Dp71 Point Mutations Induce Protein Aggregation, Loss of Nuclear Lamina Integrity and Impaired Braf35 and Ibraf Function in Neuronal Cells
Abstract
1. Introduction
2. Results
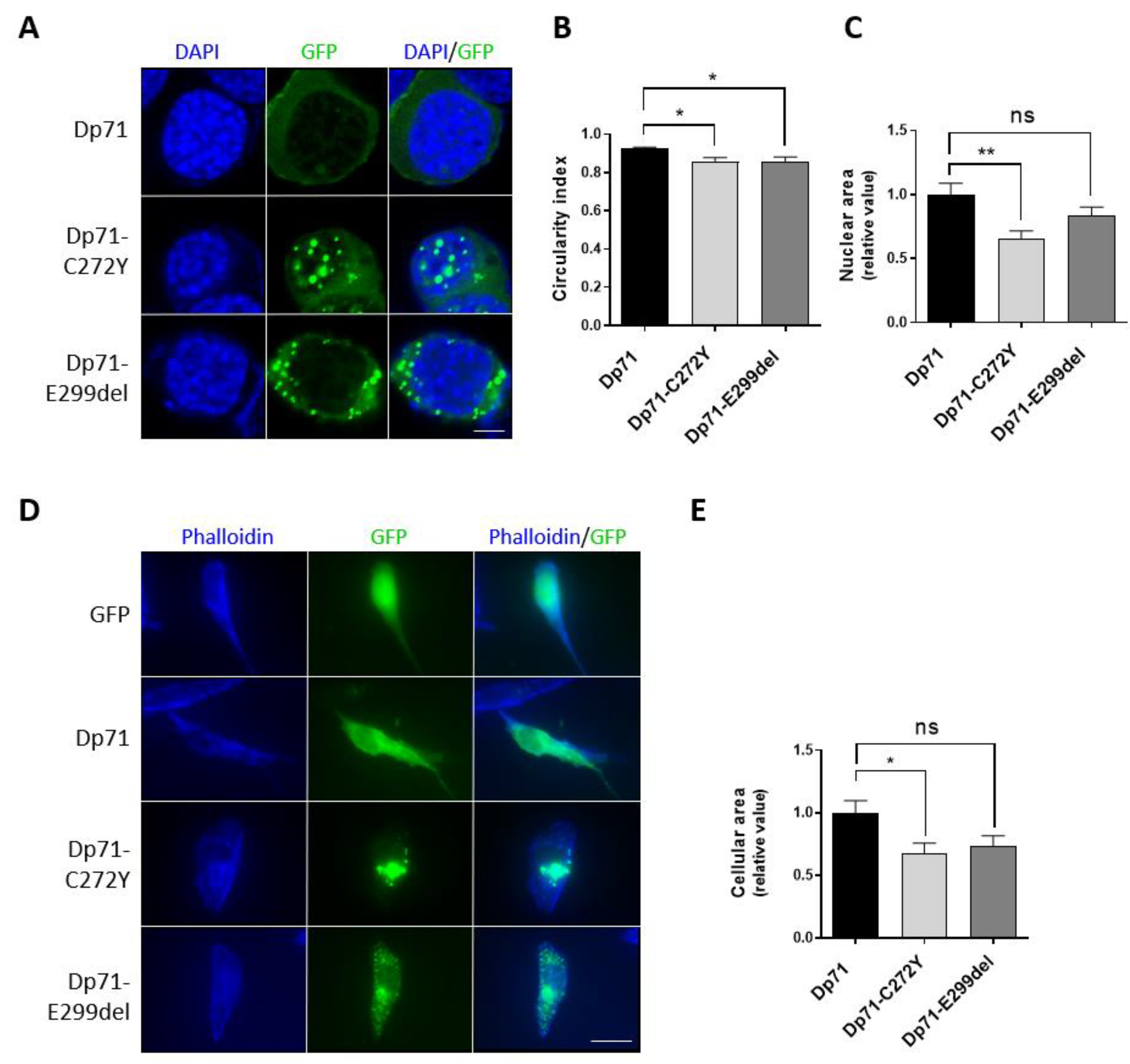
2.1. Dp71-C272Y and Dp71-E299del Mutants Form Aggregates in Neuronal Cells
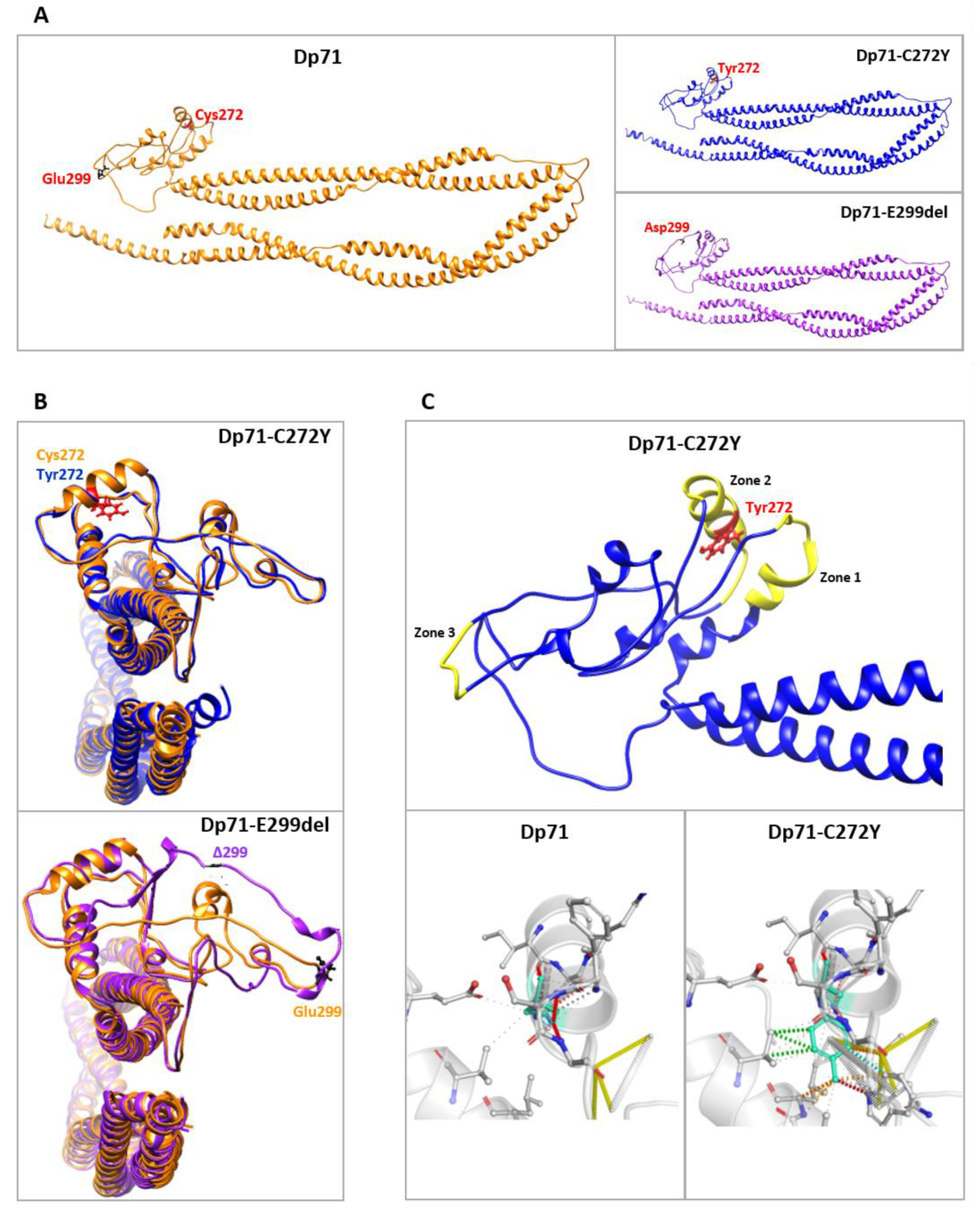
2.2. In Silico Predictions of Structure Changes in Dp71-C272Y and Dp71-E299del
2.3. Dp71-C272Y and Dp71-E299del Alter Different Morphology Parameters of Neuronal Cells
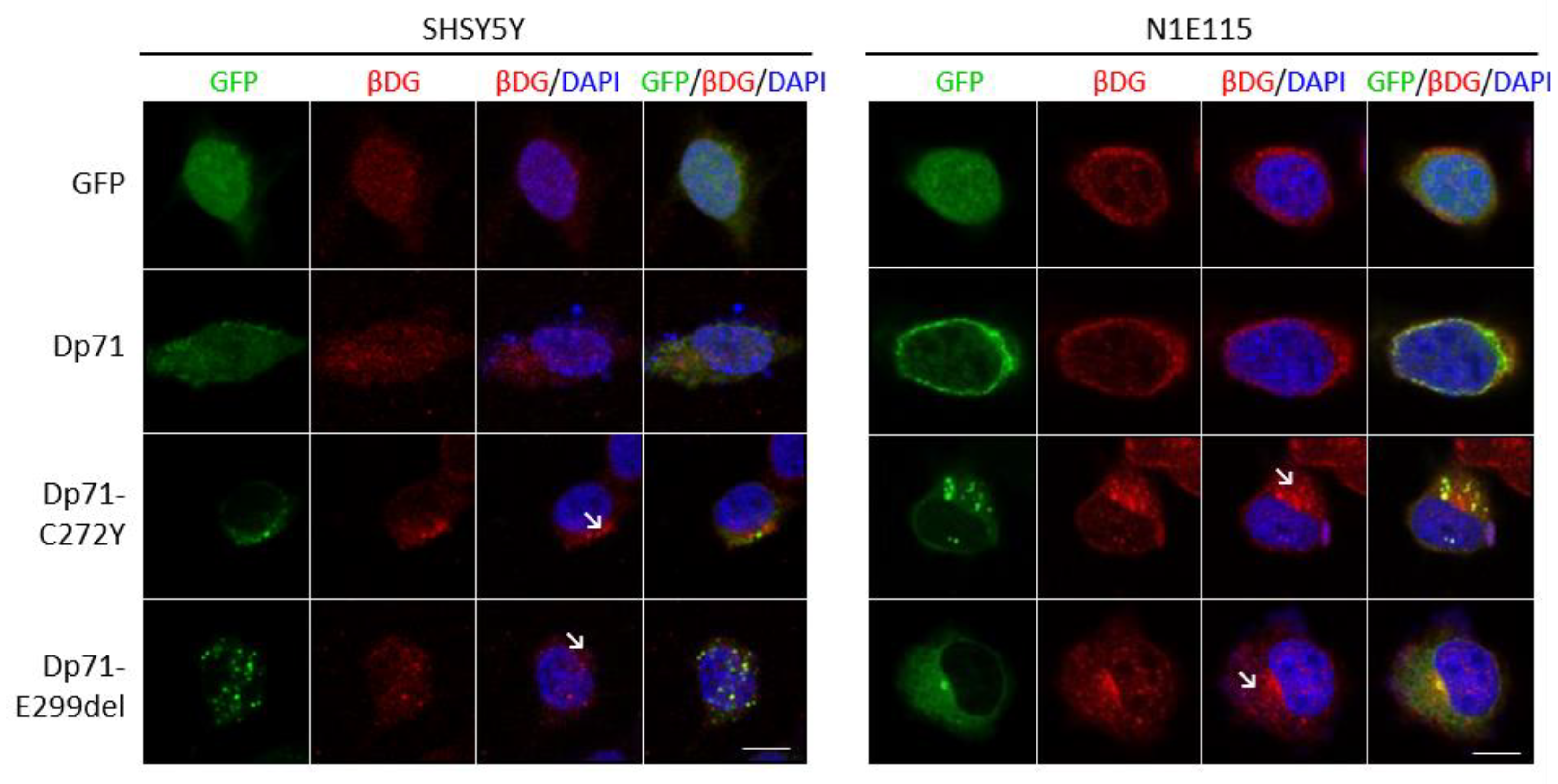
2.4. The Distribution of Β-Dg Is Affected by the Exogenous Expression of Dp71-C272Y and Dp71-E299del
2.5. Exogenous Dp71-C272Y and Dp71-E299del Expression Elicit the Mislocalization and Aggregation of Nuclear Lamins
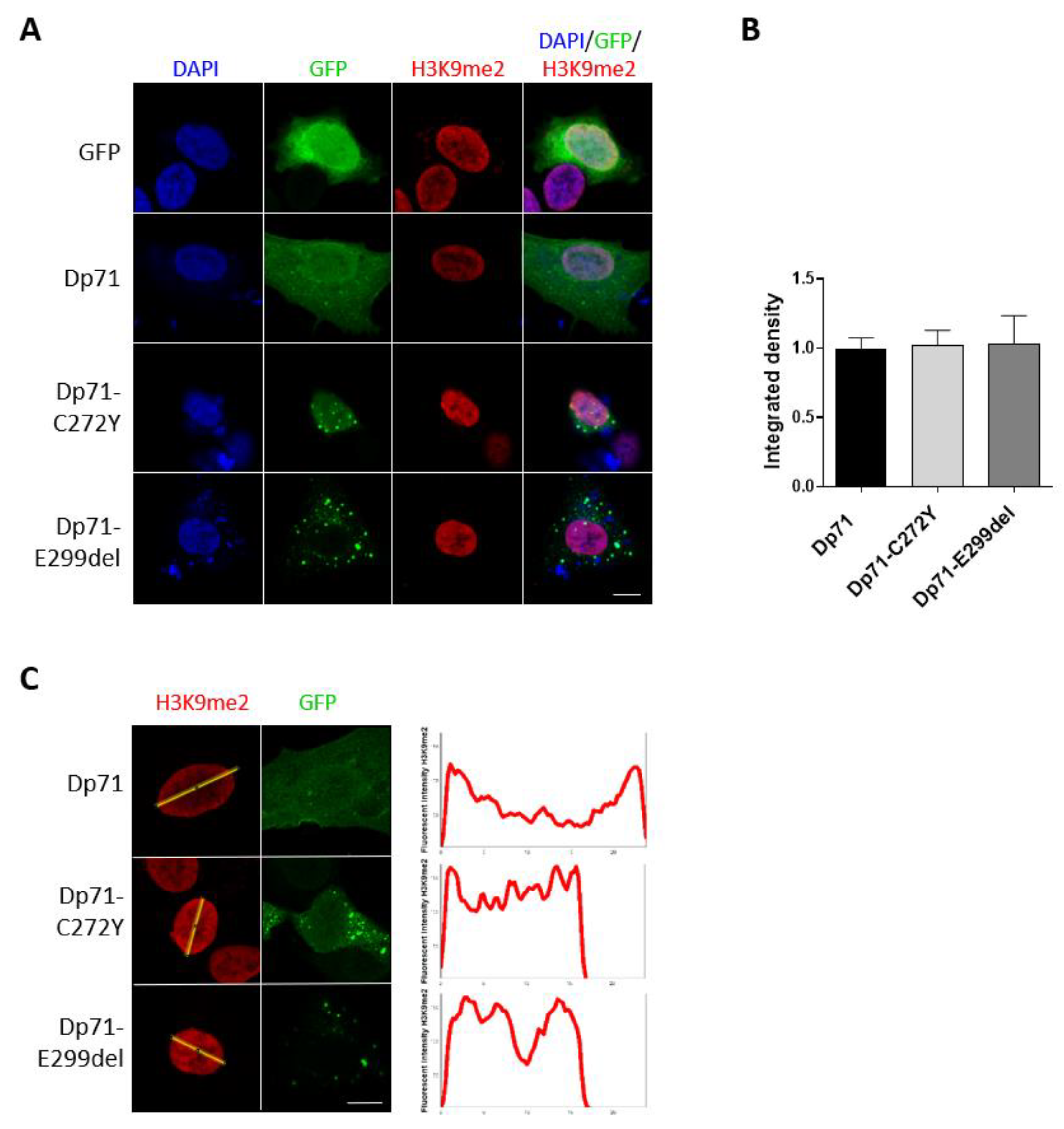
2.6. Expression of Dp71-C272Y and Dp71-E299del Change the Distribution of the Heterochromatin Marker H3K9me2
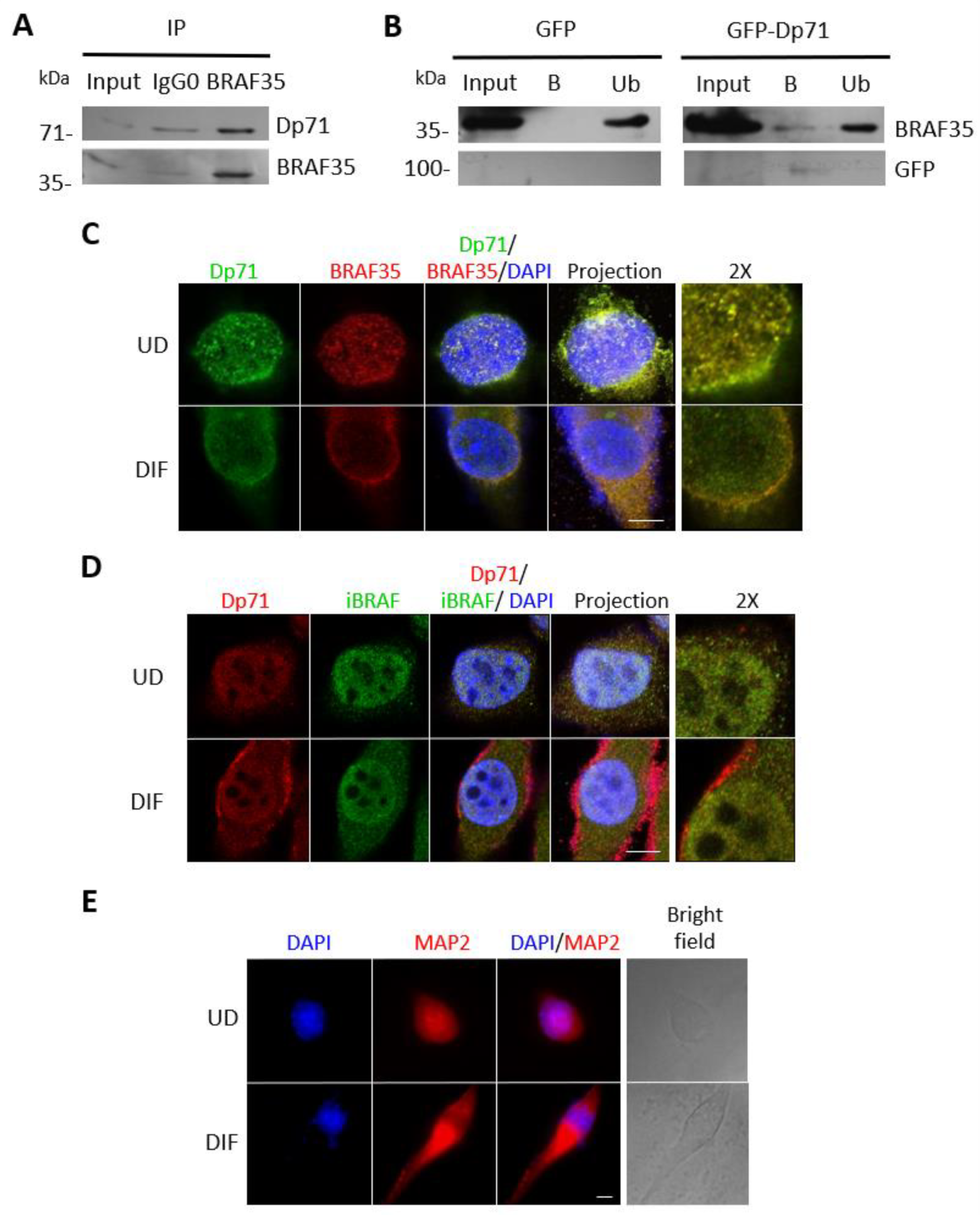
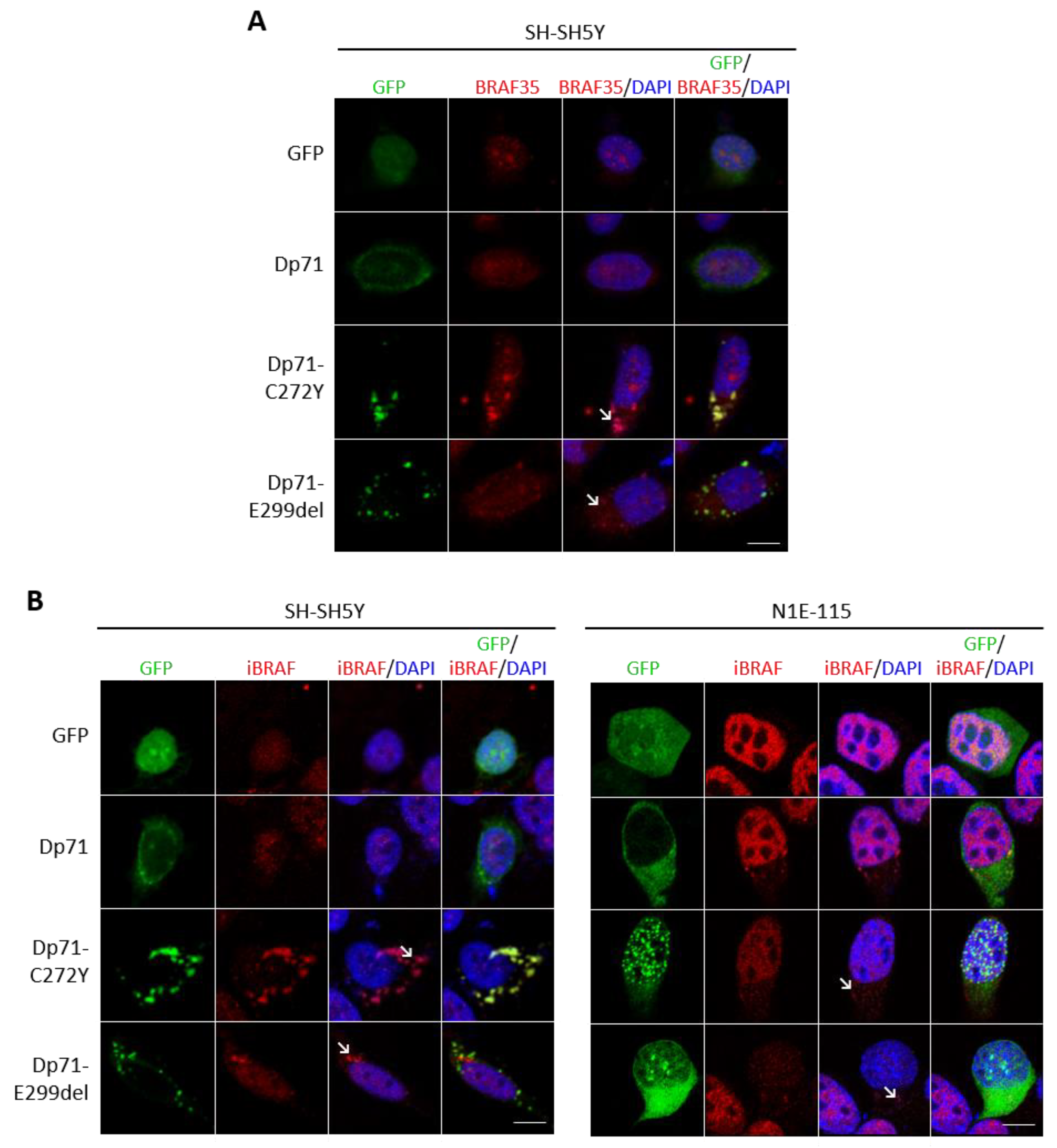
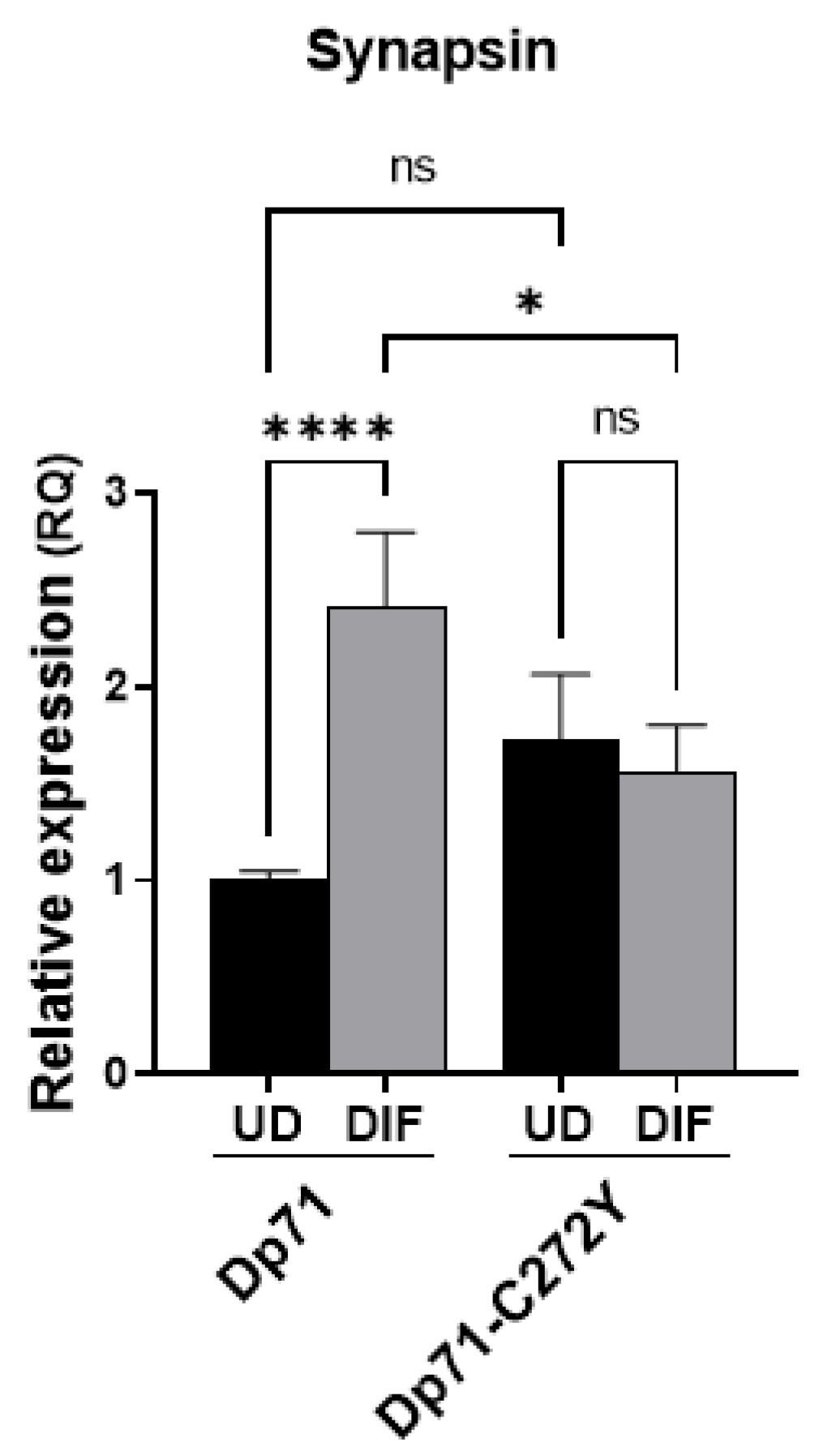
2.7. Dp71 Is Involved in RE-1 Gene Expression through Its Association with BRAF35 and iBRAF
3. Discussion
4. Materials and Methods
4.1. Plasmid Constructs
4.2. Cell Culture and Transfection
4.3. Antibodies
4.4. Immunofluorescence and Confocal Laser Scanning Microscopy
4.5. Immunoprecipitation
4.6. Western Blotting
4.7. Quantitative Reverse Transcription PCR (RT-qPCR)
4.8. Modeling and MDS analyses of Dp71, Dp71-C272Y and Dp71-E299del Structures
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crisafulli, S.; Sultana, J.; Fontana, A.; Salvo, F.; Messina, S.; Trifiro, G. Global epidemiology of Duchenne muscular dystrophy: An updated systematic review and meta-analysis. Orphanet J. Rare Dis. 2020, 15, 141. [Google Scholar] [CrossRef]
- Blake, D.J.; Weir, A.; Newey, S.E.; Davies, K.E. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol. Rev. 2002, 82, 291–329. [Google Scholar] [CrossRef]
- Boyce, F.M.; Beggs, A.H.; Feener, C.; Kunkel, L.M. Dystrophin is transcribed in brain from a distant upstream promoter. Proc. Natl. Acad. Sci. USA 1991, 88, 1276–1280. [Google Scholar] [CrossRef]
- D’Souza, V.N.; Nguyen, T.M.; Morris, G.E.; Karges, W.; Pillers, D.A.; Ray, P.N. A novel dystrophin isoform is required for normal retinal electrophysiology. Hum. Mol. Genet. 1995, 4, 837–842. [Google Scholar] [CrossRef]
- Klamut, H.J.; Gangopadhyay, S.B.; Worton, R.G.; Ray, P.N. Molecular and functional analysis of the muscle-specific promoter region of the Duchenne muscular dystrophy gene. Mol. Cell. Biol. 1990, 10, 193–205. [Google Scholar] [CrossRef]
- Lidov, H.G.; Selig, S.; Kunkel, L.M. Dp140: A novel 140 kDa CNS transcript from the dystrophin locus. Hum. Mol. Genet. 1995, 4, 329–335. [Google Scholar] [CrossRef]
