Small Heat Shock Proteins Collaborate with FAIM to Prevent Accumulation of Misfolded Protein Aggregates
Abstract
1. Introduction
2. Results
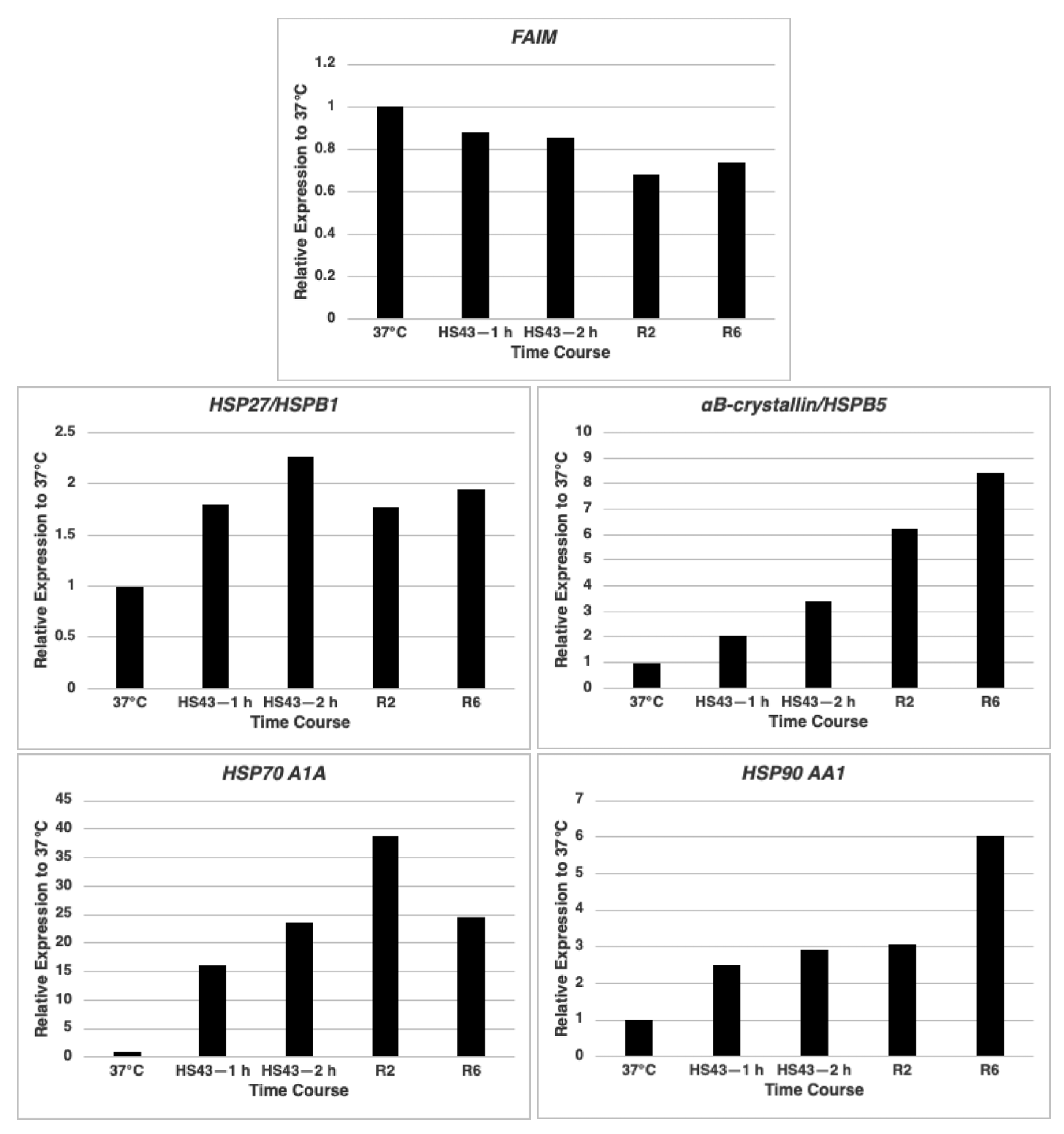
2.1. FAIM mRNA Expression Is Not Up-Regulated after Cellular Stress Induction in HeLa Cells
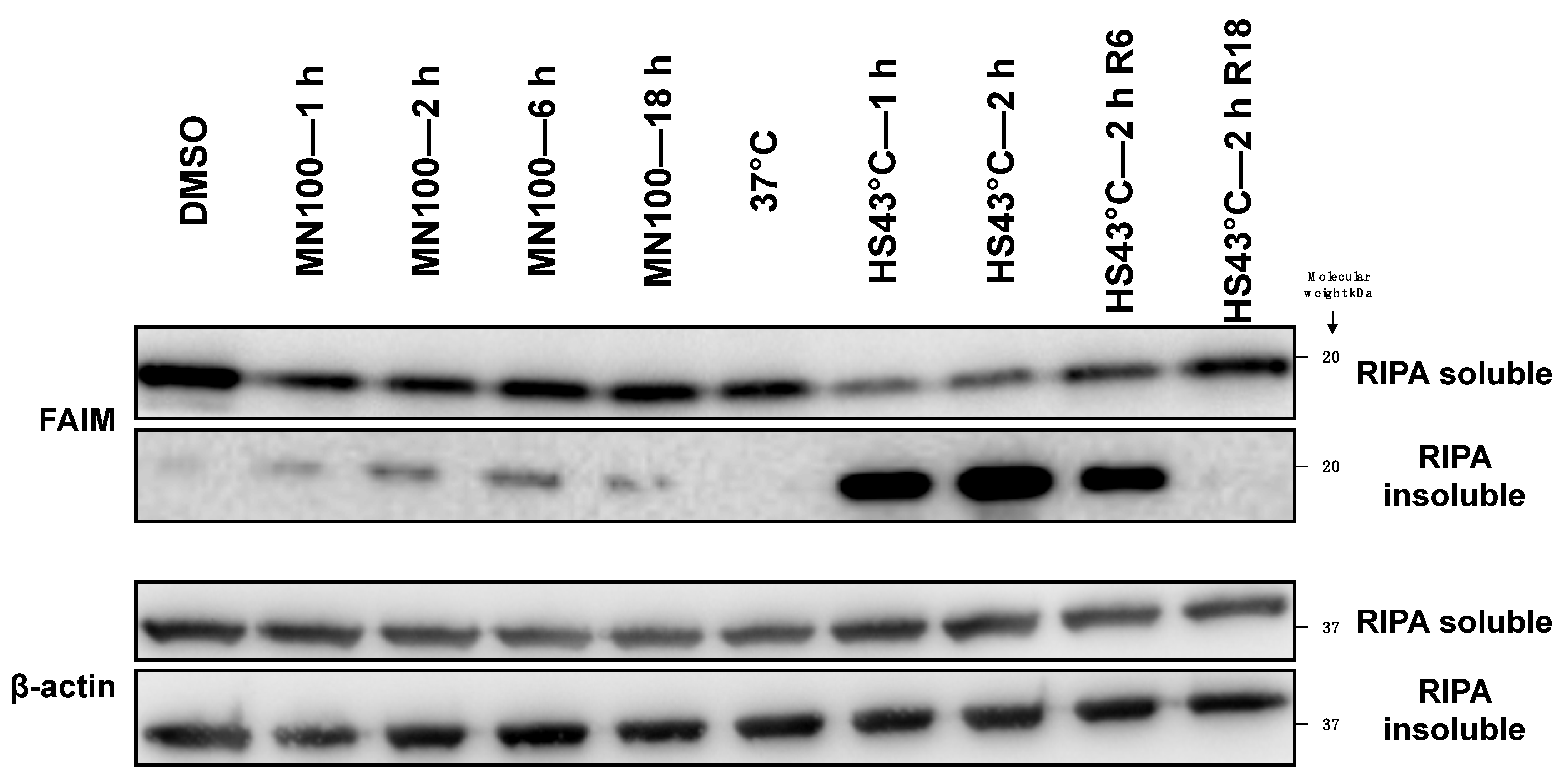
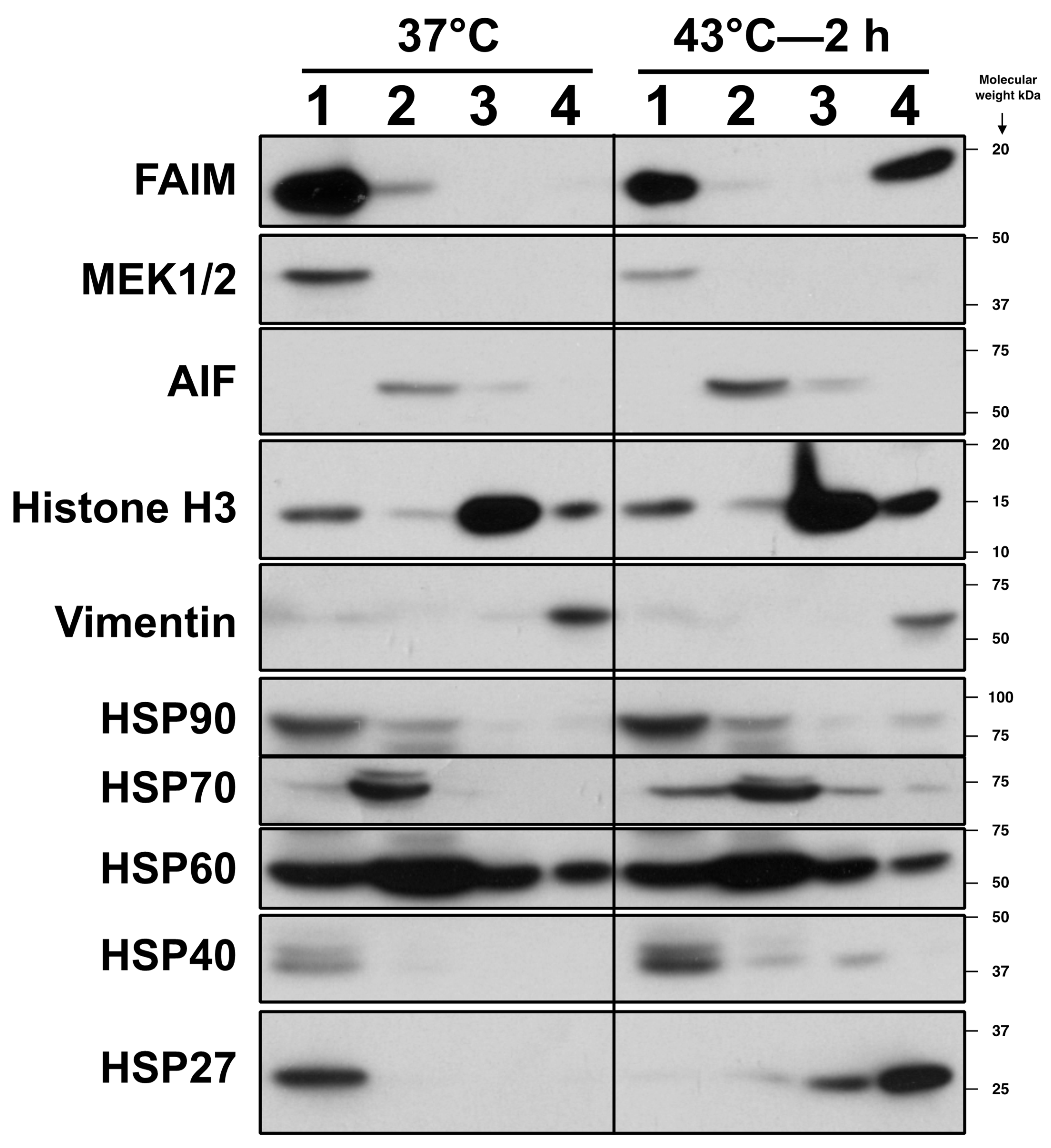
2.2. FAIM Protein Shifts to the Detergent-Insoluble Fraction after Stress Induction
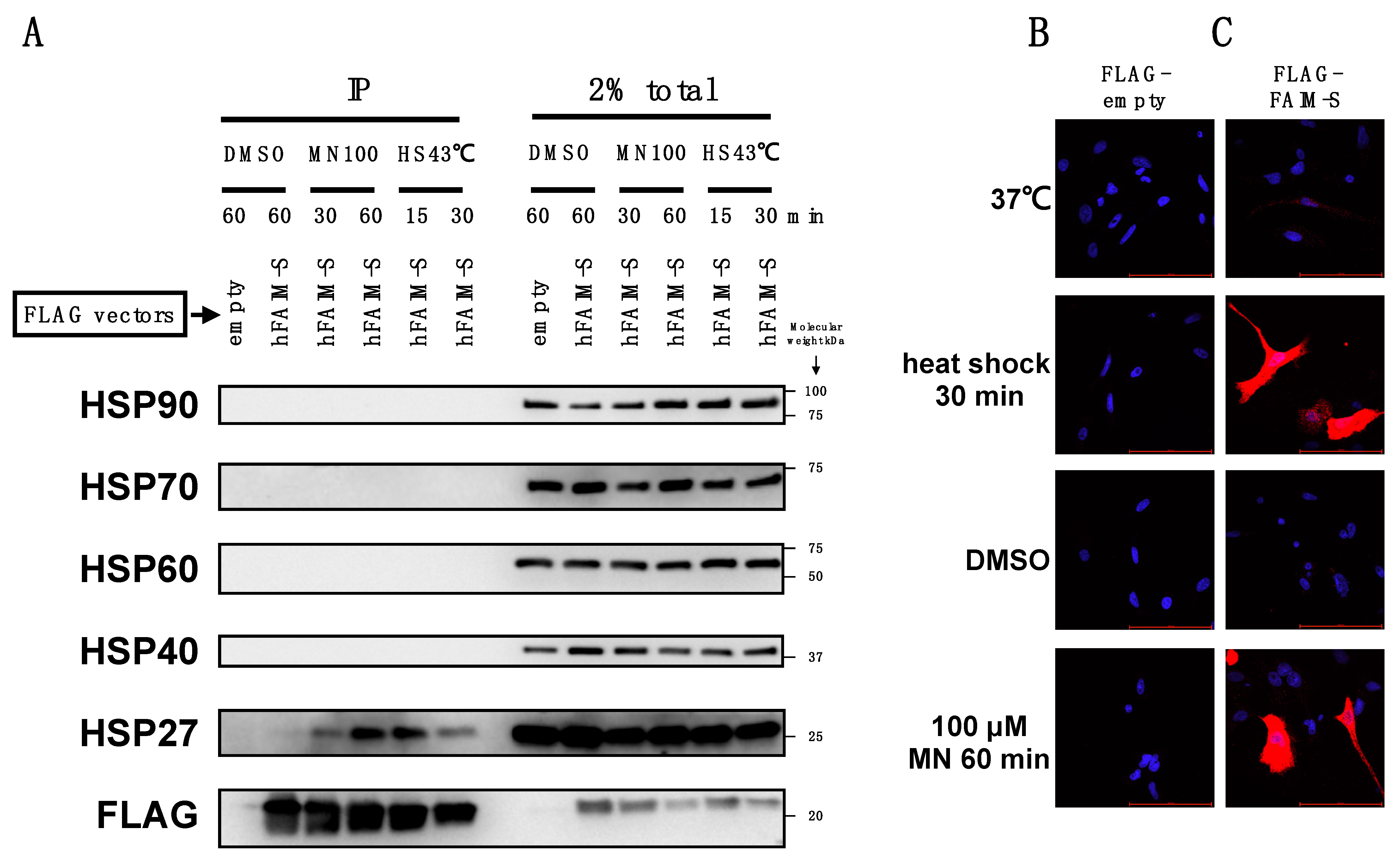
2.3. FAIM Is Recruited to the Complex Containing sHSPs after Cellular Stress Induction
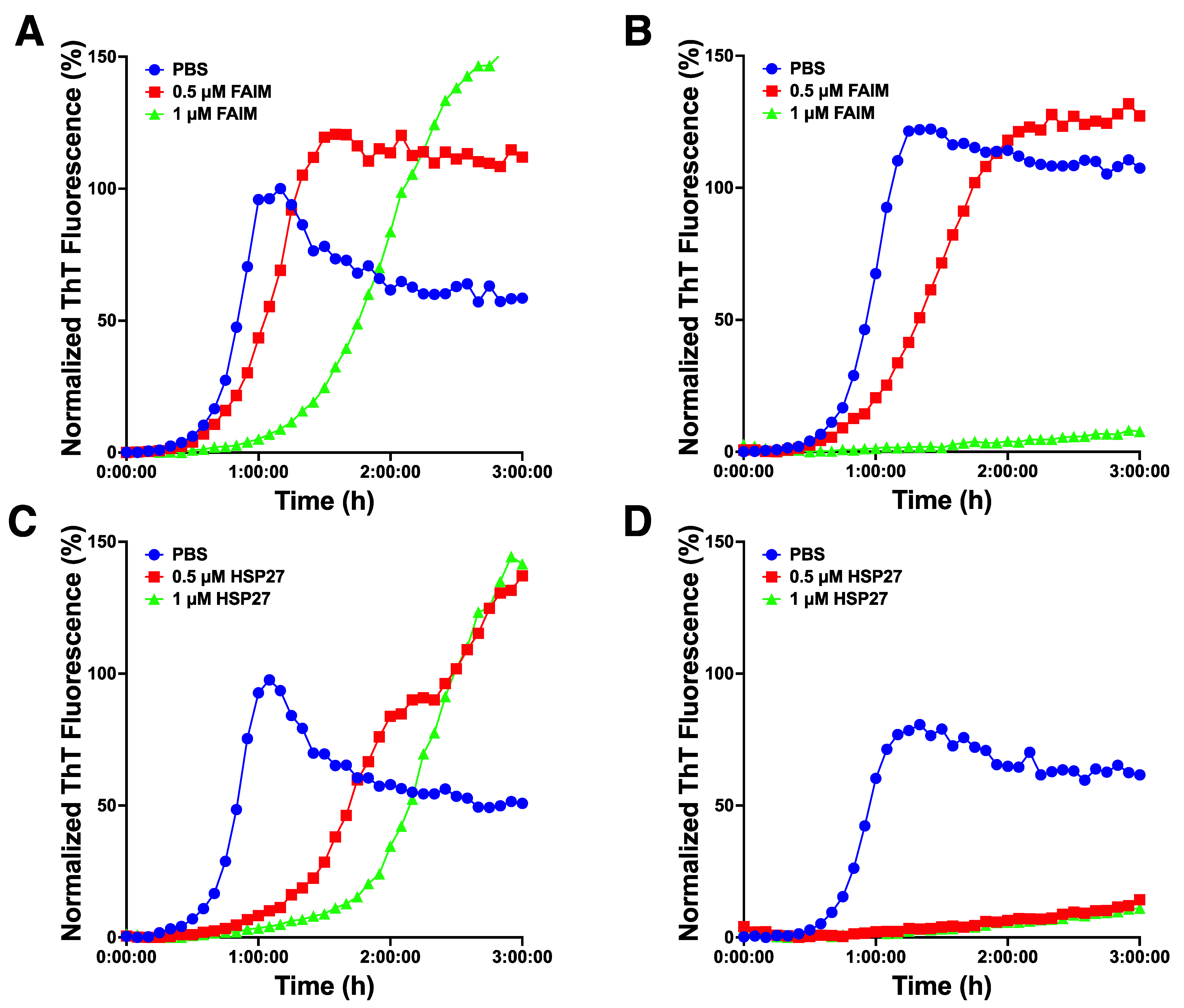
2.4. Recombinant FAIM Inhibits β-Amyloid Aggregation/Fibrillization in Collaboration with HSP27 in an In Vitro Cell-Free System
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Gene Expression Analysis by qPCR
4.3. In Vitro Cellular Stress Induction
4.4. Cell Culture and Transfection
4.5. His-Tag Recombinant Protein Production
4.6. Construction of FLAG-Tag FAIM-S Expression Vector
4.7. Thioflavin T Fluorescence Assay
4.8. Western Blotting
4.9. Immunoprecipitation
4.10. In Situ Proximity Ligation Assay (PLA)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zuo, D.; Subjeck, J.; Wang, X.-Y. Unfolding the Role of Large Heat Shock Proteins: New Insights and Therapeutic Implications. Front. Immunol. 2016, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Rothstein, T.L. FAIM Is a Non-redundant Defender of Cellular Viability in the Face of Heat and Oxidative Stress and Interferes with Accumulation of Stress-Induced Protein Aggregates. Front. Mol. Biosci. 2020, 7, 32. [Google Scholar] [CrossRef]
