LPCAT4 Knockdown Alters Barrier Integrity and Cellular Bioenergetics in Human Urothelium
Abstract
1. Introduction
2. Results
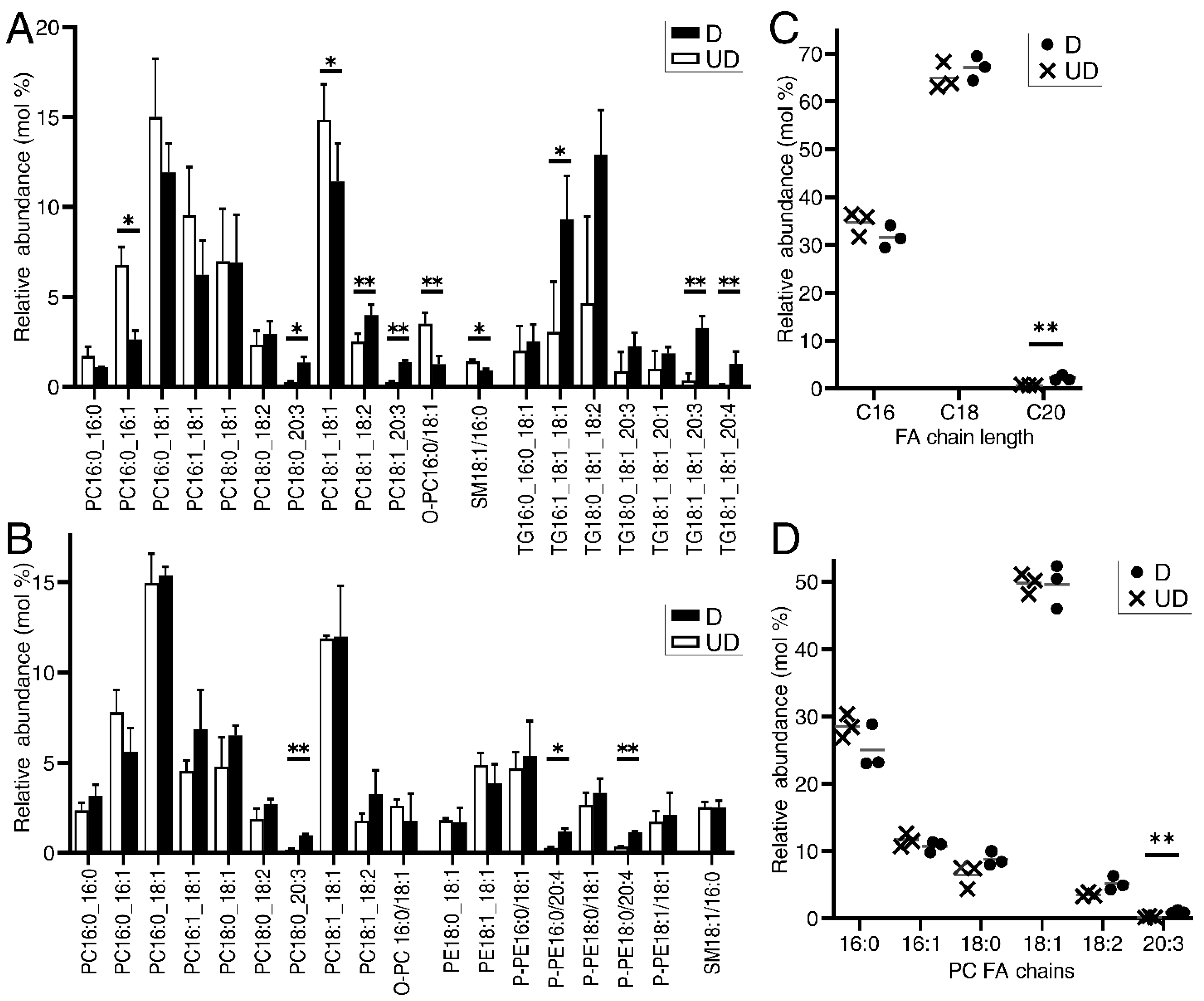
2.1. Urothelial Differentiation Induces Glycerophospholipid Remodelling and Triglyceride Accumulation
2.2. LPCAT4 Identified as a Differentiation-Associated Remodeller of Phosphatidylcholine in Urothelium
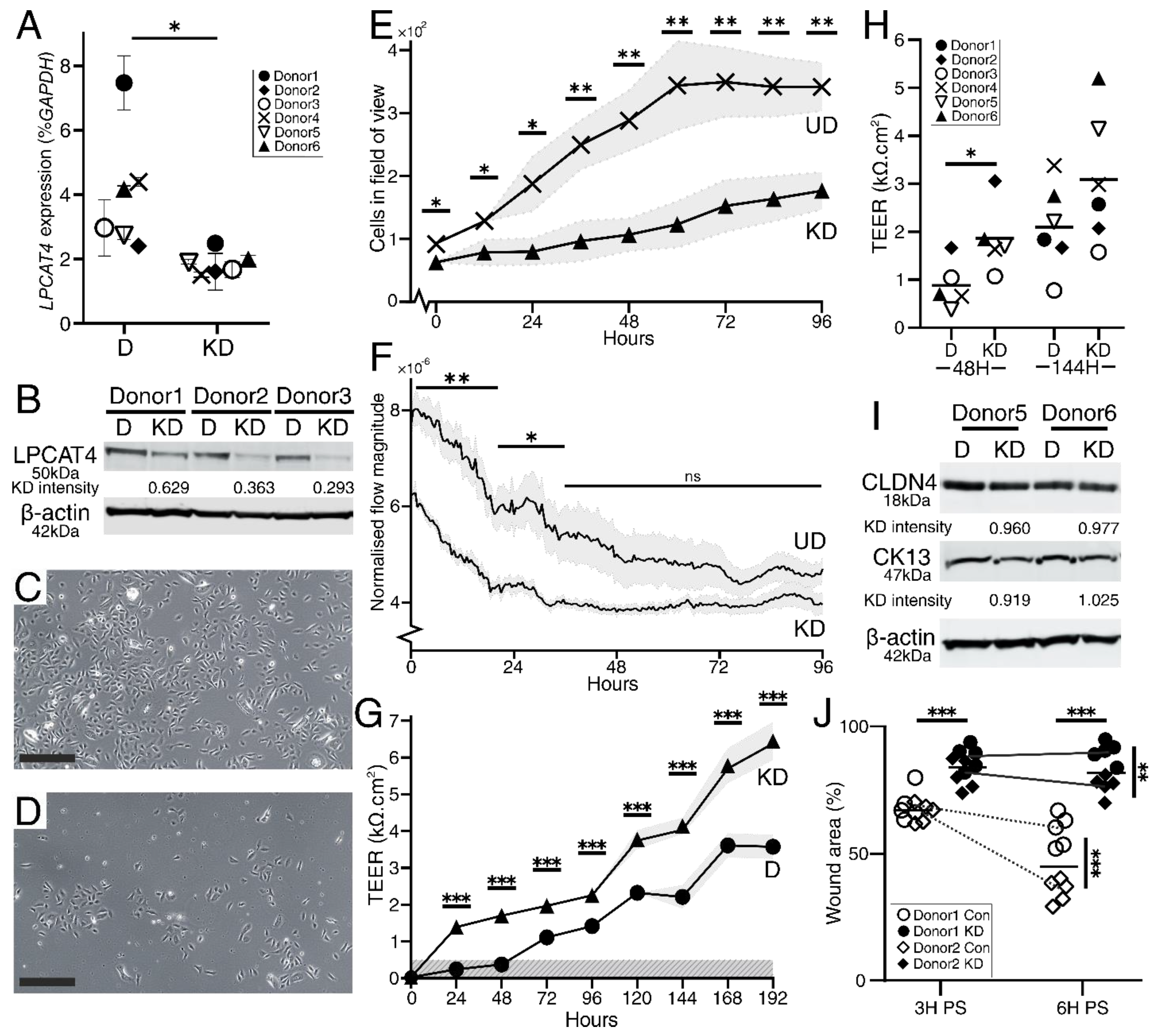
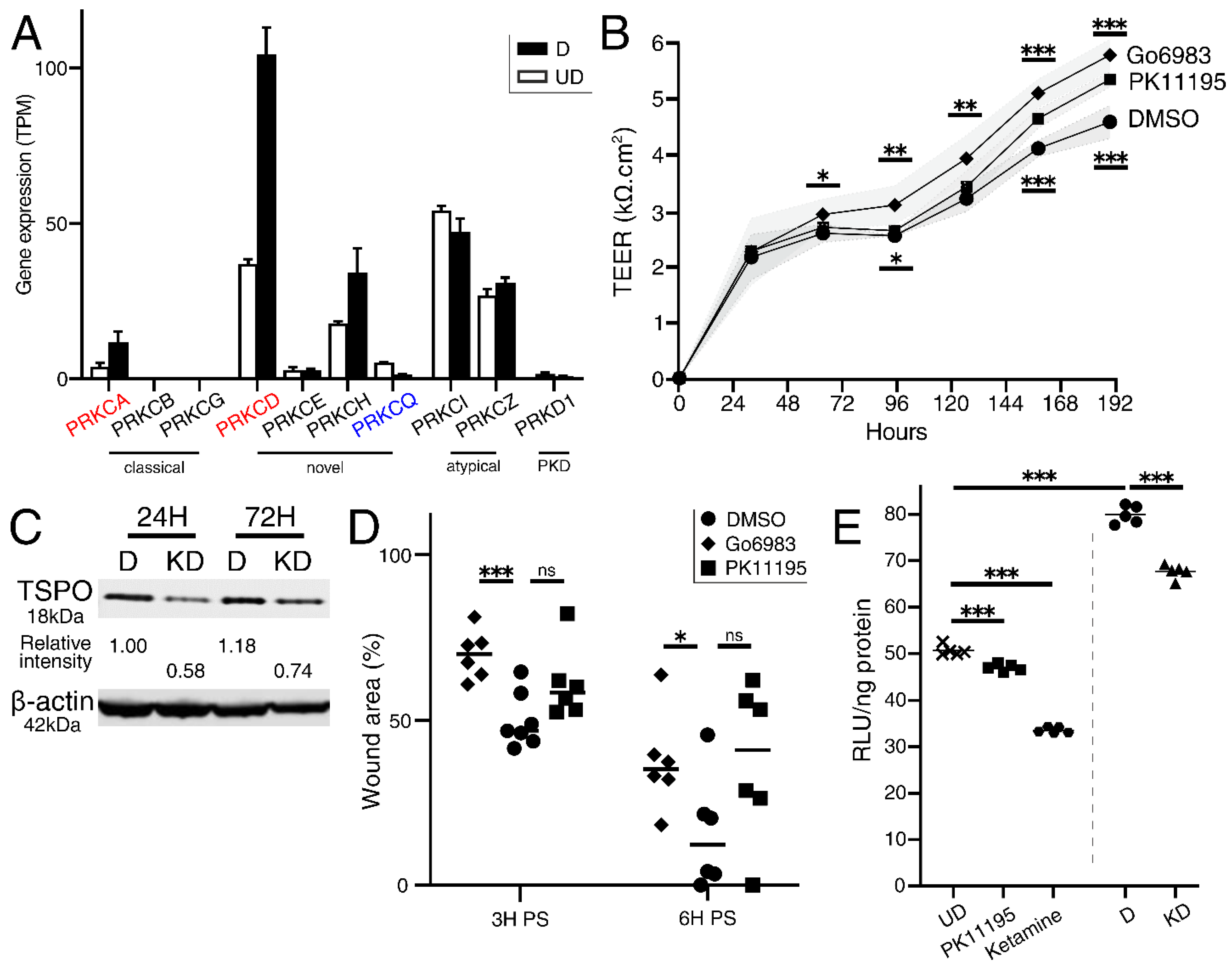
2.3. LPCAT4 Knockdown Reduces Growth Rate but Elevates Urothelial Barrier Resistance
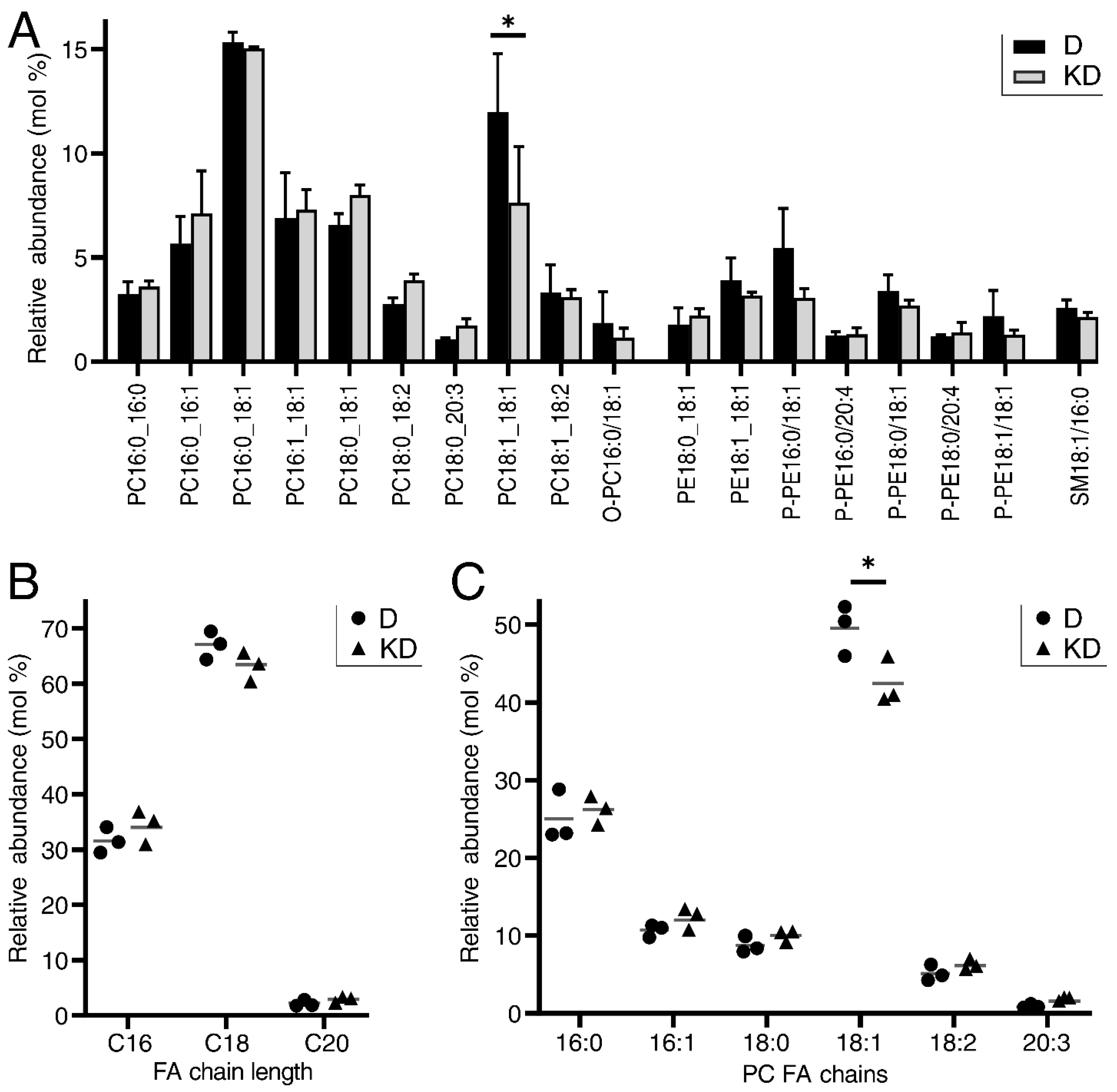
2.4. LPCAT4 Knockdown Reduces 18:1 Fatty Acyl Incorporation into Phosphatidylcholine
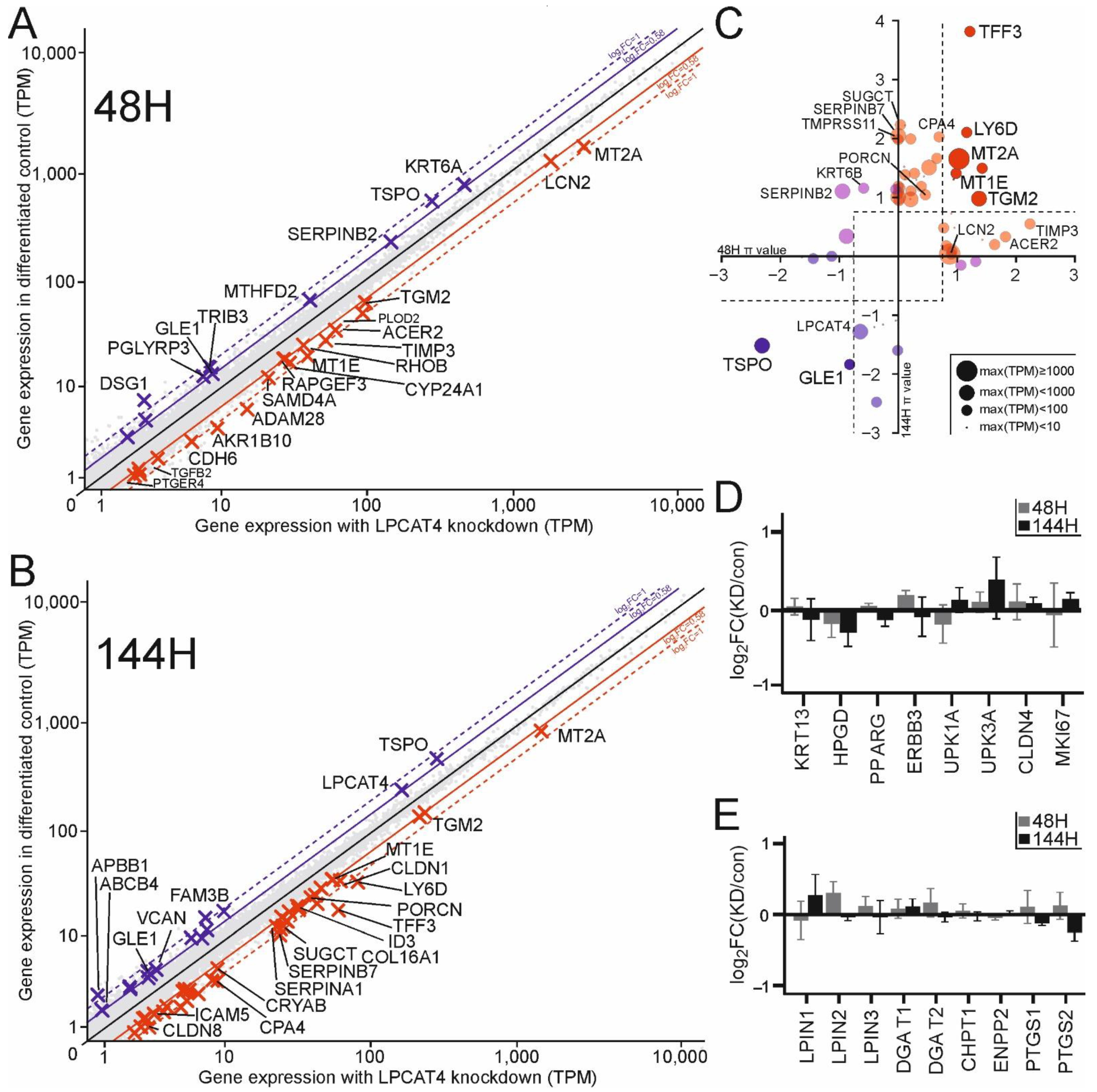
2.5. LPCAT4 Knockdown Elicits Specific Transcriptomic Changes in Lipid Regulation, Cell-Cell and Cell-Matrix Interactions
2.6. LPCAT4 Knockdown Phenotypes Are Recapitulated by Independent Inhibition of TSPO and PKC
3. Discussion
4. Materials and Methods
4.1. Tissues and Ethical Approval
4.2. Normal Human Urothelial (NHU) Cell Culture and In Vitro Differentiation
4.3. Transduction of NHU Cell Lines with shRNA Constructs
4.4. Identification of Lipid Regulators Associated with Urothelial Differentiation
4.5. Derivation of LPCAT4 Knockdown Cell Lines
4.6. Assessment of Culture Proliferation Rate
4.7. Assessment of Culture Migration Rate
4.8. RT-qPCR
4.9. Immunoblotting
4.10. Estimating Repair Capacity by Scratch-Wound Restitution
4.11. Lipid Profiling of Transduced Urothelial Cell Lines
4.12. Transcriptomic Profiling of Transduced NHU Cell Lines
4.13. Phenocopying Effects of LPCAT4 Knockdown by Inhibition of PKC and TSPO
4.14. Quantification of Cellular ATP
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Escribá, P.V.; Busquets, X.; Inokuchi, J.-I.; Balogh, G.; Török, Z.; Horváth, I.; Harwood, J.L.; Vígh, L. Membrane lipid therapy: Modulation of the cell membrane composition and structure as a molecular base for drug discovery and new disease treatment. Prog. Lipid Res. 2015, 59, 38–53. [Google Scholar] [CrossRef]