- Belhasan, D.C.; Akaaboune, M. The role of the dystrophin glycoprotein complex on the neuromuscular system. Neurosci. Lett. 2020, 722, 134833. [Google Scholar] [CrossRef]
- Ervasti, J.M.; Sonnemann, K.J. Biology of the striated muscle dystrophin-glycoprotein complex. Int. Rev. Cytol. 2008, 265, 191–225. [Google Scholar] [CrossRef]
- Deconinck, N.; Dan, B. Pathophysiology of duchenne muscular dystrophy: Current hypotheses. Pediatr. Neurol. 2007, 36, 1–7. [Google Scholar] [CrossRef]
- Hugnot, J.P.; Gilgenkrantz, H.; Vincent, N.; Chafey, P.; Morris, G.E.; Monaco, A.P.; Berwald-Netter, Y.; Koulakoff, A.; Kaplan, J.C.; Kahn, A.; et al. Distal transcript of the dystrophin gene initiated from an alternative first exon and encoding a 75-kDa protein widely distributed in nonmuscle tissues. Proc. Natl. Acad. Sci. USA 1992, 89, 7506–7510. [Google Scholar] [CrossRef]
- Bar, S.; Barnea, E.; Levy, Z.; Neuman, S.; Yaffe, D.; Nudel, U. A novel product of the Duchenne muscular dystrophy gene which greatly differs from the known isoforms in its structure and tissue distribution. Biochem. J. 1990, 272, 557–560. [Google Scholar] [CrossRef]
- Blake, D.J.; Love, D.R.; Tinsley, J.; Morris, G.E.; Turley, H.; Gatter, K.; Dickson, G.; Edwards, Y.H.; Davies, K.E. Characterization of a 4.8kb transcript from the Duchenne muscular dystrophy locus expressed in Schwannoma cells. Hum. Mol. Genet. 1992, 1, 103–109. [Google Scholar] [CrossRef]
- Aragon, J.; Gonzalez-Reyes, M.; Romo-Yanez, J.; Vacca, O.; Aguilar-Gonzalez, G.; Rendon, A.; Vaillend, C.; Montanez, C. Dystrophin Dp71 Isoforms Are Differentially Expressed in the Mouse Brain and Retina: Report of New Alternative Splicing and a Novel Nomenclature for Dp71 Isoforms. Mol. Neurobiol. 2018, 55, 1376–1386. [Google Scholar] [CrossRef]
- Lederfein, D.; Levy, Z.; Augier, N.; Mornet, D.; Morris, G.; Fuchs, O.; Yaffe, D.; Nudel, U. A 71-kilodalton protein is a major product of the Duchenne muscular dystrophy gene in brain and other nonmuscle tissues. Proc. Natl. Acad. Sci. USA 1992, 89, 5346–5350. [Google Scholar] [CrossRef]
- Jones, L.; Naidoo, M.; Machado, L.R.; Anthony, K. The Duchenne muscular dystrophy gene and cancer. Cell. Oncol. 2021, 44, 19–32. [Google Scholar] [CrossRef]
- Tadayoni, R.; Rendon, A.; Soria-Jasso, L.E.; Cisneros, B. Dystrophin Dp71: The smallest but multifunctional product of the Duchenne muscular dystrophy gene. Mol. Neurobiol. 2012, 45, 43–60. [Google Scholar] [CrossRef]
- Naidoo, M.; Anthony, K. Dystrophin Dp71 and the Neuropathophysiology of Duchenne Muscular Dystrophy. Mol. Neurobiol. 2020, 57, 1748–1767. [Google Scholar] [CrossRef]
- Pane, M.; Messina, S.; Bruno, C.; D’Amico, A.; Villanova, M.; Brancalion, B.; Sivo, S.; Bianco, F.; Striano, P.; Battaglia, D.; et al. Duchenne muscular dystrophy and epilepsy. Neuromuscul. Disord. 2013, 23, 313–315. [Google Scholar] [CrossRef]