- Kaku, H.; Ludlow, A.V.; Gutknecht, M.F.; Rothstein, T.L. FAIM Opposes Aggregation of Mutant SOD1 That Typifies Some Forms of Familial Amyotrophic Lateral Sclerosis. Front. Neurosci. 2020, 14, 110. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Ludlow, A.V.; Gutknecht, M.F.; Rothstein, T.L. Fas Apoptosis Inhibitory Molecule Blocks and Dissolves Pathological Amyloid-β Species. Front. Mol. Neurosci. 2021, 14, 750578. [Google Scholar] [CrossRef] [PubMed]
- Schneider, T.J.; Fischer, G.M.; Donohoe, T.J.; Colarusso, T.P.; Rothstein, T.L. A Novel Gene Coding for a Fas Apoptosis Inhibitory Molecule (FAIM) Isolated from Inducibly Fas-resistant B Lymphocytes. J. Exp. Med. 1999, 189, 949–956. [Google Scholar] [CrossRef]
- Zhong, X.; Schneider, T.J.; Cabral, D.S.; Donohoe, T.J.; Rothstein, T.L. An alternatively Spliced Long Form of Fas Apoptosis Inhibitory Molecule (FAIM) With Tissue-Specific Expression in the Brain. Mol. Immunol. 2001, 38, 65–72. [Google Scholar] [CrossRef]
- Li, S.; Armstrong, C.M.; Bertin, N.; Ge, H.; Milstein, S.; Boxem, M.; Vidalain, P.O.; Han, J.D.; Chesneau, A.; Vidal, M.; et al. A Map of the Interactome Network of the Metazoan C. Elegans. Science 2004, 303, 540–543. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Andley, U.P.; Song, Z.; Wawrousek, E.F.; Fleming, T.P.; Bassnett, S. Differential Protective Activity of Alpha a- and Alphab-Crystallin in Lens Epithelial Cells. J. Biol. Chem. 2000, 275, 36823–36831. [Google Scholar] [CrossRef]
- Wang-Su, S.T.; McCormack, A.L.; Yang, S.; Hosler, M.R.; Mixon, A.; Riviere, M.A.; Wilmarth, P.A.; Andley, U.P.; Garland, D.; Li, H.; et al. Proteome Analysis of Lens Epithelia, Fibers, and The HLE B-3 Cell Line. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4829–4836. [Google Scholar] [CrossRef][Green Version]