- Kennedy, E.P.; Weiss, S.B. The function of cytidine coenzymes in the biosynthesis of phospholipids. J. Biol. Chem. 1956, 222, 193–214. [Google Scholar] [CrossRef]
- Harayama, T.; Eto, M.; Shindou, H.; Kita, Y.; Otsubo, E.; Hishikawa, D.; Ishii, S.; Sakimura, K.; Mishina, M.; Shimizu, T. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 2014, 20, 295–305. [Google Scholar] [CrossRef]
- Yamashita, A.; Sugiura, T.; Waku, K. Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J. Biochem. 1997, 122, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; de Kroon, A.I.P.M. Lipid map of the mammalian cell. J. Cell Sci. 2011, 124, 5–8. [Google Scholar] [CrossRef]
- Lands, W.E. Metabolism of glycerolipids; a comparison of lecithin and triglyceride synthesis. J. Biol. Chem. 1958, 231, 883–888. [Google Scholar] [CrossRef]
- Hishikawa, D.; Hashidate, T.; Shimizu, T.; Shindou, H. Diversity and function of membrane glycerophospholipids generated by the remodeling pathway in mammalian cells. J. Lipid Res. 2014, 55, 799–807. [Google Scholar] [CrossRef]
- Varley, C.L.; Bacon, E.J.; Holder, J.C.; Southgate, J. FOXA1 and IRF-1 intermediary transcriptional regulators of PPARgamma-induced urothelial cytodifferentiation. Cell Death Differ. 2009, 16, 103–114. [Google Scholar] [CrossRef]
- Varley, C.L.; Stahlschmidt, J.; Lee, W.-C.; Holder, J.; Diggle, C.; Selby, P.J.; Trejdosiewicz, L.K. Role of PPAR γ and EGFR signalling in the urothelial terminal differentiation programme. J. Cell Sci. 2004, 117, 2029–2036. [Google Scholar] [CrossRef]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPARγ Is Required for Placental, Cardiac, and Adipose Tissue Development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef]
