1. Introduction
RNA is the template that codes for the proteins required to create cellular functions. RNA is structurally similar to DNA, but its function and chemical composition are fundamentally different. At a higher level, RNA is divided into two main groups: coding RNA that accounts for approximately 2% of all RNAs, and non-coding RNA (ncRNA) that accounts for the majority (>90%) of RNAs [
1]. Non-coding RNA refers to all RNAs that are transcribed from DNA but do not code for proteins. Additionally, ncRNA can be categorized into two groups according to the size of the sequence: long non-coding RNAs (lncRNAs) with sequences >200 nucleotides and small non-coding RNAs (sncRNAs) with sequences shorter than 200 nucleotides [
2]. In previous research, ncRNAs have frequently been referred to as “useless genes” or transcriptional “noise” [
3,
4]. In contrast, a growing number of experiments have demonstrated that ncRNAs play important biological roles in a variety of biological processes, including gene regulation/expression, gene silencing, RNA modification and processing, as well as multiple important roles in life activities [
5,
6,
7]. Numerous plant-specific biological processes, including the regulation of plant nutrient homeostasis, development, and stress responses, have been linked to ncRNAs [
8,
9,
10]. MiRNAs and trans-acting siRNAs, for instance, contribute to leaf senescence in
Arabidopsis; miR164 and its target ORE1 control leaf senescence in
Arabidopsis, and as miR164 expression declines, ORE1 expression eventually increases [
11]. In addition, overexpression of miR398b has been shown to decrease the transcript levels of genes encoding superoxide dismutase (CSD1, CSD2, SODX, and CCSD), which resulted in the production of reactive oxygen species (ROS) and increased rice resistance to
Magnaporthe oryzae [
12,
13]. In recent years, to facilitate subsequent analyses and research of transcripts, ncRNA identification has been one of the tasks that needs to be addressed. Numerous bioinformatics methods and experiments have been developed for ncRNA identification and to evaluate their functions [
14,
15]. Genomic SELEX, microarray analysis, and chemical RNA-Seq are the most commonly used experimental techniques [
16]; however, they are costly and time-consuming. Therefore, bioinformatics may be a more effective means of addressing the biological aspects of the problem.
Kong et al. developed the Coding Potential Calculator (CPC) in 2007 [
17]. The CPC selected a number of biologically significant features, including ORF quality, coverage, and integrity. These features were incorporated into a support vector machine for coding potential identity, but its performance was dependent on sequence comparisons. CPC was revised in 2017 with the release of CPC2 [
18]. CPC2 is faster and more accurate than CPC, and, as an input to the SVM model, it uses ORF size and integrity, a Fickett score, and the isoelectric point extracted from the original RNA sequence. CPC2 is a relatively neutral tool, which makes it somewhat more applicable to transcriptomes of non-model organisms. CPAT, developed by Wang et al. in 2013, is a logistic-regression-model-based ncRNA identification tool that classifies ncRNAs and cRNAs based on features such as ORF size and coverage, Fickett score, and hexamer score [
19]. CNCI was proposed by Liang et al. in 2013, and while it is also based on the same SVM classifier as CPC2, it uses different features, categorizing ncRNA and cRNA based on ANT features [
20]. CNIT is an updated version of CNCI that was released in 2019. CNIT employs the more robust integrated machine model XGBoost for classification [
21]. Tong et al. introduced CPPred in 2019 [
22] as an SVM-based tool. This tool distinguishes between ncRNAs and coding RNAs using the same ORF features as CPC2, as well as the isoelectric point, stability index, gravity three peptide, hexamer score, CTD, and Fickett score features. A number of tools have been published that can distinguish between ncRNA and coding RNA; however, the tools have some limitations, for example, their application is mainly limited to vertebrates and mammals. In addition, these tools rarely consider using plants for model training. Most tools only use the model plant
Arabidopsis, and rarely involve other non-model plants. Moreover, since ncRNAs of animals are mainly transcribed by polymerase II, while ncRNAs of plants are mainly transcribed by RNA polymerase II, IV, and V [
23], and ncRNAs are characterized by low-level expression and cross-species conservation [
24], these tools for ncRNA identification in animals cannot guarantee the reliability in plants. Therefore, it is necessary to construct a powerful tool for ncRNA identification in plants.
