Assessment of DDAH1 and DDAH2 Contributions to Psychiatric Disorders via In Silico Methods
Abstract
1. Introduction
2. Results
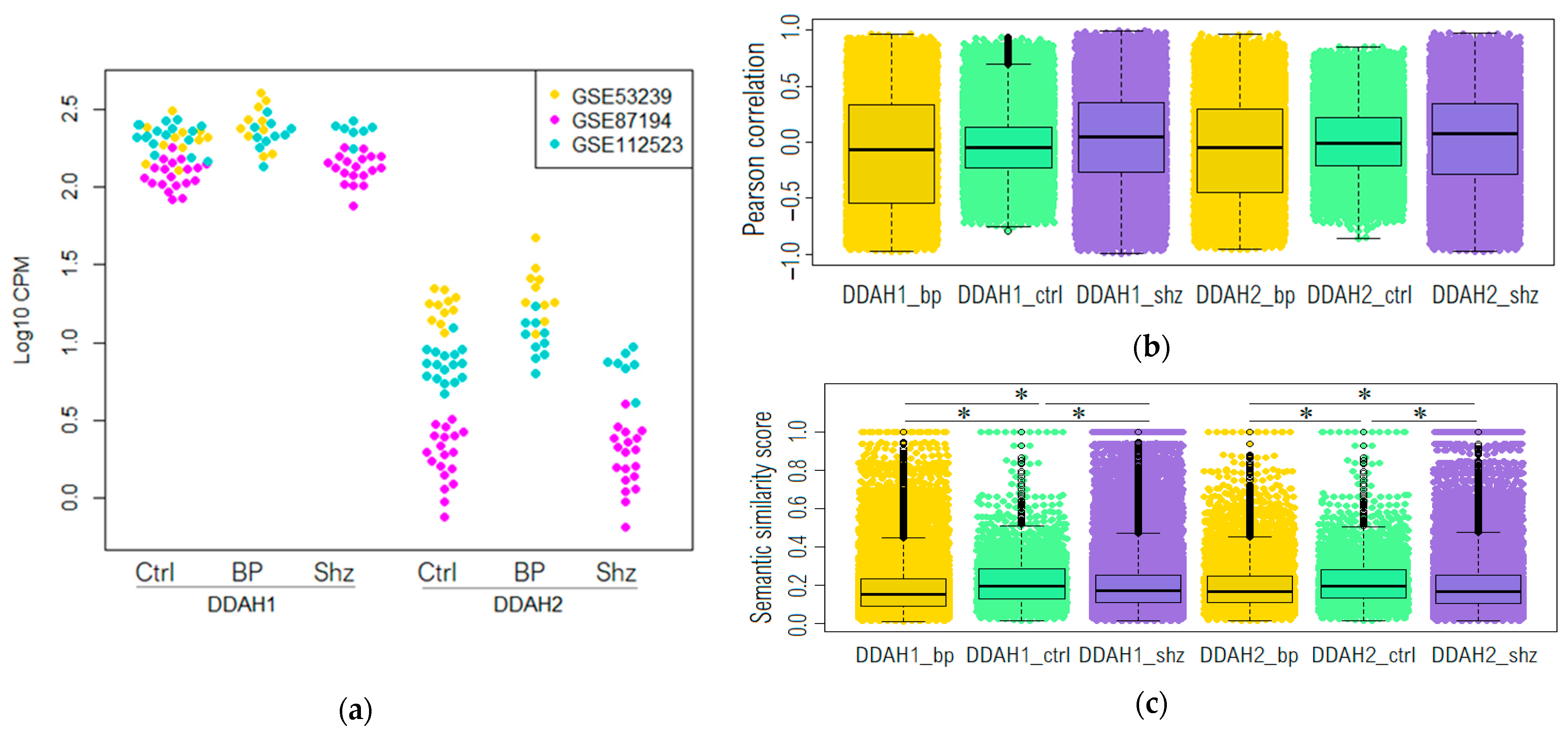
2.1. DDAH1 and DDAH2 mRNAs Are Represented in the Dorsolateral Prefrontal Cortex in Non-Psychiatric Controls and Psychotic Patients
2.2. Genome-Wide Co-Expression Analysis of DDAH1 and DDAH2 Co-Expressed Genes
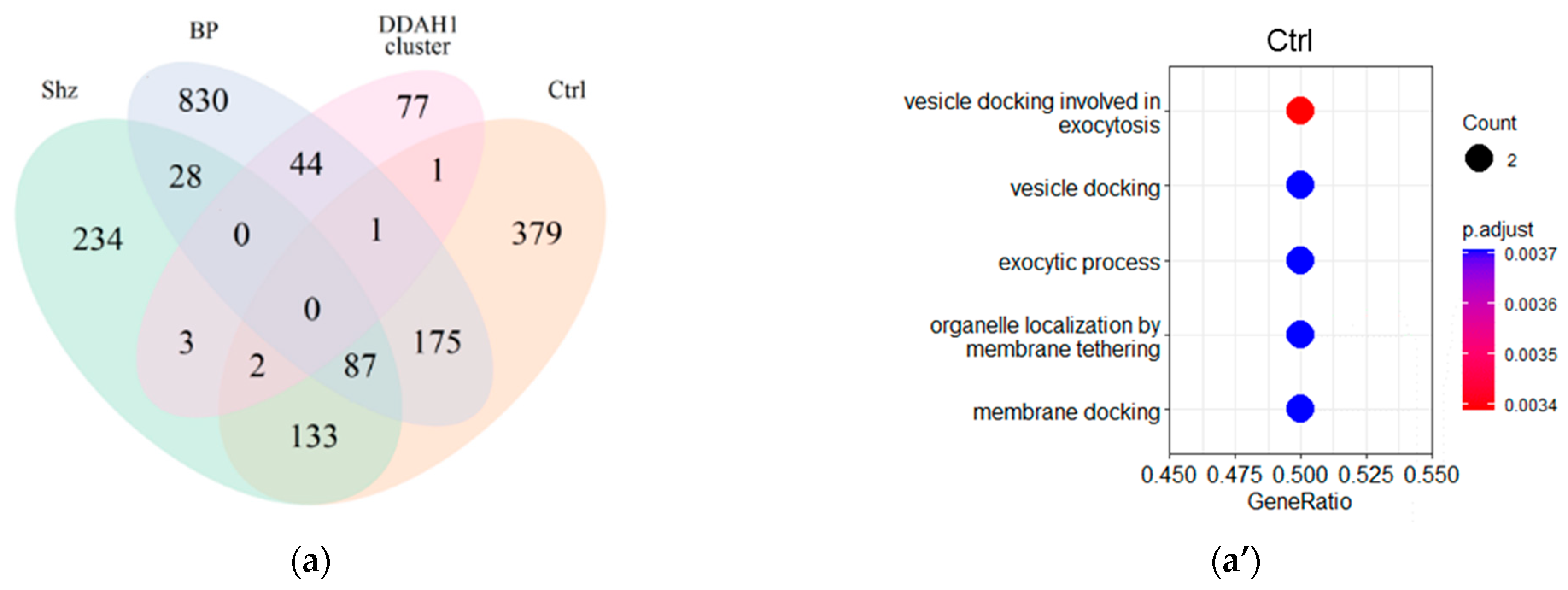
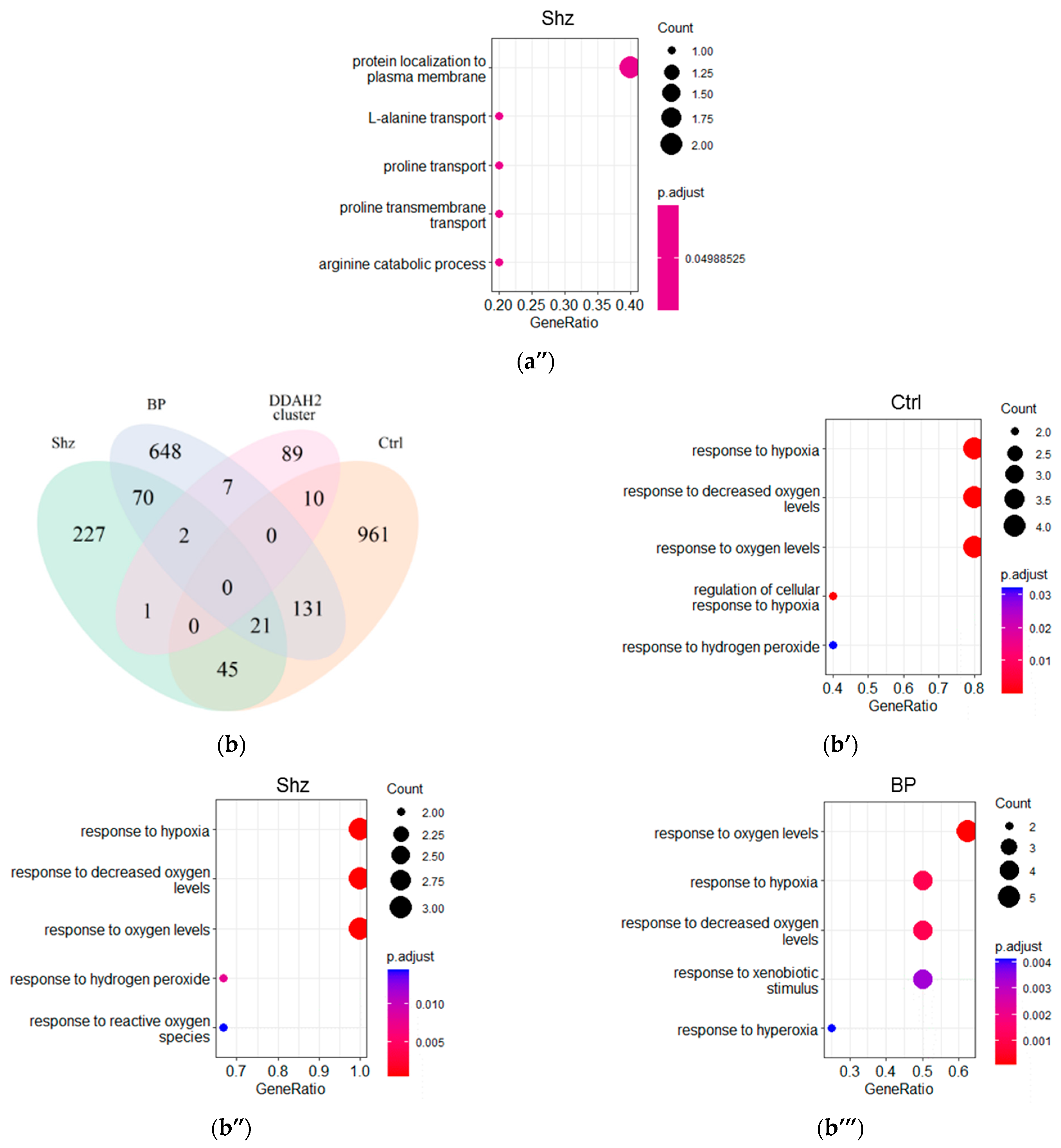
2.3. Functional Analysis of DDAH1 and DDAH2 Co-Expressed Genes
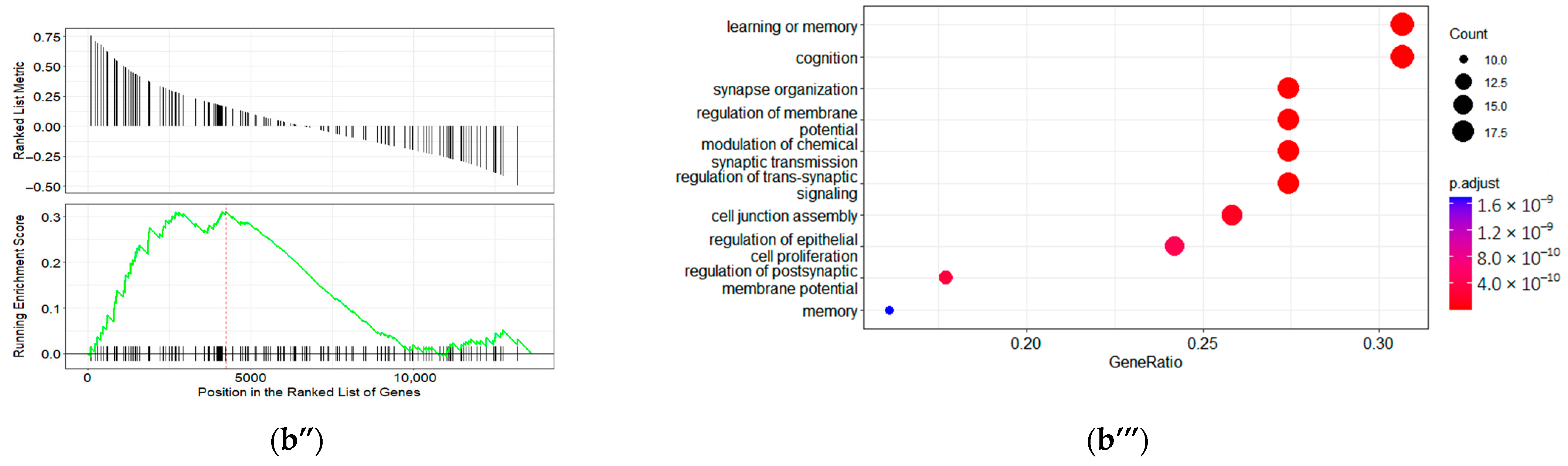
2.4. GO Term Enrichment Results
2.5. Identification of Enriched Transcription Factors and Other Protein Binding Motives in Promoters of DDAH1/DDAH2 Co-Expressed Genes
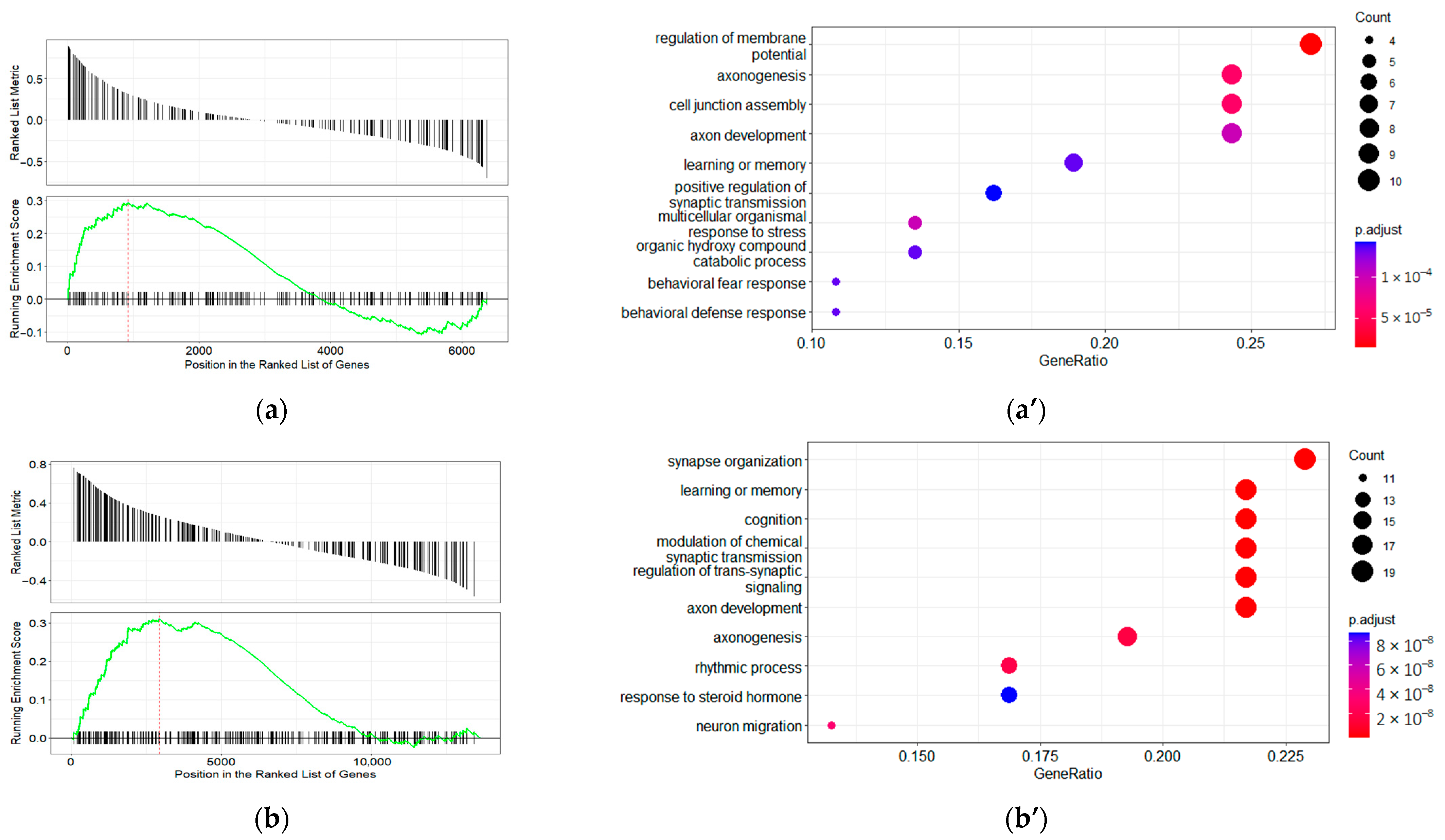
2.6. Disease Ontology Gene Set Enrichment Analysis
3. Discussion
4. Materials and Methods
4.1. Public Resources and Databases
4.2. Data Normalization and Statistical Analysis
4.3. Measurement of Co-Expression
4.4. Analysis of Functional Semantic Similarity between Genes
4.5. Function and Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- GBD 2019 Mental Disorders Collaborators. Global, Regional, and National Burden of 12 Mental Disorders in 204 Countries and Territories, 1990–2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Lancet Psychiatry 2022, 9, 137–150. [Google Scholar] [CrossRef]
- McNeill, R.V.; Kehrwald, C.; Brum, M.; Knopf, K.; Brunkhorst-Kanaan, N.; Etyemez, S.; Koreny, C.; Bittner, R.A.; Freudenberg, F.; Herterich, S.; et al. Uncovering Associations between Mental Illness Diagnosis, Nitric Oxide Synthase Gene Variation, and Peripheral Nitric Oxide Concentration. Brain. Behav. Immun. 2022, 101, 275–283. [Google Scholar] [CrossRef]
- Jagannathan, K.; Calhoun, V.D.; Gelernter, J.; Stevens, M.C.; Liu, J.; Bolognani, F.; Windemuth, A.; Ruaño, G.; Assaf, M.; Pearlson, G.D. Genetic Associations of Brain Structural Networks in Schizophrenia: A Preliminary Study. Biol. Psychiatry 2010, 68, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Kittel-Schneider, S.; Reuß, M.; Meyer, A.; Weber, H.; Gessner, A.; Leistner, C.; Kopf, J.; Schmidt, B.; Hempel, S.; Volkert, J.; et al. Multi-Level Biomarker Analysis of Nitric Oxide Synthase Isoforms in Bipolar Disorder and Adult ADHD. J. Psychopharmacol. 2015, 29, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Minutolo, G.; Petralia, A.; Dipasquale, S.; Aguglia, E. Nitric Oxide in Patients with Schizophrenia: The Relationship with the Severity of Illness and the Antipsychotic Treatment. Expert Opin. Pharmacother. 2012, 13, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Morales-Medina, J.C.; Aguilar-Alonso, P.; Di Cerbo, A.; Iannitti, T.; Flores, G. New Insights on Nitric Oxide: Focus on Animal Models of Schizophrenia. Behav. Brain Res. 2021, 409, 113304. [Google Scholar] [CrossRef] [PubMed]
- Garthwaite, J.; Garthwaite, G.; Palmer, R.M.; Moncada, S. NMDA Receptor Activation Induces Nitric Oxide Synthesis from Arginine in Rat Brain Slices. Eur. J. Pharmacol. 1989, 172, 413–416. [Google Scholar] [CrossRef]
- Kiss, J.P.; Vizi, E.S. Nitric Oxide: A Novel Link between Synaptic and Nonsynaptic Transmission. Trends Neurosci. 2001, 24, 211–215. [Google Scholar] [CrossRef]
- Nasyrova, R.F.; Ivashchenko, D.V.; Ivanov, M.V.; Neznanov, N.G. Role of Nitric Oxide and Related Molecules in Schizophrenia Pathogenesis: Biochemical, Genetic and Clinical Aspects. Front. Physiol. 2015, 6, 139. [Google Scholar] [CrossRef]