- Varley, C.L.; Garthwaite, M.A.E.; Cross, W.; Hinley, J.; Trejdosiewicz, L.K.; Southgate, J. PPARgamma-regulated tight junction development during human urothelial cytodifferentiation. J. Cell Physiol. 2006, 208, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Keay, S.; De Deyne, P.G.; Chai, T.C. Augmented stretch activated adenosine triphosphate release from bladder uroepithelial cells in patients with interstitial cystitis. J. Urol. 2001, 166, 1951–1956. [Google Scholar] [PubMed]
- Fellows, G.J.; Marshall, D.H. The permeability of human bladder epithelium to water and sodium. Investig. Urol. 1972, 9, 339–344. [Google Scholar]
- Tripathi, P.; Somashekar, B.S.; Ponnusamy, M.; Gursky, A.; Dailey, S.; Kunju, P.; Lee, C.T.; Chinnaiyan, A.M.; Rajendiran, T.M.; Ramamoorthy, A. HR-MAS NMR tissue metabolomic signatures cross-validated by mass spectrometry distinguish bladder cancer from benign disease. J. Proteome Res. 2013, 12, 3519–3528. [Google Scholar] [CrossRef] [PubMed]
- Issaq, H.J.; Nativ, O.; Waybright, T.; Luke, B.; Veenstra, T.D.; Issaq, E.J.; Kravstov, A.; Mullerad, M. Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J. Urol. 2008, 179, 2422–2426. [Google Scholar] [CrossRef]
- Pasikanti, K.K.; Esuvaranathan, K.; Ho, P.C.; Mahendran, R.; Kamaraj, R.; Wu, Q.H.; Chiong, E.; Chan, E.C.Y. Noninvasive urinary metabonomic diagnosis of human bladder cancer. J. Proteome Res. 2010, 9, 2988–2995. [Google Scholar]
- Southgate, J.; Hutton, K.A.; Thomas, D.F.; Trejdosiewicz, L.K. Normal human urothelial cells in vitro: Proliferation and induction of stratification. Lab. Investig. 1994, 71, 583–594. [Google Scholar]
- Cross, W.R.; Eardley, I.; Leese, H.J.; Southgate, J. A biomimetic tissue from cultured normal human urothelial cells: Analysis of physiological function. Am. J. Physiol. Renal. Physiol. 2005, 289, F459–F468. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Maceyka, M.; Sankala, H.; Hait, N.C.; Le Stunff, H.; Liu, H.; Toman, R.; Collier, C.; Zhang, M.; Satin, L.S.; Merrill, A.H., Jr.; et al. SphK1 and SphK2, sphingosine kinase isoenzymes with opposing functions in sphingolipid metabolism. J. Biol. Chem. 2005, 280, 37118–37129. [Google Scholar] [CrossRef]