- Ricotti, V.; Mandy, W.P.; Scoto, M.; Pane, M.; Deconinck, N.; Messina, S.; Mercuri, E.; Skuse, D.H.; Muntoni, F. Neurodevelopmental, emotional, and behavioural problems in Duchenne muscular dystrophy in relation to underlying dystrophin gene mutations. Dev. Med. Child. Neurol. 2016, 58, 77–84. [Google Scholar] [CrossRef]
- Magri, F.; Govoni, A.; D’Angelo, M.G.; Del Bo, R.; Ghezzi, S.; Sandra, G.; Turconi, A.C.; Sciacco, M.; Ciscato, P.; Bordoni, A.; et al. Genotype and phenotype characterization in a large dystrophinopathic cohort with extended follow-up. J. Neurol. 2011, 258, 1610–1623. [Google Scholar] [CrossRef]
- Rasic, V.M.; Vojinovic, D.; Pesovic, J.; Mijalkovic, G.; Lukic, V.; Mladenovic, J.; Kosac, A.; Novakovic, I.; Maksimovic, N.; Romac, S.; et al. Intellectual ability in the duchenne muscular dystrophy and dystrophin gene mutation location. Balkan J. Med. Genet. 2014, 17, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.J.; Betts, G.A.; Maroulis, S.; Gilissen, C.; Pedersen, R.L.; Mowat, D.R.; Johnston, H.M.; Buckley, M.F. Dystrophin gene mutation location and the risk of cognitive impairment in Duchenne muscular dystrophy. PLoS ONE 2010, 5, e8803. [Google Scholar] [CrossRef] [PubMed]
- Daoud, F.; Candelario-Martinez, A.; Billard, J.M.; Avital, A.; Khelfaoui, M.; Rozenvald, Y.; Guegan, M.; Mornet, D.; Jaillard, D.; Nudel, U.; et al. Role of mental retardation-associated dystrophin-gene product Dp71 in excitatory synapse organization, synaptic plasticity and behavioral functions. PLoS ONE 2009, 4, e6574. [Google Scholar] [CrossRef] [PubMed]
- Miranda, R.; Nudel, U.; Laroche, S.; Vaillend, C. Altered presynaptic ultrastructure in excitatory hippocampal synapses of mice lacking dystrophins Dp427 or Dp71. Neurobiol. Dis. 2011, 43, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Connors, N.C.; Adams, M.E.; Froehner, S.C.; Kofuji, P. The potassium channel Kir4.1 associates with the dystrophin-glycoprotein complex via alpha-syntrophin in glia. J. Biol. Chem. 2004, 279, 28387–28392. [Google Scholar] [CrossRef] [PubMed]
- Perronnet, C.; Vaillend, C. Dystrophins, utrophins, and associated scaffolding complexes: Role in mammalian brain and implications for therapeutic strategies. J. Biomed. Biotechnol. 2010, 2010, 849426. [Google Scholar] [CrossRef]
- Artegiani, B.; Labbaye, C.; Sferra, A.; Quaranta, M.T.; Torreri, P.; Macchia, G.; Ceccarini, M.; Petrucci, T.C.; Macioce, P. The interaction with HMG20a/b proteins suggests a potential role for beta-dystrobrevin in neuronal differentiation. J. Biol. Chem. 2010, 285, 24740–24750. [Google Scholar] [CrossRef]
- Bianchi, M.E.; Agresti, A. HMG proteins: Dynamic players in gene regulation and differentiation. Curr. Opin. Genet. Dev. 2005, 15, 496–506. [Google Scholar] [CrossRef]