- Voorter, C.E.; Wintjes, L.; Bloemendal, H.; de Jong, W.W. Relocalization of αB-Crystallin by Heat Shock in Ovarian Carcinoma Cells. FEBS Lett. 1992, 309, 111–114. [Google Scholar] [CrossRef]
- van de Klundert, F.A.; Gijsen, M.L.; van den IJssel, P.R.; Snoeckx, L.H.; de Jong, W.W. αB-Crystallin and Hsp25 in Neonatal Cardiac Cells—Differences in Cellular Localization Under Stress Conditions. Eur. J. Cell Biol. 1998, 75, 38–45. [Google Scholar] [CrossRef]
- Söderberg, O.; Gullberg, M.; Jarvius, M.; Ridderstråle, K.; Leuchowius, K.-J.; Jarvius, J.; Wester, K.; Hydbring, P.; Bahram, F.; Larsson, L.-G.; et al. Direct Observation of Individual Endogenous Protein Complexes In Situ by Proximity Ligation. Nat. Methods 2006, 3, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.; Skalli, O. Synemin: Molecular Features and the Use of Proximity Ligation Assay to Study Its Interactions. Methods Enzymol. 2016, 568, 537–555. [Google Scholar] [CrossRef] [PubMed]
- Raman, B.; Ban, T.; Sakai, M.; Pasta, S.Y.; Ramakrishna, T.; Naiki, H.; Goto, Y.; Rao, C.M. αB-crystallin, a Small Heat-Shock Protein, Prevents the Amyloid Fibril Growth of an Amyloid Β-Peptide and Β2-Microglobulin. Biochem. J. 2005, 392, 573–581. [Google Scholar] [CrossRef]
- Fonte, V.; Kipp, D.R.; Yerg, J.; Merin, D.; Forrestal, M.; Wagner, E.; Roberts, C.M.; Link, C.D. Suppression of In Vivo Β-Amyloid Peptide Toxicity by Overexpression of the HSP-16.2 Small Chaperone Protein. J. Biol. Chem. 2008, 283, 784–791. [Google Scholar] [CrossRef]