- Ye, G.-M.; Chen, C.; Huang, S.; Han, D.-D.; Guo, J.-H.; Wan, B.; Long, Y. Cloning and characterization a novel human 1-acyl-sn-glycerol-3-phosphate acyltransferase gene AGPAT7. DNA Seq. 2005, 16, 386–390. [Google Scholar] [CrossRef]
- Cao, J.; Shan, D.; Revett, T.; Li, D.; Wu, L.; Liu, W.; Tobin, J.F.; Gimeno, R.E. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 2008, 283, 19049–19057. [Google Scholar] [CrossRef] [PubMed]
- Valentine, W.J.; Yanagida, K.; Kawana, H.; Kono, N.; Noda, N.N.; Aoki, J.; Shindou, H. Update and nomenclature proposal for mammalian lysophospholipid acyltransferases, which create membrane phospholipid diversity. J. Biol. Chem. 2022, 298, 101470. [Google Scholar] [CrossRef] [PubMed]
- Hishikawa, D.; Shindou, H.; Kobayashi, S.; Nakanishi, H.; Taguchi, R.; Shimizu, T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA 2008, 105, 2830–2835. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Renigunta, A.; Yang, J.; Waldegger, S. Claudin-4 forms paracellular chloride channel in the kidney and requires claudin-8 for tight junction localization. Proc. Natl. Acad. Sci. USA 2010, 107, 18010–18015. [Google Scholar] [CrossRef] [PubMed]
- Hiti, A.L.; Rideout WM 3rd Laug, W.E.; Jones, P.A. Plasminogen activator regulation by transforming growth factor-beta in normal and neoplastic human urothelium. Cancer Commun. 1990, 2, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.A.; Kennedy, W.J.; Harnden, P.; Selby, P.J.; Trejdosiewicz, L.K.; Southgate, J. Identification of genes involved in human urothelial cell-matrix interactions: Implications for the progression pathways of malignant urothelium. Cancer Res. 2001, 61, 1678–1685. [Google Scholar] [PubMed]