- Ceballos-Chavez, M.; Rivero, S.; Garcia-Gutierrez, P.; Rodriguez-Paredes, M.; Garcia-Dominguez, M.; Bhattacharya, S.; Reyes, J.C. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc. Natl. Acad. Sci. USA 2012, 109, 8085–8090. [Google Scholar] [CrossRef]
- Hakimi, M.A.; Bochar, D.A.; Chenoweth, J.; Lane, W.S.; Mandel, G.; Shiekhattar, R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA 2002, 99, 7420–7425. [Google Scholar] [CrossRef]
- Wynder, C.; Hakimi, M.A.; Epstein, J.A.; Shilatifard, A.; Shiekhattar, R. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell. Biol. 2005, 7, 1113–1117. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Ginjaar, I.B.; Bushby, K. The importance of genetic diagnosis for Duchenne muscular dystrophy. J. Med. Genet. 2016, 53, 145–151. [Google Scholar] [CrossRef]
- Mendell, J.R.; Buzin, C.H.; Feng, J.; Yan, J.; Serrano, C.; Sangani, D.S.; Wall, C.; Prior, T.W.; Sommer, S.S. Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology 2001, 57, 645–650. [Google Scholar] [CrossRef]
- Becker, K.; Robb, S.A.; Hatton, Z.; Yau, S.C.; Abbs, S.; Roberts, R.G. Loss of a single amino acid from dystrophin resulting in Duchenne muscular dystrophy with retention of dystrophin protein. Hum. Mutat. 2003, 21, 651. [Google Scholar] [CrossRef]
- Lenk, U.; Oexle, K.; Voit, T.; Ancker, U.; Hellner, K.A.; Speer, A.; Hubner, C. A cysteine 3340 substitution in the dystroglycan-binding domain of dystrophin associated with Duchenne muscular dystrophy, mental retardation and absence of the ERG b-wave. Hum. Mol. Genet. 1996, 5, 973–975. [Google Scholar] [CrossRef]
- Vulin, A.; Wein, N.; Strandjord, D.M.; Johnson, E.K.; Findlay, A.R.; Maiti, B.; Howard, M.T.; Kaminoh, Y.J.; Taylor, L.E.; Simmons, T.R.; et al. The ZZ Domain of Dystrophin in DMD: Making Sense of Missense Mutations. Hum. Mutat. 2014, 35, 257–264. [Google Scholar] [CrossRef]
- Gonzalez-Ramirez, R.; Morales-Lazaro, S.L.; Tapia-Ramirez, V.; Mornet, D.; Cisneros, B. Nuclear and nuclear envelope localization of dystrophin Dp71 and dystrophin-associated proteins (DAPs) in the C2C12 muscle cells: DAPs nuclear localization is modulated during myogenesis. J. Cell. Biochem. 2008, 105, 735–745. [Google Scholar] [CrossRef]
- Villarreal-Silva, M.; Suarez-Sanchez, R.; Rodriguez-Munoz, R.; Mornet, D.; Cisneros, B. Dystrophin Dp71 is critical for stability of the DAPs in the nucleus of PC12 cells. Neurochem. Res. 2010, 35, 366–373. [Google Scholar] [CrossRef]
- Fujimoto, T.; Yaoi, T.; Nakano, K.; Arai, T.; Okamura, T.; Itoh, K. Generation of dystrophin short product-specific tag-insertion mouse: Distinct Dp71 glycoprotein complexes at inhibitory postsynapse and glia limitans. Cell. Mol. Life. Sci. 2022, 79, 109. [Google Scholar] [CrossRef]