- Jovcevski, B.; Kelly, M.A.; Rote, A.P.; Berg, T.; Gastall, H.Y.; Benesch, J.L.; Aquilina, J.A.; Ecroyd, H. Phosphomimics Destabilize Hsp27 Oligomeric Assemblies and Enhance Chaperone Activity. Chem. Biol. 2015, 22, 186–195. [Google Scholar] [CrossRef]
- Mymrikov, E.; Daake, M.; Richter, B.; Haslbeck, M.; Buchner, J. The Chaperone Activity and Substrate Spectrum of Human Small Heat Shock Proteins. J. Biol. Chem. 2017, 292, 672–684. [Google Scholar] [CrossRef]
- Taylor, R.P.; Benjamin, I.J. Small Heat Shock Proteins: A New Classification Scheme in Mammals. J. Mol. Cell. Cardiol. 2005, 38, 433–444. [Google Scholar] [CrossRef]
- Garrido, C.; Paul, C.; Seigneuric, R.; Kampinga, H.H. The Small Heat Shock Proteins Family: The Long Forgotten Chaperones. Int. J. Biochem. Cell Biol. 2012, 44, 1588–1592. [Google Scholar] [CrossRef]
- Sun, Y.; Macrae, T.H. Small Heat Shock Proteins: Molecular Structure and Chaperone Function. Cell. Mol. Life Sci. CMLS 2005, 62, 2460–2476. [Google Scholar] [CrossRef] [PubMed]
- Jakob, U.; Gaestel, M.; Engel, K.; Buchner, J. Small Heat Shock Proteins Are Molecular Chaperones. J. Biol. Chem. 1993, 268, 1517–1520. [Google Scholar] [CrossRef]
- Mogk, A.; Bukau, B. Role of sHsps in Organizing Cytosolic Protein Aggregation and Disaggregation. Cell Stress Chaperon 2017, 22, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Delbecq, S.P.; Jehle, S.; Klevit, R. Binding Determinants of the Small Heat Shock Protein, Ab-Crystallin: Recognition of the ‘Ixi’motif. EMBO J. 2012, 31, 4587–4594. [Google Scholar] [CrossRef]