- Alamri, A.; Biswas, L.; Watson, D.G.; Shu, X. Deletion of TSPO Resulted in Change of Metabolomic Profile in Retinal Pigment Epithelial Cells. Int. J. Mol. Sci. 2019, 20, 1387. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.N.; Lomasney, J.W.; Mak, E.C.; Lorand, L. Interactions of G(h)/transglutaminase with phospholipase Cdelta1 and with GTP. Proc. Natl. Acad. Sci. USA 1999, 96, 11815–11819. [Google Scholar] [CrossRef]
- Duquesnes, N.; Lezoualc’h, F.; Crozatier, B. PKC-delta and PKC-epsilon: Foes of the same family or strangers? J. Mol. Cell. Cardiol. 2011, 51, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Eckert, J.J.; McCallum, A.; Mears, A.; Rumsby, M.G.; Cameron, I.T.; Fleming, T.P. PKC signalling regulates tight junction membrane assembly in the pre-implantation mouse embryo. Reproduction 2004, 127, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.C.; Shabir, S.; Georgopoulos, N.T.; Southgate, J. Ketamine-Induced Apoptosis in Normal Human Urothelial Cells. Am. J. Pathol. 2016, 21, 1267–1277. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.-J.; Middleton, R.J.; Kam, W.W.-Y.; Chin, D.Y.; Hatty, C.R.; Chan, R.H.Y.; Banati, R.B. Functional gains in energy and cell metabolism after TSPO gene insertion. Cell Cycle 2017, 16, 436–447. [Google Scholar] [CrossRef]
- Yao, R.; Pan, R.; Shang, C.; Li, X.; Cheng, J.; Xu, J.; Li, Y. Translocator Protein 18 kDa (TSPO) Deficiency Inhibits Microglial Activation and Impairs Mitochondrial Function. Front. Pharmacol. 2020, 11, 986. [Google Scholar] [CrossRef] [PubMed]
- Somashekar, B.S.; Kamarajan, P.; Danciu, T.; Kapila, Y.L.; Chinnaiyan, A.M.; Rajendiran, T.M.; Ramamoorthy, A. Magic angle spinning NMR-based metabolic profiling of head and neck squamous cell carcinoma tissues. J. Proteome Res. 2011, 10, 5232–5241. [Google Scholar] [CrossRef]
- Leoni, G.; Neumann, P.-A.; Sumagin, R.; Denning, T.L.; Nusrat, A. Wound repair: Role of immune-epithelial interactions. Mucosal. Immunol. 2015, 8, 959–968. [Google Scholar] [CrossRef]