- Huang, X.; Poy, F.; Zhang, R.; Joachimiak, A.; Sudol, M.; Eck, M.J. Structure of a WW domain containing fragment of dystrophin in complex with beta-dystroglycan. Nat. Struct. Biol. 2000, 7, 634–638. [Google Scholar] [CrossRef]
- Fuentes-Mera, L.; Rodriguez-Munoz, R.; Gonzalez-Ramirez, R.; Garcia-Sierra, F.; Gonzalez, E.; Mornet, D.; Cisneros, B. Characterization of a novel Dp71 dystrophin-associated protein complex (DAPC) present in the nucleus of HeLa cells: Members of the nuclear DAPC associate with the nuclear matrix. Exp. Cell. Res. 2006, 312, 3023–3035. [Google Scholar] [CrossRef]
- Hnia, K.; Zouiten, D.; Cantel, S.; Chazalette, D.; Hugon, G.; Fehrentz, J.A.; Masmoudi, A.; Diment, A.; Bramham, J.; Mornet, D.; et al. ZZ domain of dystrophin and utrophin: Topology and mapping of a beta-dystroglycan interaction site. Biochem. J. 2007, 401, 667–677. [Google Scholar] [CrossRef]
- Fujimoto, T.; Yaoi, T.; Tanaka, H.; Itoh, K. Dystroglycan regulates proper expression, submembranous localization and subsequent phosphorylation of Dp71 through physical interaction. Hum. Mol. Genet. 2020, 29, 3312–3326. [Google Scholar] [CrossRef]
- Villarreal-Silva, M.; Centeno-Cruz, F.; Suarez-Sanchez, R.; Garrido, E.; Cisneros, B. Knockdown of dystrophin Dp71 impairs PC12 cells cycle: Localization in the spindle and cytokinesis structures implies a role for Dp71 in cell division. PLoS ONE 2011, 6, e23504. [Google Scholar] [CrossRef]
- Wong, X.; Melendez-Perez, A.J.; Reddy, K.L. The Nuclear Lamina. Cold Spring Harb. Perspect. Biol. 2022, 14, a040113. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef]
- Towbin, B.D.; Gonzalez-Aguilera, C.; Sack, R.; Gaidatzis, D.; Kalck, V.; Meister, P.; Askjaer, P.; Gasser, S.M. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 2012, 150, 934–947. [Google Scholar] [CrossRef]
- Roopra, A.; Qazi, R.; Schoenike, B.; Daley, T.J.; Morrison, J.F. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 2004, 14, 727–738. [Google Scholar] [CrossRef]
- Qureshi, I.A.; Gokhan, S.; Mehler, M.F. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle 2010, 9, 4477–4486. [Google Scholar] [CrossRef]
- Tsai, Y.; Cutts, J.; Kimura, A.; Varun, D.; Brafman, D.A. A chemically defined substrate for the expansion and neuronal differentiation of human pluripotent stem cell-derived neural progenitor cells. Stem Cell Res. 2015, 15, 75–87. [Google Scholar] [CrossRef]
- Zimowski, J.; Fidzianska, E.; Holding, M.; Zaremba, J. Two mutations in one dystrophin gene. Neurol. Neurochir. Pol. 2013, 47, 131–137. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Monaco, A.P.; Bertelson, C.J.; Liechti-Gallati, S.; Moser, H.; Kunkel, L.M. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics 1988, 2, 90–95. [Google Scholar] [CrossRef]