- Lelj-Garolla, B.; Mauk, A.G. Roles of the N- and C-Terminal Sequences in Hsp27 Self-Association and Chaperone Activity. Protein Sci. 2011, 21, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Pasta, S.Y.; Raman, B.; Ramakrishna, T.; Rao, C.M. The IXI/V Motif in the C-Terminal Extension of Alpha-Crystallins: Alternative Interactions and Oligomeric Assemblies. Mol. Vis. 2004, 10, 655–662. [Google Scholar] [PubMed]
- Bakthisaran, R.; Tangirala, R.; Rao, C.M. Small Heat Shock Proteins: Role in Cellular Functions and Pathology. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2015, 1854, 291–319. [Google Scholar] [CrossRef]
- Sole, C.; Dolcet, X.; Segura, M.F.; Gutierrez, H.; Diaz-Meco, M.T.; Gozzelino, R.; Sanchis, J.R.; Gallego, C.; Moscat, J.; Davies, A.M.; et al. The Death Receptor Antagonist FAIM Promotes Neurite Outgrowth by a Mechanism That Depends on ERK and NF-Κb Signaling. J. Cell Biol. 2004, 167, 479–492. [Google Scholar] [CrossRef]
- Williams, K.L.; Rahimtula, M.; Mearow, K.M. Heat Shock Protein 27 Is Involved in Neurite Extension and Branching of Dorsal Root Ganglion Neurons in Vitro. J. Neurosci. Res. 2006, 84, 716–723. [Google Scholar] [CrossRef]
- Segura, M.F.; Sole, C.; Pascual, M.; Moubarak, R.S.; Perez-Garcia, M.J.; Gozzelino, R.; Iglesias, V.; Badiola, N.; Bayascas, J.R.; Llecha, N.; et al. The Long Form of Fas Apoptotic Inhibitory Molecule Is Expressed Specifically in Neurons and Protects Them Against Death Receptor-Triggered Apoptosis. J. Neurosci. 2007, 27, 11228–11241. [Google Scholar] [CrossRef]