- Green, K.A.; Almholt, K.; Ploug, M.; Rønø, B.; Castellino, F.J.; Johnsen, M.; Bugge, T.H.; Rømer, J.; Lund, L.R. Profibrinolytic effects of metalloproteinases during skin wound healing in the absence of plasminogen. J. Investig. Dermatol. 2008, 128, 2092–2101. [Google Scholar] [CrossRef]
- Tabe, S.; Hikiji, H.; Ariyoshi, W.; Hashidate-Yoshida, T.; Shindou, H.; Shimizu, T.; Okinaga, T.; Seta, Y.; Tominaga, K.; Nishihara, T. Lysophosphatidylcholine acyltransferase 4 is involved in chondrogenic differentiation of ATDC5 cells. Sci. Rep. 2017, 7, 16701. [Google Scholar] [CrossRef]
- Ren, S.; Shatadal, S.; Shen, G.X. Protein kinase C-beta mediates lipoprotein-induced generation of PAI-1 from vascular endothelial cells. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E656–E662. [Google Scholar] [CrossRef]
- Congote, L.F.; Temmel, N. The C-terminal 26-residue peptide of serpin A1 stimulates proliferation of breast and liver cancer cells: Role of protein kinase C and CD47. FEBS Lett. 2004, 576, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Takeuchi, K.; Csaki, L.S.; Reue, K. Lipin-1 phosphatidic phosphatase activity modulates phosphatidate levels to promote peroxisome proliferator-activated receptor γ (PPARγ) gene expression during adipogenesis. J. Biol. Chem. 2012, 287, 3485–3494. [Google Scholar] [CrossRef] [PubMed]
- Batarseh, A.; Li, J.; Papadopoulos, V. Protein kinase C epsilon regulation of translocator protein (18 kDa) Tspo gene expression is mediated through a MAPK pathway targeting STAT3 and c-Jun transcription factors. Biochemistry 2010, 49, 4766–4778. [Google Scholar] [CrossRef] [PubMed]
- Shabir, S.; Cross, W.; Kirkwood, L.A.; Pearson, J.F.; Appleby, P.A.; Walker, D.; Eardley, I.; Southgate, J. Functional expression of purinergic P2 receptors and transient receptor potential channels by the human urothelium. Am. J. Physiol. Renal. Physiol. 2013, 305, F396–F406. [Google Scholar] [CrossRef]