Automatic machine learning (AutoML) is the process of applying machine learning to real-world problems in an automated manner. Since 2013, frameworks have been developed that have been based on the AutoML concept. AutoWEKA was the first AutoML framework to emerge [
25]; it automatically selected models and hyperparameters. Additionally, H2O [
26] and TPOT [
27] were created. H2O is a JAVA-based framework that supports multiple types of grid searches to identify the optimal parameters following the generation of an integrated model. At its core, TPOT is a tree-based process optimization tool based on a genetic algorithm. Today, more and more frameworks, such as AutoGluon [
28] and AutoKeras [
29], have been developed based on the concept of AutoML. These frameworks have also been applied to Alzheimer’s disease diagnosis [
30], biomedical big data [
31], and additional bioinformatics fields [
32].
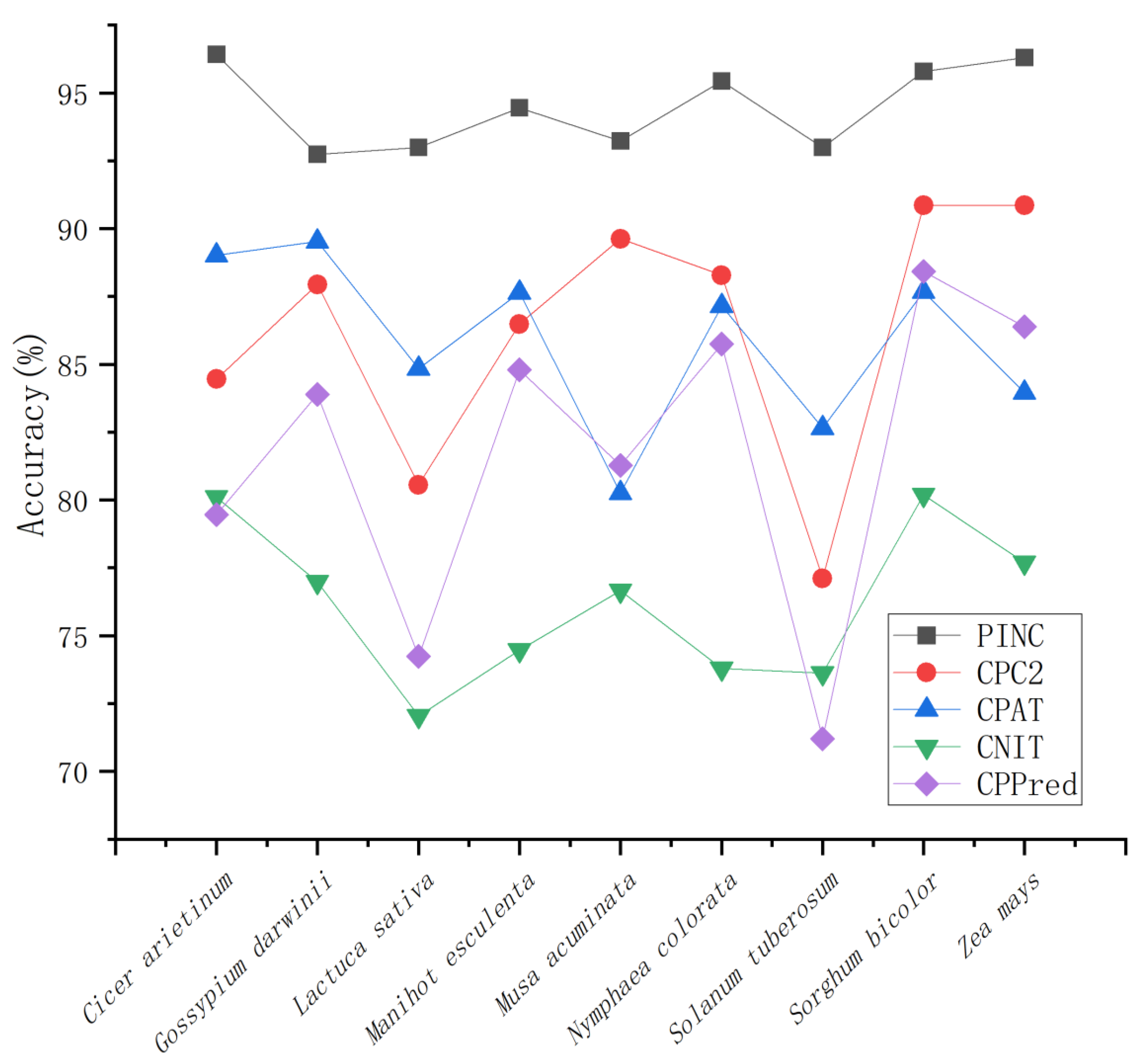
In this experiment, we developed PINC, an AutoML-based instrument for the identification of ncRNAs and cRNAs in plants. The AutoML framework does not require a great deal of effort and time to optimize the model; it simply accepts the processed data as an input, tunes and sets the framework’s parameters, and then outputs the model automatically. Our experimental results include a number of significant contributions: (1) By combining the F-test and variance threshold, 10 out of 91 features were identified as being able to strongly distinguish between ncRNAs and coding RNA in plants. (2) Using the AutoML framework, a neutral model for non-coding RNA identification was obtained. (3) We combined the two previous points and developed a tool