- Flanigan, K.M.; von Niederhausern, A.; Dunn, D.M.; Alder, J.; Mendell, J.R.; Weiss, R.B. Rapid direct sequence analysis of the dystrophin gene. Am. J. Hum. Genet. 2003, 72, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Flanigan, K.M.; Dunn, D.M.; von Niederhausern, A.; Soltanzadeh, P.; Gappmaier, E.; Howard, M.T.; Sampson, J.B.; Mendell, J.R.; Wall, C.; King, W.M.; et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: Application of modern diagnostic techniques to a large cohort. Hum. Mutat. 2009, 30, 1657–1666. [Google Scholar] [CrossRef]
- Juan-Mateu, J.; Gonzalez-Quereda, L.; Rodriguez, M.J.; Baena, M.; Verdura, E.; Nascimento, A.; Ortez, C.; Baiget, M.; Gallano, P. DMD Mutations in 576 Dystrophinopathy Families: A Step Forward in Genotype-Phenotype Correlations. PLoS ONE 2015, 10, e0135189. [Google Scholar] [CrossRef]
- Suarez-Sanchez, R.; Aguilar, A.; Wagstaff, K.M.; Velez, G.; Azuara-Medina, P.M.; Gomez, P.; Vasquez-Limeta, A.; Hernandez-Hernandez, O.; Lieu, K.G.; Jans, D.A.; et al. Nucleocytoplasmic shuttling of the Duchenne muscular dystrophy gene product dystrophin Dp71d is dependent on the importin alpha/beta and CRM1 nuclear transporters and microtubule motor dynein. Biochim. Biophys. Acta 2014, 1843, 985–1001. [Google Scholar] [CrossRef]
- Ruggieri, S.; Viggiano, L.; Annese, T.; Rubolino, C.; Gerbino, A.; De Zio, R.; Corsi, P.; Tamma, R.; Ribatti, D.; Errede, M.; et al. DP71 and SERCA2 alteration in human neurons of a Duchenne muscular dystrophy patient. Stem Cell Res. Ther. 2019, 10, 29. [Google Scholar] [CrossRef]
- Imamura, M.; Ozawa, E. Differential expression of dystrophin isoforms and utrophin during dibutyryl-cAMP-induced morphological differentiation of rat brain astrocytes. Proc. Natl. Acad. Sci. USA 1998, 95, 6139–6144. [Google Scholar] [CrossRef]
- Cisneros, B.; Rendon, A.; Genty, V.; Aranda, G.; Marquez, F.; Mornet, D.; Montanez, C. Expression of dystrophin Dp71 during PC12 cell differentiation. Neurosci. Lett. 1996, 213, 107–110. [Google Scholar] [CrossRef]
- Marquez, F.G.; Cisneros, B.; Garcia, F.; Ceja, V.; Velazquez, F.; Depardon, F.; Cervantes, L.; Rendon, A.; Mornet, D.; Rosas-vargas, H.; et al. Differential expression and subcellular distribution of dystrophin Dp71 isoforms during differentiation process. Neuroscience 2003, 118, 957–966. [Google Scholar] [CrossRef]
- Henderson, D.M.; Lee, A.; Ervasti, J.M. Disease-causing missense mutations in actin binding domain 1 of dystrophin induce thermodynamic instability and protein aggregation. Proc. Natl. Acad. Sci. USA 2010, 107, 9632–9637. [Google Scholar] [CrossRef]
- Acosta, R.; Montanez, C.; Fuentes-Mera, L.; Gonzalez, E.; Gomez, P.; Quintero-Mora, L.; Mornet, D.; Alvarez-Salas, L.M.; Cisneros, B. Dystrophin Dp71 is required for neurite outgrowth in PC12 cells. Exp. Cell Res. 2004, 296, 265–275. [Google Scholar] [CrossRef]