- Liu, P.; Jing, Y.; Collie, N.D.; Dean, B.; Bilkey, D.K.; Zhang, H. Altered Brain Arginine Metabolism in Schizophrenia. Transl. Psychiatry. 2016, 6, e871. [Google Scholar] [CrossRef]
- Quan, L.; Uyeda, A.; Muramatsu, R. Central Nervous System Regeneration: The Roles of Glial Cells in the Potential Molecular Mechanism Underlying Remyelination. Inflamm. Regen. 2022, 42, 7. [Google Scholar] [CrossRef]
- Rose, E.J.; Greene, C.; Kelly, S.; Morris, D.W.; Robertson, I.H.; Fahey, C.; Jacobson, S.; O’Doherty, J.; Newell, F.N.; McGrath, J.; et al. The NOS1 Variant Rs6490121 Is Associated with Variation in Prefrontal Function and Grey Matter Density in Healthy Individuals. NeuroImage 2012, 60, 614–622. [Google Scholar] [CrossRef]
- Freudenberg, F.; Alttoa, A.; Reif, A. Neuronal Nitric Oxide Synthase (NOS1) and Its Adaptor, NOS1AP, as a Genetic Risk Factors for Psychiatric Disorders. Genes Brain Behav. 2015, 14, 46–63. [Google Scholar] [CrossRef]
- Ginsberg, S.D.; Hemby, S.E.; Smiley, J.F. Expression Profiling in Neuropsychiatric Disorders: Emphasis on Glutamate Receptors in Bipolar Disorder. Pharmacol. Biochem. Behav. 2012, 100, 705–711. [Google Scholar] [CrossRef] [PubMed]
- González-Castro, T.B.; Genis-Mendoza, A.D.; Tovilla-Zárate, C.A.; Juárez-Rojop, I.E.; López-Narvaez, M.L.; Pérez-Hernández, N.; Rodríguez-Pérez, J.M.; Martínez-Magaña, J.J. Association between Polymorphisms of NOS1, NOS2 and NOS3 Genes and Suicide Behavior: A Systematic Review and Meta-Analysis. Metab. Brain Dis. 2019, 34, 967–977. [Google Scholar] [CrossRef]
- Oliveira, J.; Debnath, M.; Etain, B.; Bennabi, M.; Hamdani, N.; Lajnef, M.; Bengoufa, D.; Fortier, C.; Boukouaci, W.; Bellivier, F.; et al. Violent Suicidal Behaviour in Bipolar Disorder Is Associated with Nitric Oxide Synthase 3 Gene Polymorphism. Acta Psychiatr. Scand. 2015, 132, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Reif, A.; Herterich, S.; Strobel, A.; Ehlis, A.-C.; Saur, D.; Jacob, C.P.; Wienker, T.; Töpner, T.; Fritzen, S.; Walter, U.; et al. A Neuronal Nitric Oxide Synthase (NOS-I) Haplotype Associated with Schizophrenia Modifies Prefrontal Cortex Function. Mol. Psychiatry 2006, 11, 286–300. [Google Scholar] [CrossRef] [PubMed]
- Reif, A.; Strobel, A.; Jacob, C.P.; Herterich, S.; Freitag, C.M.; Töpner, T.; Mössner, R.; Fritzen, S.; Schmitt, A.; Lesch, K.-P. A NOS-III Haplotype That Includes Functional Polymorphisms Is Associated with Bipolar Disorder. Int. J. Neuropsychopharmacol. 2006, 9, 13–20. [Google Scholar] [CrossRef]
- Sarginson, J.E.; Deakin, J.W.; Anderson, I.M.; Downey, D.; Thomas, E.; Elliott, R.; Juhasz, G. Neuronal Nitric Oxide Synthase (NOS1) Polymorphisms Interact with Financial Hardship to Affect Depression Risk. Neuropsychopharmacology 2014, 39, 2857–2866. [Google Scholar] [CrossRef]
- Wigner, P.; Czarny, P.; Synowiec, E.; Bijak, M.; Białek, K.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Variation of Genes Involved in Oxidative and Nitrosative Stresses in Depression. Eur. Psychiatry 2018, 48, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Wockner, L.F.; Noble, E.P.; Lawford, B.R.; Young, R.M.; Morris, C.P.; Whitehall, V.L.J.; Voisey, J. Genome-Wide DNA Methylation Analysis of Human Brain Tissue from Schizophrenia Patients. Transl. Psychiatry 2014, 4, e339. [Google Scholar] [CrossRef]
- Weber, H.; Klamer, D.; Freudenberg, F.; Kittel-Schneider, S.; Rivero, O.; Scholz, C.-J.; Volkert, J.; Kopf, J.; Heupel, J.; Herterich, S.; et al. The Genetic Contribution of the NO System at the Glutamatergic Post-Synapse to Schizophrenia: Further Evidence and Meta-Analysis. Eur. Neuropsychopharmacol. 2014, 24, 65–85. [Google Scholar] [CrossRef]