- Frömter, E.; Diamond, J. Route of passive ion permeation in epithelia. Nat. New Biol. 1972, 235, 9–13. [Google Scholar] [CrossRef]
- Shaw, N.J.; Georgopoulos, N.T.; Southgate, J.; Trejdosiewicz, L.K. Effects of loss of p53 and p16 function on life span and survival of human urothelial cells. Int. J. Cancer 2005, 116, 634–639. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bray, N.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal RNA-Seq quantification with kallisto. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Res 2015, 4, 1521. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef]
- Fang, Z. GSEAPY: Gene Set Enrichment Analysis in Python. Zenodo. 2020. Available online: https://zenodo.org/record/3748085#.Yy2eLddByUk (accessed on 9 November 2020).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef] [PubMed]
- Vert, J.-P.; Foveau, N.; Lajaunie, C.; Vandenbrouck, Y. An accurate and interpretable model for siRNA efficacy prediction. BMC Bioinform. 2006, 7, 520. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.; Springthorpe, V.; Rossoni, L.; Minde, D.-P.; Langer, S.; Walker, H.; Alstrom-Moore, A.; Larson, T.; Lilley, K.; Eastham, G.; et al. Systems Analyses Reveal the Resilience of Escherichia coli Physiology during Accumulation and Export of the Nonnative Organic Acid Citramalate. mSystems 2019, 4, e00187-19. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef]
- Koelmel, J.P.; Kroeger, N.M.; Ulmer, C.Z.; Bowden, J.A.; Patterson, R.E.; Cochran, J.A.; Beecher, C.W.W.; Garrett, T.J.; Yost, R.A. LipidMatch: An automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinform. 2017, 18, 331. [Google Scholar] [CrossRef]
- Kind, T.; Liu, K.-H.; Lee, D.Y.; DeFelice, B.; Meissen, J.K.; Fiehn, O. LipidBlast in silico tandem mass spectrometry database for lipid identification. Nat. Methods 2013, 10, 755–758. [Google Scholar] [CrossRef]