- Li, G.; Qu, L.; Ma, S.; Wu, Y.; Jin, C.; Zheng, X. Structure Determination of Human Fas Apoptosis Inhibitory Molecule and Identification of the Critical Residues Linking the Interdomain Interaction to the Anti-Apoptotic Activity. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Mehlen, P.; Schulze-Osthoff, K.; Arrigo, A.P. Small Stress Proteins as Novel Regulators of Apoptosis: Heat Shock Protein 27 Blocks Fas/APO-1-And Staurosporine-Induced Cell Death. J. Biol. Chem. 1996, 271, 16510–16514. [Google Scholar] [CrossRef] [PubMed]
- Kaku, H.; Rothstein, T.L. Fas Apoptosis Inhibitory Molecule Enhances CD40 Signaling in B Cells and Augments the Plasma Cell Compartment. J. Immunol. 2009, 183, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Parcellier, A.; Schmitt, E.; Gurbuxani, S.; Seigneurin-Berny, D.; Pance, A.; Chantôme, A.; Plenchette, S.; Khochbin, S.; Solary, E.; Garrido, C. HSP27 is a Ubiquitin-Binding Protein Involved in I-Κbα Proteasomal Degradation. Mol. Cell. Biol. 2003, 23, 5790–5802. [Google Scholar] [CrossRef]
- Vavouri, T.; Semple, J.I.; Garcia-Verdugo, R.; Lehner, B. Intrinsic Protein Disorder and Interaction Promiscuity are Widely Associated with Dosage Sensitivity. Cell 2009, 138, 198–208. [Google Scholar] [CrossRef]
- Gibson, T.J.; Seiler, M.; Veitia, R.A. The Transience of Transient Overexpression. Nat. Methods 2013, 10, 715–721. [Google Scholar] [CrossRef]
- Veitia, R.A.; Potier, M.C. Gene Dosage Imbalances: Action, Reaction, And Models. Trends Biochem. Sci. 2015, 40, 309–317. [Google Scholar] [CrossRef]
- Carra, S.; Alberti, S.; Arrigo, P.A.; Benesch, J.L.; Benjamin, I.J.; Boelens, W.; Bartelt-Kirbach, B.; Brundel, B.J.J.M.; Buchner, J.; Bukau, B.; et al. The Growing World of Small Heat Shock Proteins: From Structure to Functions. Cell Stress Chaperon 2017, 22, 601–611. [Google Scholar] [CrossRef]