- Bruenig, D.; Morris, C.P.; Mehta, D.; Harvey, W.; Lawford, B.; Young, R.M.; Voisey, J. Nitric Oxide Pathway Genes (NOS1AP and NOS1) Are Involved in PTSD Severity, Depression, Anxiety, Stress and Resilience. Gene 2017, 625, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Anirudh, C.S.; Pathak, A.K.; Sinha, P.; Jainarayanan, A.K.; Jain, S.; Brahmachari, S.K. Multi-Scale Analysis of Schizophrenia Risk Loci: Integrating Centenarian Genomes and Spatio-Temporal Expression Profiles Suggests the Need for Adjunctive Therapeutic Interventions for Neuropsychiatric Disorders. bioRxiv 2018. [Google Scholar] [CrossRef]
- Shinkai, T.; Ohmori, O.; Hori, H.; Nakamura, J. Allelic Association of the Neuronal Nitric Oxide Synthase (NOS1) Gene with Schizophrenia. Mol. Psychiatry 2002, 7, 560–563. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhou, Q.-G.; Zhu, X.-H.; Nemes, A.D.; Zhu, D.-Y. Neuronal Nitric Oxide Synthase and Affective Disorders. IBRO Rep. 2018, 5, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Tran, C.T.L.; Fox, M.F.; Vallance, P.; Leiper, J.M. Chromosomal Localization, Gene Structure, and Expression Pattern of DDAH1: Comparison with DDAH2 and Implications for Evolutionary Origins. Genomics 2000, 68, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Kozlova, A.A.; Ragavan, V.N.; Jarzebska, N.; Lukianova, I.V.; Bikmurzina, A.E.; Rubets, E.; Suzuki-Yamamoto, T.; Kimoto, M.; Mangoni, A.A.; Gainetdinov, R.R.; et al. Divergent Dimethylarginine Dimethylaminohydrolase Isoenzyme Expression in the Central Nervous System. Cell Mol. Neurobiol. 2021, 42, 2273–2288. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Samuels, J.F.; Wang, Y.; Cao, H.; Ritter, M.; Nestadt, P.S.; Krasnow, J.; Greenberg, B.D.; Fyer, A.J.; McCracken, J.T.; et al. Polygenic Risk Score and Heritability Estimates Reveals a Genetic Relationship between ASD and OCD. Eur. Neuropsychopharmacol. 2017, 27, 657–666. [Google Scholar] [CrossRef]
- Cieślik, P.; Siekierzycka, A.; Radulska, A.; Płoska, A.; Burnat, G.; Brański, P.; Kalinowski, L.; Wierońska, J.M. Nitric Oxide-Dependent Mechanisms Underlying MK-801- or Scopolamine-Induced Memory Dysfunction in Animals: Mechanistic Studies. Int. J. Mol. Sci. 2021, 22, 12282. [Google Scholar] [CrossRef] [PubMed]
- Cortelazzo, A.; De Felice, C.; Guy, J.; Timperio, A.M.; Zolla, L.; Guerranti, R.; Leoncini, S.; Signorini, C.; Durand, T.; Hayek, J. Brain Protein Changes in Mecp2 Mouse Mutant Models: Effects on Disease Progression of Mecp2 Brain Specific Gene Reactivation. J. Proteom. 2020, 210, 103537. [Google Scholar] [CrossRef] [PubMed]
- Whittle, N.; Li, L.; Chen, W.-Q.; Yang, J.-W.; Sartori, S.B.; Lubec, G.; Singewald, N. Changes in Brain Protein Expression Are Linked to Magnesium Restriction-Induced Depression-like Behavior. Amino Acids 2011, 40, 1231–1248. [Google Scholar] [CrossRef]
- Clark, D.; Dedova, I.; Cordwell, S.; Matsumoto, I. A Proteome Analysis of the Anterior Cingulate Cortex Gray Matter in Schizophrenia. Mol. Psychiatry 2006, 11, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Narayan, S.; Tang, B.; Head, S.R.; Gilmartin, T.J.; Sutcliffe, J.G.; Dean, B.; Thomas, E.A. Molecular Profiles of Schizophrenia in the CNS at Different Stages of Illness. Brain Res. 2008, 1239, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Hass, J.; Walton, E.; Wright, C.; Beyer, A.; Scholz, M.; Turner, J.; Liu, J.; Smolka, M.N.; Roessner, V.; Sponheim, S.R.; et al. Associations between DNA Methylation and Schizophrenia-Related Intermediate Phenotypes—A Gene Set Enrichment Analysis. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2015, 59, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Cirera, L.; Cabana-Domínguez, J.; Lee, P.H.; Fernàndez-Castillo, N.; Cormand, B. Identification of Genetic Variants Influencing Methylation in Brain with Pleiotropic Effects on Psychiatric Disorders. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2022, 113, 110454. [Google Scholar] [CrossRef]