- Böck, M.; Hinley, J.; Schmitt, C.; Wahlicht, T.; Kramer, S.; Southgate, J. Identification of ELF3 as an early transcriptional regulator of human urothelium. Dev. Biol. 2014, 386, 321–330. [Google Scholar] [CrossRef]

| Antigen | Supplier | Reference | Host Species | Concentration |
|---|---|---|---|---|
| LPCAT4 | Stratech, Ely, UK. | LS-C749318 | Rabbit | 1:3000 |
| Claudin 4 | Invitrogen Europe Ltd. | 3E2C1 | Mouse | 1:1000 |
| CK13 | OriGene supplied by Cambridge Bioscience, Cambridge, UK | IC7 | Mouse | 1:500 |
| TSPO | Abcam, Cambridge, UK | EPR5384 | Rabbit | 1:10,000 |
| β-actin | Sigma, Merck UK | A5441 | Mouse | 1:10,000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mason, A.S.; Varley, C.L.; Foody, O.M.; Li, X.; Skinner, K.; Walker, D.; Larson, T.R.; Wakamatsu, D.; Baker, S.C.; Southgate, J. LPCAT4 Knockdown Alters Barrier Integrity and Cellular Bioenergetics in Human Urothelium. Int. J. Mol. Sci. 2022, 23, 11871. https://doi.org/10.3390/ijms231911871
Mason AS, Varley CL, Foody OM, Li X, Skinner K, Walker D, Larson TR, Wakamatsu D, Baker SC, Southgate J. LPCAT4 Knockdown Alters Barrier Integrity and Cellular Bioenergetics in Human Urothelium. International Journal of Molecular Sciences. 2022; 23(19):11871. https://doi.org/10.3390/ijms231911871
Chicago/Turabian StyleMason, Andrew S., Claire L. Varley, Olivia M. Foody, Xiang Li, Katie Skinner, Dawn Walker, Tony R. Larson, Daisuke Wakamatsu, Simon C. Baker, and Jennifer Southgate. 2022. "LPCAT4 Knockdown Alters Barrier Integrity and Cellular Bioenergetics in Human Urothelium" International Journal of Molecular Sciences 23, no. 19: 11871. https://doi.org/10.3390/ijms231911871
APA StyleMason, A. S., Varley, C. L., Foody, O. M., Li, X., Skinner, K., Walker, D., Larson, T. R., Wakamatsu, D., Baker, S. C., & Southgate, J. (2022). LPCAT4 Knockdown Alters Barrier Integrity and Cellular Bioenergetics in Human Urothelium. International Journal of Molecular Sciences, 23(19), 11871. https://doi.org/10.3390/ijms231911871