- Hemond, M.; Rothstein, T.L.; Wagner, G. Fas Apoptosis Inhibitory Molecule Contains a Novel Β-Sandwich in Contact with a Partially Ordered Domain. J. Mol. Biol. 2009, 386, 1024–1037. [Google Scholar] [CrossRef]
- Bardwell, J.C.A.; Jakob, U. Conditional Disorder in Chaperone Action. Trends Biochem. Sci. 2012, 37, 517–525. [Google Scholar] [CrossRef]
- Kim, K.K.; Kim, R.; Kim, S.-H. Crystal Structure of a Small Heat-Shock Protein. Nature 1998, 394, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Kriehuber, T.; Rattei, T.; Weinmaier, T.; Bepperling, A.; Haslbeck, M.; Buchner, J. Independent Evolution of the Core Domain and Its Flanking Sequences in Small Heat Shock Proteins. FASEB J. 2010, 24, 3633–3642. [Google Scholar] [CrossRef] [PubMed]
- Carriba, P.; Jimenez, S.; Navarro, V.; Moreno-Gonzalez, I.; Barneda-Zahonero, B.; Moubarak, R.S.; Lopez-Soriano, J.; Gutierrez, A.; Vitorica, J.; Comella, J.X. Amyloid-β Reduces the Expression of Neuronal FAIM-L, Thereby Shifting the Inflammatory Response Mediated by Tnfα from Neuronal Protection to Death. Cell Death Dis. 2015, 6, e1639. [Google Scholar] [CrossRef] [PubMed]
- Renkawek, K.; Bosman, G.I.C.G.M.; De Jong, W.W. Expression of Small Heat-Shock Protein Hsp 27 in Reactive Gliosis in Alzheimer Disease and Other Types of Dementia. Acta Neuropathol. 1994, 87, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Renkawek, K.; Voorter, C.E.M.; Bosman, G.J.C.G.M.; Van Workum, F.P.A.; De Jong, W.W. Expression of αB-Crystallin in Alzheimer’s Disease. Acta Neuropathol. 1994, 87, 155–160. [Google Scholar] [CrossRef]