- Wu, X.; Ye, J.; Wang, Z.; Zhao, C. Epigenetic Age Acceleration Was Delayed in Schizophrenia. Schizophr. Bull. 2021, 47, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Bani-Fatemi, A.; Jeremian, R.; Wang, K.Z.; Silveira, J.; Zai, C.; Kolla, N.J.; Graff, A.; Gerretsen, P.; Strauss, J.; De Luca, V. Epigenome-Wide Association Study of Suicide Attempt in Schizophrenia. J. Psychiatr. Res. 2018, 104, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Reif, A.; Schecklmann, M.; Eirich, E.; Jacob, C.P.; Jarczok, T.A.; Kittel-Schneider, S.; Lesch, K.-P.; Fallgatter, A.J.; Ehlis, A.-C. A Functional Promoter Polymorphism of Neuronal Nitric Oxide Synthase Moderates Prefrontal Functioning in Schizophrenia. Int. J. Neuropsychopharmacol. 2011, 14, 887–897. [Google Scholar] [CrossRef]
- Connor, C.M.; Crawford, B.C.; Akbarian, S. White Matter Neuron Alterations in Schizophrenia and Related Disorders. Int. J. Dev. Neurosci. 2011, 29, 325–334. [Google Scholar] [CrossRef]
- Pai, S.; Li, P.; Killinger, B.; Marshall, L.; Jia, P.; Liao, J.; Petronis, A.; Szabó, P.E.; Labrie, V. Differential methylation of enhancer at IGF2 is associated with abnormal dopamine synthesis in major psychosis. Nat. Commun. 2019, 10, 2046. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID Database: A Comprehensive Biomedical Resource of Curated Protein, Genetic, and Chemical Interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Licata, L.; Briganti, L.; Peluso, D.; Perfetto, L.; Iannuccelli, M.; Galeota, E.; Sacco, F.; Palma, A.; Nardozza, A.P.; Santonico, E.; et al. MINT, the Molecular Interaction Database: 2012 Update. Nucleic Acids Res. 2012, 40, D857–D861. [Google Scholar] [CrossRef]
- Keshava Prasad, T.S.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database—2009 Update. Nucleic Acids Res. 2009, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Peri, S.; Navarro, J.D.; Amanchy, R.; Kristiansen, T.Z.; Jonnalagadda, C.K.; Surendranath, V.; Niranjan, V.; Muthusamy, B.; Gandhi, T.K.B.; Gronborg, M.; et al. Development of Human Protein Reference Database as an Initial Platform for Approaching Systems Biology in Humans. Genome Res. 2003, 13, 2363–2371. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qu, S.; Wang, W.; Guo, L.; Zhang, K.; Chang, S.; Wang, J. A Combined Analysis of Genome-Wide Expression Profiling of Bipolar Disorder in Human Prefrontal Cortex. J. Psychiatr. Res. 2016, 82, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.-F.; Qi, X.-R.; Zhao, J.; Balesar, R.; Bao, A.-M.; Swaab, D.F. Decreased NOS1 Expression in the Anterior Cingulate Cortex in Depression. Cereb. Cortex 2013, 23, 2956–2964. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster Analysis and Display of Genome-Wide Expression Patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Furlotte, N.A.; Kang, H.M.; Ye, C.; Eskin, E. Mixed-Model Coexpression: Calculating Gene Coexpression While Accounting for Expression Heterogeneity. Bioinformatics 2011, 27, i288–i294. [Google Scholar] [CrossRef]