- Romo-Yanez, J.; Ceja, V.; Ilarraza-Lomeli, R.; Coral-Vazquez, R.; Velazquez, F.; Mornet, D.; Rendon, A.; Montanez, C. Dp71ab/DAPs complex composition changes during the differentiation process in PC12 cells. J. Cell Biochem. 2007, 102, 82–97. [Google Scholar] [CrossRef]
- Cerna, J.; Cerecedo, D.; Ortega, A.; Garcia-Sierra, F.; Centeno, F.; Garrido, E.; Mornet, D.; Cisneros, B. Dystrophin Dp71f associates with the beta1-integrin adhesion complex to modulate PC12 cell adhesion. J. Mol. Biol. 2006, 362, 954–965. [Google Scholar] [CrossRef]
- Enriquez-Aragon, J.A.; Cerna-Cortes, J.; Bermudez de Leon, M.; Garcia-Sierra, F.; Gonzalez, E.; Mornet, D.; Cisneros, B. Dystrophin Dp71 in PC12 cell adhesion. Neuroreport 2005, 16, 235–238. [Google Scholar] [CrossRef]
- Ishikawa-Sakurai, M.; Yoshida, M.; Imamura, M.; Davies, K.E.; Ozawa, E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to beta-dystroglycan. Hum. Mol. Genet. 2004, 13, 693–702. [Google Scholar] [CrossRef][Green Version]
- Rodriguez-Munoz, R.; Villarreal-Silva, M.; Gonzalez-Ramirez, R.; Garcia-Sierra, F.; Mondragon, M.; Mondragon, R.; Cerna, J.; Cisneros, B. Neuronal differentiation modulates the dystrophin Dp71d binding to the nuclear matrix. Biochem. Biophys. Res. Commun. 2008, 375, 303–307. [Google Scholar] [CrossRef]
- Poleshko, A.; Katz, R.A. Specifying peripheral heterochromatin during nuclear lamina reassembly. Nucleus 2014, 5, 32–39. [Google Scholar] [CrossRef]
- Poleshko, A.; Mansfield, K.M.; Burlingame, C.C.; Andrake, M.D.; Shah, N.R.; Katz, R.A. The human protein PRR14 tethers heterochromatin to the nuclear lamina during interphase and mitotic exit. Cell Rep. 2013, 5, 292–301. [Google Scholar] [CrossRef]
- Romo-Yanez, J.; Rodriguez-Martinez, G.; Aragon, J.; Siqueiros-Marquez, L.; Herrera-Salazar, A.; Velasco, I.; Montanez, C. Characterization of the expression of dystrophins and dystrophin-associated proteins during embryonic neural stem/progenitor cell differentiation. Neurosci. Lett. 2020, 736, 135247. [Google Scholar] [CrossRef]
- Garcia-Cruz, C.; Merino-Jimenez, C.; Ceja, V.; Aragon, J.; Siqueiros-Marquez, L.; Reyes-Grajeda, J.P.; Montanez, C. The dystrophin isoform Dp71eDelta71 is involved in neurite outgrowth and neuronal differentiation of PC12 cells. J. Proteom. 2019, 191, 80–87. [Google Scholar] [CrossRef]
- Merino-Jimenez, C.; Aragon, J.; Ceja, V.; Rodriguez-Martinez, G.; Cazares-Raga, F.E.; Chardonnet, S.; Pionneau, C.; Rendon, A.; Montanez, C. Dp71Delta78-79 dystrophin mutant stimulates neurite outgrowth in PC12 cells via upregulation and phosphorylation of HspB1. Proteomics 2016, 16, 1331–1340. [Google Scholar] [CrossRef]