- Kaku, H.; Rothstein, T.L. Fas Apoptosis Inhibitory Molecule Expression in B Cells Is Regulated through IRF4 in a Feed-Forward Mechanism. J. Immunol. 2009, 183, 5575–5581. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Fu, J.C.-M.; Lee, Y.-C.; Lu, P.-J. Hyperthermia Stress Activates Heat Shock Protein Expression via Propyl Isomerase 1 Regulation with Heat Shock Factor 1. Mol. Cell. Biol. 2013, 33, 4889–4899. [Google Scholar] [CrossRef]
- Koczor, C.A.; Shokolenko, I.N.; Boyd, A.K.; Balk, S.P.; Wilson, G.L.; LeDoux, S.P. Mitochondrial DNA Damage Initiates a Cell Cycle Arrest by a Chk2-associated Mechanism in Mammalian Cells. J. Biol. Chem. 2009, 284, 36191–36201. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaku, H.; Balaj, A.R.; Rothstein, T.L. Small Heat Shock Proteins Collaborate with FAIM to Prevent Accumulation of Misfolded Protein Aggregates. Int. J. Mol. Sci. 2022, 23, 11841. https://doi.org/10.3390/ijms231911841
Kaku H, Balaj AR, Rothstein TL. Small Heat Shock Proteins Collaborate with FAIM to Prevent Accumulation of Misfolded Protein Aggregates. International Journal of Molecular Sciences. 2022; 23(19):11841. https://doi.org/10.3390/ijms231911841
Chicago/Turabian StyleKaku, Hiroaki, Allison R. Balaj, and Thomas L. Rothstein. 2022. "Small Heat Shock Proteins Collaborate with FAIM to Prevent Accumulation of Misfolded Protein Aggregates" International Journal of Molecular Sciences 23, no. 19: 11841. https://doi.org/10.3390/ijms231911841
APA StyleKaku, H., Balaj, A. R., & Rothstein, T. L. (2022). Small Heat Shock Proteins Collaborate with FAIM to Prevent Accumulation of Misfolded Protein Aggregates. International Journal of Molecular Sciences, 23(19), 11841. https://doi.org/10.3390/ijms231911841