- Gan, M. Correlating Information Contents of Gene Ontology Terms to Infer Semantic Similarity of Gene Products. Comput. Math. Methods Med. 2014, 2014, e891842. [Google Scholar] [CrossRef] [PubMed]
- Dho, S.E.; Silva-Gagliardi, N.; Morgese, F.; Coyaud, E.; Lamoureux, E.; Berry, D.M.; Raught, B.; McGlade, C.J. Proximity interactions of the ubiquitin ligase Mind bomb 1 reveal a role in regulation of epithelial polarity complex proteins. Sci. Rep. 2019, 9, 12471. [Google Scholar] [CrossRef]
- Sumithra, B.; Saxena, U.; Das, A.B. A comprehensive study on genome-wide coexpression network of KHDRBS1/Sam68 reveals its cancer and patient-specific association. Sci Rep 2019, 9, 11083. [Google Scholar] [CrossRef]
- Martins, T.; Burgoyne, T.; Kenny, B.-A.; Hudson, N.; Futter, C.E.; Ambrósio, A.F.; Silva, A.P.; Greenwood, J.; Turowski, P. Methamphetamine-Induced Nitric Oxide Promotes Vesicular Transport in Blood–Brain Barrier Endothelial Cells. Neuropharmacology 2013, 65, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Picón-Pagès, P.; Garcia-Buendia, J.; Muñoz, F.J. Functions and Dysfunctions of Nitric Oxide in Brain. Biochim. Biophys. Acta BBA Mol. Basis Dis. 2019, 1865, 1949–1967. [Google Scholar] [CrossRef] [PubMed]
- Pong, S.; Lizano, P.; Karmacharya, R. Investigating Blood-Brain Barrier Dysfunction in Schizophrenia Using Brain Microvascular Endothelial Cells Derived From Patient-Specific Stem Cells. Biol. Psychiatry 2020, 87, S189–S190. [Google Scholar] [CrossRef]
- Najjar, S.; Pahlajani, S.; De Sanctis, V.; Stern, J.N.H.; Najjar, A.; Chong, D. Neurovascular Unit Dysfunction and Blood–Brain Barrier Hyperpermeability Contribute to Schizophrenia Neurobiology: A Theoretical Integration of Clinical and Experimental Evidence. Front Psychiatry 2017, 8, 83. [Google Scholar] [CrossRef]
- Lambden, S.; Martin, D.; Tomlinson, J.; Mythen, M.; Leiper, J. Role of Dimethylarginine Dimethylaminohydrolase 2 in the Regulation of Nitric Oxide Synthesis in Animal and Observational Human Models of Normobaric Hypoxia. Lancet 2016, 387, S62. [Google Scholar] [CrossRef]
- Lambden, S.; Martin, D.; Vanezis, K.; Lee, B.; Tomlinson, J.; Piper, S.; Boruc, O.; Mythen, M.; Leiper, J. Hypoxia Causes Increased Monocyte Nitric Oxide Synthesis Which Is Mediated by Changes in Dimethylarginine Dimethylaminohydrolase 2 Expression in Animal and Human Models of Normobaric Hypoxia. Nitric Oxide 2016, 58, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Harbaum, L.; Glatzel, A.; Klose, H.; Böger, R.H.; Lüneburg, N. Modulation of Symmetric Dimethyarginine Formation by Apelin in Human Pulmonary Endothelial Cells. Eur. Respir. J. 2015, 46, PA859. [Google Scholar] [CrossRef]
- Williams, G.; Shi-Wen, X.; Abraham, D.; Selvakumar, S.; Baker, D.M.; Tsui, J.C.S. Nitric Oxide Manipulation: A Therapeutic Target for Peripheral Arterial Disease? Cardiol. Res. Pract. 2012, 2012, e656247. [Google Scholar] [CrossRef] [PubMed]
- Turek, M.; Lewandrowski, I.; Bringmann, H. An AP2 Transcription Factor Is Required for a Sleep-Active Neuron to Induce Sleep-like Quiescence in C. Elegans. Curr. Biol. 2013, 23, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Korovaichuk, A.; Astiz, M.; Schroeder, H.; Islam, R.; Barrenetxea, J.; Fischer, A.; Oster, H.; Bringmann, H. Functional Divergence of Mammalian TFAP2a and TFAP2b Transcription Factors for Bidirectional Sleep Control. Genetics 2020, 216, 735–752. [Google Scholar] [CrossRef] [PubMed]