- Oh, J.E.; Karlmark, K.R.; Shin, J.H.; Pollak, A.; Freilinger, A.; Hengstschlager, M.; Lubec, G. Differentiation of neuroblastoma cell line N1E-115 involves several signaling cascades. Neurochem. Res. 2005, 30, 333–348. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Anderson, R.J.; Weng, Z.; Campbell, R.K.; Jiang, X. Main-chain conformational tendencies of amino acids. Proteins 2005, 60, 679–689. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Rigsby, R.E.; Parker, A.B. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 2016, 44, 433–437. [Google Scholar] [CrossRef]
- Lindahl, E.; Abraham, M.; Hess, B.; Van Der Spoel, D. GROMACS 2020.1 Manual (2020.1); Zenodo, 2020; CERN: Meyrin, Switzerland, 2020. [Google Scholar] [CrossRef]
- Robertson, M.J.; Tirado-Rives, J.; Jorgensen, W.L. Improved Peptide and Protein Torsional Energetics with the OPLSAA Force Field. J. Chem. Theory Comput. 2015, 11, 3499–3509. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High. performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rugerio-Martínez, C.I.; Ramos, D.; Segura-Olvera, A.; Murillo-Melo, N.M.; Tapia-Guerrero, Y.S.; Argüello-García, R.; Leyva-García, N.; Hernández-Hernández, O.; Cisneros, B.; Suárez-Sánchez, R. Dp71 Point Mutations Induce Protein Aggregation, Loss of Nuclear Lamina Integrity and Impaired Braf35 and Ibraf Function in Neuronal Cells. Int. J. Mol. Sci. 2022, 23, 11876. https://doi.org/10.3390/ijms231911876
Rugerio-Martínez CI, Ramos D, Segura-Olvera A, Murillo-Melo NM, Tapia-Guerrero YS, Argüello-García R, Leyva-García N, Hernández-Hernández O, Cisneros B, Suárez-Sánchez R. Dp71 Point Mutations Induce Protein Aggregation, Loss of Nuclear Lamina Integrity and Impaired Braf35 and Ibraf Function in Neuronal Cells. International Journal of Molecular Sciences. 2022; 23(19):11876. https://doi.org/10.3390/ijms231911876
Chicago/Turabian StyleRugerio-Martínez, Claudia Ivette, Daniel Ramos, Abel Segura-Olvera, Nadia Mireya Murillo-Melo, Yessica Sarai Tapia-Guerrero, Raúl Argüello-García, Norberto Leyva-García, Oscar Hernández-Hernández, Bulmaro Cisneros, and Rocío Suárez-Sánchez. 2022. "Dp71 Point Mutations Induce Protein Aggregation, Loss of Nuclear Lamina Integrity and Impaired Braf35 and Ibraf Function in Neuronal Cells" International Journal of Molecular Sciences 23, no. 19: 11876. https://doi.org/10.3390/ijms231911876
APA StyleRugerio-Martínez, C. I., Ramos, D., Segura-Olvera, A., Murillo-Melo, N. M., Tapia-Guerrero, Y. S., Argüello-García, R., Leyva-García, N., Hernández-Hernández, O., Cisneros, B., & Suárez-Sánchez, R. (2022). Dp71 Point Mutations Induce Protein Aggregation, Loss of Nuclear Lamina Integrity and Impaired Braf35 and Ibraf Function in Neuronal Cells. International Journal of Molecular Sciences, 23(19), 11876. https://doi.org/10.3390/ijms231911876





