for Effective Genetic Engineering of Methanosarcina mazei Allowed First Insights in Plasmid Replication Mechanism in the Methanoarchaeon. International Journal of Molecular Sciences. 2022; 23(19):11910. https://doi.org/10.3390/ijms231911910
Chicago/Turabian StyleThomsen, Johanna, and Ruth A. Schmitz. 2022. "Generating a Small Shuttle Vector for Effective Genetic Engineering of Methanosarcina mazei Allowed First Insights in Plasmid Replication Mechanism in the Methanoarchaeon" International Journal of Molecular Sciences 23, no. 19: 11910. https://doi.org/10.3390/ijms231911910
APA StyleThomsen, J., & Schmitz, R. A. (2022). Generating a Small Shuttle Vector for Effective Genetic Engineering of Methanosarcina mazei Allowed First Insights in Plasmid Replication Mechanism in the Methanoarchaeon. International Journal of Molecular Sciences, 23(19), 11910. https://doi.org/10.3390/ijms231911910