- Hensch, T.; Wozniak, D.; Spada, J.; Sander, C.; Ulke, C.; Wittekind, D.A.; Thiery, J.; Löffler, M.; Jawinski, P.; Hegerl, U. Vulnerability to Bipolar Disorder Is Linked to Sleep and Sleepiness. Transl. Psychiatry 2019, 9, 294. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liu, K.; Lei, M.; Yang, A.; Li, Y.; Hughes, T.R.; Min, J. DNA Sequence Recognition of Human CXXC Domains and Their Structural Determinants. Structure 2018, 26, 85–95.e3. [Google Scholar] [CrossRef]
- Lee, J.-H.; Voo, K.S.; Skalnik, D.G. Identification and Characterization of the DNA Binding Domain of CpG-Binding Protein. J. Biol. Chem. 2001, 276, 44669–44676. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, Y. Mechanisms and Functions of Tet Protein-Mediated 5-Methylcytosine Oxidation. Genes Dev. 2011, 25, 2436–2452. [Google Scholar] [CrossRef]
- Dong, E.; Gavin, D.P.; Chen, Y.; Davis, J. Upregulation of TET1 and Downregulation of APOBEC3A and APOBEC3C in the Parietal Cortex of Psychotic Patients. Transl. Psychiatry 2012, 2, e159. [Google Scholar] [CrossRef]
- Gaboli, M.; Kotsi, P.A.; Gurrieri, C.; Cattoretti, G.; Ronchetti, S.; Cordon-Cardo, C.; Broxmeyer, H.E.; Hromas, R.; Pandolfi, P.P. Mzf1 Controls Cell Proliferation and Tumorigenesis. Genes Dev. 2001, 15, 1625–1630. [Google Scholar] [CrossRef]
- Bellora, N.; Farré, D.; Albà, M.M. Positional Bias of General and Tissue-Specific Regulatory Motifs in Mouse Gene Promoters. BMC Genom. 2007, 8, 459. [Google Scholar] [CrossRef]
- Rowe, D.D.; Leonardo, C.C.; Hall, A.A.; Shahaduzzaman, M.D.; Collier, L.A.; Willing, A.E.; Pennypacker, K.R. Cord Blood Administration Induces Oligodendrocyte Survival through Alterations in Gene Expression. Brain Res. 2010, 1366, 172–188. [Google Scholar] [CrossRef]
- Shahaduzzaman, M.D.; Mehta, V.; Golden, J.E.; Rowe, D.D.; Green, S.; Tadinada, R.; Foran, E.A.; Sanberg, P.R.; Pennypacker, K.R.; Willing, A.E. Human Umbilical Cord Blood Cells Induce Neuroprotective Change in Gene Expression Profile in Neurons after Ischemia through Activation of Akt Pathway. Cell Transplant. 2015, 24, 721–735. [Google Scholar] [CrossRef]
- Chandran, V.; Coppola, G.; Nawabi, H.; Omura, T.; Versano, R.; Huebner, E.A.; Zhang, A.; Costigan, M.; Yekkirala, A.; Barrett, L.; et al. A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron 2016, 89, 956–970. [Google Scholar] [CrossRef]
- Zhaonan, Z.; Tazro, O.; Fumihito, M.; Shinya, O. ChIP-Atlas 2021 Update: A Data-Mining Suite for Exploring Epigenomic Landscapes by Fully Integrating ChIP-Seq, ATAC-Seq and Bisulfite-Seq Data. Nucleic Acids Res. 2022, 50, W175–W182. [Google Scholar] [CrossRef]
- Ochsner, S.A.; Abraham, D.; Martin, K.; Ding, W.; McOwiti, A.; Kankanamge, W.; Wang, Z.; Andreano, K.; Hamilton, R.A.; Chen, Y.; et al. The Signaling Pathways Project, an Integrated ‘omics Knowledgebase for Mammalian Cellular Signaling Pathways. Sci. Data 2019, 6, 252. [Google Scholar] [CrossRef] [PubMed]
- Osborne, J.D.; Flatow, J.; Holko, M.; Lin, S.M.; Kibbe, W.A.; Zhu, L.; Danila, M.I.; Feng, G.; Chisholm, R.L. Annotating the Human Genome with Disease Ontology. BMC Genom. 2009, 10, S6. [Google Scholar] [CrossRef]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.-W.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A Backbone for Disease Semantic Integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef]
- Guan, J.; Cai, J.J.; Ji, G.; Sham, P.C. Commonality in Dysregulated Expression of Gene Sets in Cortical Brains of Individuals with Autism, Schizophrenia, and Bipolar Disorder. Transl. Psychiatry 2019, 9, 152. [Google Scholar] [CrossRef] [PubMed]
- Bora, E. Developmental Trajectory of Cognitive Impairment in Bipolar Disorder: Comparison with Schizophrenia. Eur. Neuropsychopharmacol. 2015, 25, 158–168. [Google Scholar] [CrossRef]
- Cristino, A.S.; Williams, S.M.; Hawi, Z.; An, J.-Y.; Bellgrove, M.A.; Schwartz, C.E.; Costa, L.d.F.; Claudianos, C. Neurodevelopmental and Neuropsychiatric Disorders Represent an Interconnected Molecular System. Mol. Psychiatry 2014, 19, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Nomura, J.; Mardo, M.; Takumi, T. Molecular Signatures from Multi-Omics of Autism Spectrum Disorders and Schizophrenia. J. Neurochem. 2021, 159, 647–659. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, K.S.; McGregor, N.W.; Lochner, C.; Emsley, R.; Warnich, L. The Genetic Architecture of Schizophrenia, Bipolar Disorder, Obsessive-Compulsive Disorder and Autism Spectrum Disorder. Mol. Cell. Neurosci. 2018, 88, 300–307. [Google Scholar] [CrossRef] [PubMed]
- De Silva, P.N. Do Patterns of Synaptic Pruning Underlie Psychoses, Autism and ADHD? BJPsych Adv. 2018, 24, 212–217. [Google Scholar] [CrossRef]
- Jensen, M.; Girirajan, S. Mapping a Shared Genetic Basis for Neurodevelopmental Disorders. Genome Med. 2017, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Parellada, M.; Gomez-Vallejo, S.; Burdeus, M.; Arango, C. Developmental Differences Between Schizophrenia and Bipolar Disorder. Schizophr. Bull. 2017, 43, 1176–1189. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, M. The Future of Psychiatry and Neurodevelopmental Disorders: A Paradigm Shift; IntechOpen: London, UK, 2019; ISBN 978-1-78923-826-6. [Google Scholar]
- Selley, M.L. Increased (E)-4-Hydroxy-2-Nonenal and Asymmetric Dimethylarginine Concentrations and Decreased Nitric Oxide Concentrations in the Plasma of Patients with Major Depression. J. Affect. Disord. 2004, 80, 249–256. [Google Scholar] [CrossRef]
- Telo, S.; Gurok, M.G. Asymmetric Dimethylarginine (ADMA), 4-OH-Nonenal and Vitamin E Levels in Chronic Schizophrenic Patients. Psychiatry Res. 2016, 240, 295–299. [Google Scholar] [CrossRef]
- Yang, Y.-J.; Xiong, J.-W.; Zhao, Y.; Zhan, J.-Q.; Chen, H.-B.; Yan, K.; Hu, M.-R.; Yu, B.; Wei, B. Increased Plasma Asymmetric Dimethylarginine Is Associated with Cognitive Deficits in Patients with Schizophrenia. Psychiatry Res. 2016, 246, 480–484. [Google Scholar] [CrossRef]
- Yu, Z.; Zhao, Y.; Zhan, J.; Luo, T.; Xiong, J.; Yu, B.; Wei, B.; Yang, Y. Treatment Responses of Cognitive Function and Plasma Asymmetric Dimethylarginine to Atypical Antipsychotic in Patients With Schizophrenia. Front. Psychiatry 2019, 9, 733. [Google Scholar] [CrossRef]
- Kielstein, H.; Suntharalingam, M.; Perthel, R.; Song, R.; Schneider, S.M.; Martens-Lobenhoffer, J.; Jäger, K.; Bode-Böger, S.M.; Kielstein, J.T. Role of the Endogenous Nitric Oxide Inhibitor Asymmetric Dimethylarginine (ADMA) and Brain-Derived Neurotrophic Factor (BDNF) in Depression and Behavioural Changes: Clinical and Preclinical Data in Chronic Kidney Disease. Nephrol. Dial. Transplant. 2015, 30, 1699–1705. [Google Scholar] [CrossRef]
- Fan, Y.; Gao, Q.; Guan, J.-X.; Liu, L.; Hong, M.; Jun, L.; Wang, L.; Ding, H.-F.; Jiang, L.-H.; Hou, B.-Y.; et al. DDAH2 (-449 G/C) G Allele Is Positively Associated with Leukoaraiosis in Northeastern China: A Double-Blind, Intergroup Comparison, Case-Control Study. Neural. Regen. Res. 2021, 16, 1592–1597. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Yan, C.; Gao, Q.; Li, J.; Wang, L.; Hong, M.; Zheng, X.; Song, Z.; Li, M.; Liu, M.; et al. Analysis of Risk Factors in Patients with Leukoaraiosis. Medicine 2017, 96, e6153. [Google Scholar] [CrossRef]
- Gao, Q.; Fan, Y.; Mu, L.-Y.; Ma, L.; Song, Z.-Q.; Zhang, Y.-N. S100B and ADMA in Cerebral Small Vessel Disease and Cognitive Dysfunction. J. Neurol. Sci. 2015, 354, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets--Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R Package for Measuring Semantic Similarity among GO Terms and Gene Products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.-H.; Lin, H.-H.; Li, Y.-F.; Tsai, W.-C.; Hueng, D.-Y. Clinical Significance and Systematic Expression Analysis of the Thyroid Receptor Interacting Protein 13 (TRIP13) as Human Gliomas Biomarker. Cancers 2021, 13, 2338. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Yan, G.-R.; He, Q.-Y. DOSE: An R/Bioconductor Package for Disease Ontology Semantic and Enrichment Analysis. Bioinformatics 2015, 31, 608–609. [Google Scholar] [CrossRef] [PubMed]
| Accession Number | Title | Diagnosis | Non-Psychiatric Controls (N) | Patients (N) |
|---|---|---|---|---|
| GSE53239 | RNA-sequencing of the brain transcriptome implicates dysregulation of neuroplasticity, circadian rhythms, and GTPase binding in bipolar disorder | Bipolar affective disorder | 11 | 10 |
| GSE87194 | Schizophrenia: post-mortem dorsolateral prefrontal cortex | Schizophrenia | 19 | 19 |
| GSE112523 | DNA methylation in neurons from post-mortem brains in schizophrenia and bipolar disorder | Bipolar affective disorder | 17 | 10 |
| Schizophrenia | 7 |
| Characteristics | Bipolar Disorder | Schizophrenia | Non-Psychiatric Controls | |
|---|---|---|---|---|
| n = 10 | n = 7 | n = 17 | ||
| Gender | Male | 7 | 6 | 12 |
| Female | 3 | 1 | 5 | |
| Age | Median | 47.7 | 45.1 | 45.8 |
| Range | 29–77 | 29–55 | 31–68 | |
| Smoker status | Yes | 8 | 2 | 5 |
| No | 1 | 1 | 9 | |
| Previous | 0 | 0 | 1 | |
| Unknown | 1 | 4 | 2 | |
| Antipsychotic therapy | Yes | 6 | 0 | 4 |
| No | 4 | 17 | 3 | |
| Mood stabilizer therapy | Yes | 6 | 0 | 0 |
| No | 4 | 7 | 17 | |
| Co-Expressed Genes | DDAH1 | DDAH2 | ||
|---|---|---|---|---|
| Number of Motives | Protein Families Which Bind the Enriched Motives | Number of Motives | Protein Families Which Bind the Enriched Motives | |
| Non-psychiatric controls | 1 | CxxC | 1 | C2H2 ZF |
| Bipolar affective disorder | No enrichment | 29 | AP-2, bHLH, C2H2 ZF, CxxC, E2F, GCM, Nuclear receptor, Paired box | |
| Schizophrenia | No enrichment | No enrichment | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozlova, A.A.; Vaganova, A.N.; Rodionov, R.N.; Gainetdinov, R.R.; Bernhardt, N. Assessment of DDAH1 and DDAH2 Contributions to Psychiatric Disorders via In Silico Methods. Int. J. Mol. Sci. 2022, 23, 11902. https://doi.org/10.3390/ijms231911902
Kozlova AA, Vaganova AN, Rodionov RN, Gainetdinov RR, Bernhardt N. Assessment of DDAH1 and DDAH2 Contributions to Psychiatric Disorders via In Silico Methods. International Journal of Molecular Sciences. 2022; 23(19):11902. https://doi.org/10.3390/ijms231911902
Chicago/Turabian StyleKozlova, Alena A., Anastasia N. Vaganova, Roman N. Rodionov, Raul R. Gainetdinov, and Nadine Bernhardt. 2022. "Assessment of DDAH1 and DDAH2 Contributions to Psychiatric Disorders via In Silico Methods" International Journal of Molecular Sciences 23, no. 19: 11902. https://doi.org/10.3390/ijms231911902
APA StyleKozlova, A. A., Vaganova, A. N., Rodionov, R. N., Gainetdinov, R. R., & Bernhardt, N. (2022). Assessment of DDAH1 and DDAH2 Contributions to Psychiatric Disorders via In Silico Methods. International Journal of Molecular Sciences, 23(19), 11902. https://doi.org/10.3390/ijms231911902