called PINC for ncRNA identification. After comparing PINC with the CPC2, CPAT, CNIT, and CPPred identification tools on nine independent test sets to validate the performance of PINC, we discovered that PINC performed exceptionally well on these independent test sets. This suggests that PINC is a reliable method for ncRNA identification in plants. In addition, users can upload their data for identification, which facilitates the study of plants that have received less attention.
3. Discussion
In the field of bioinformatics, automated machine learning methods are now beginning to be implemented. In our experiments, we compared four automatic machine learning frameworks that are good matches for the more recently introduced frameworks and the older frameworks. For all the automatic machine learning frameworks, we used the same preprocessing methods to process the data as a raw input, then, we adjust the parameters of each framework in order to find the most suitable parameters, and finally we output the model. In general, we consider automatic machine learning frameworks to be black boxes and do not examine frame-specific methods for automatically optimizing parameters and integrating the model for direct output. Automated machine learning frameworks automatically optimize models, thereby reducing the time and effort devoted by researchers and, to a certain extent, allowing non-experts in machine learning to solve bioinformatics problems.
Utilizing high-quality features is one way to improve performance in machine learning. It is necessary to find features that are suitable for ncRNA identification in the study because providing or discovering good features is one of the most important tasks in machine learning. We extracted k-mer frequency features, coding sequence features, and other features during our experiments. Despite the fact that traditional k-mer features have been used in a variety of studies, such as gene identification [
33], subcellular localization [
34], and sequence analysis, it has been demonstrated that the k-mer frequency is highly effective at detecting ncRNAs [
35]. Many tools have also used features related to coding sequences and some other features [
36]. Ninety-one extracted features were filtered using our feature selection method; the filtered features successfully identified ncRNAs and it was the most precise tool, to date, for ncRNA identification in all plant species.
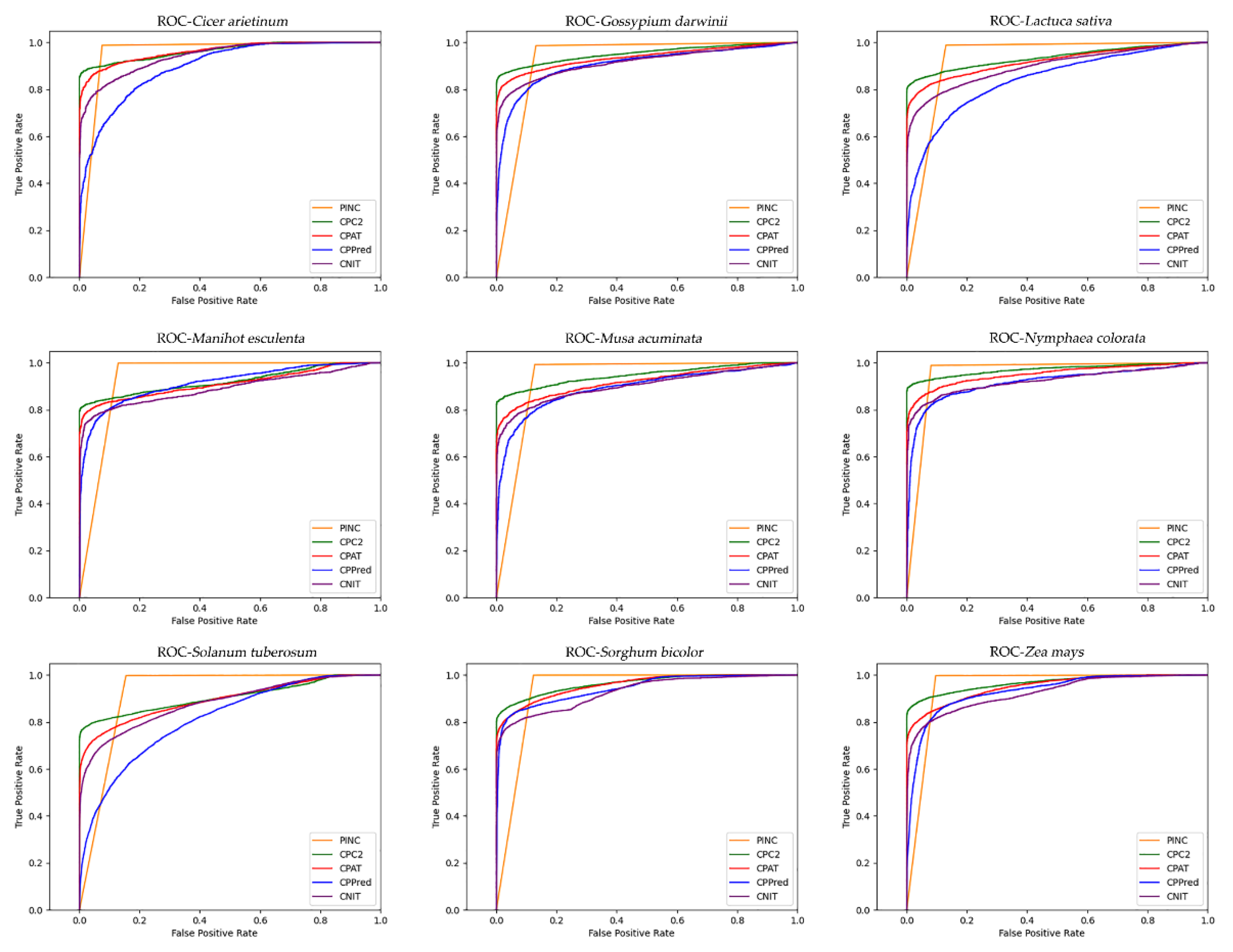
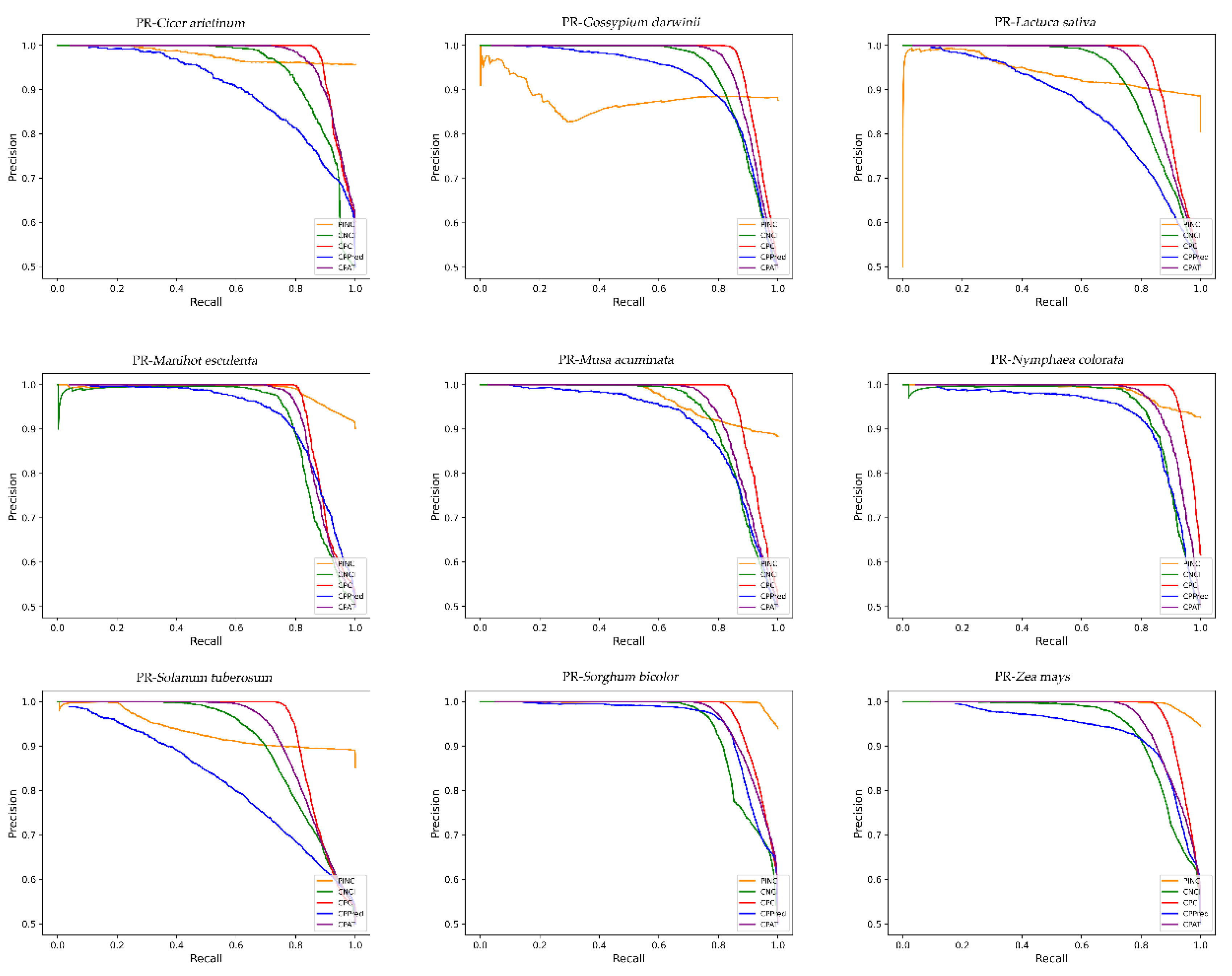
For ncRNA identification, there are additional factors to consider, such as the trade-off between sensitivity and specificity. At present, the number of ncRNAs is small as compared with the number of coding RNAs identified. To prevent ncRNAs from being missed, high sensitivity is important. Currently, CPAT, CNCI, CPPred, and CPC2 are less sensitive and focus more on identifying coding RNAs, but this requires an additional step to screen for non-coding RNAs. In contrast, the high sensitivity of PINC reduces the necessity for additional filtering processes. Moreover, PINC demonstrated a higher rate of accuracy than any other tool among the nine plants evaluated. Although some tools for non-coding RNA identification have reached over 85 percent accuracy, increased accuracy is not meaningless, as large amounts of data have become available due to advances in sequencing technology, and it is possible that for every one percent increase, hundreds of additional correct RNAs can be identified. Here, PINC achieves a high degree of ncRNA identification precision. This may be because the model in PINC adopts the stacking strategy, while other tools use single models such as SVM, logistic regression, and xgboost. For a long time, the performance of combining the predicted results of multiple models has been better than that of a single model, and the variance has been significantly reduced [
37]. In the experiment, we selected the default basic model in the AutoGluon framework. Here, the basic model is trained separately, and then the prediction of the basic model is used as a feature to train the stacked model. Stacked models can improve the shortcomings of a single-model prediction and can take advantage of their interactions to improve the prediction ability [
38]. In addition, it can be seen from the feature level distribution map described earlier that these features also have strong discrimination ability.
In addition, we plan to continue research in two areas: first, deep learning, which can automatically extract features, reduce the time required to extract features, and can improve the accuracy of cross-species recognition. In contrast, we should consider machine learning techniques to gain a deeper understanding of these RNA types and to investigate their biological significance. In addition, for plants, only a handful of ncRNA functions have been identified; once these functions are identified, new mechanisms can be explored and new features can be added to PINC to improve our tool further.
4. Materials and Methods
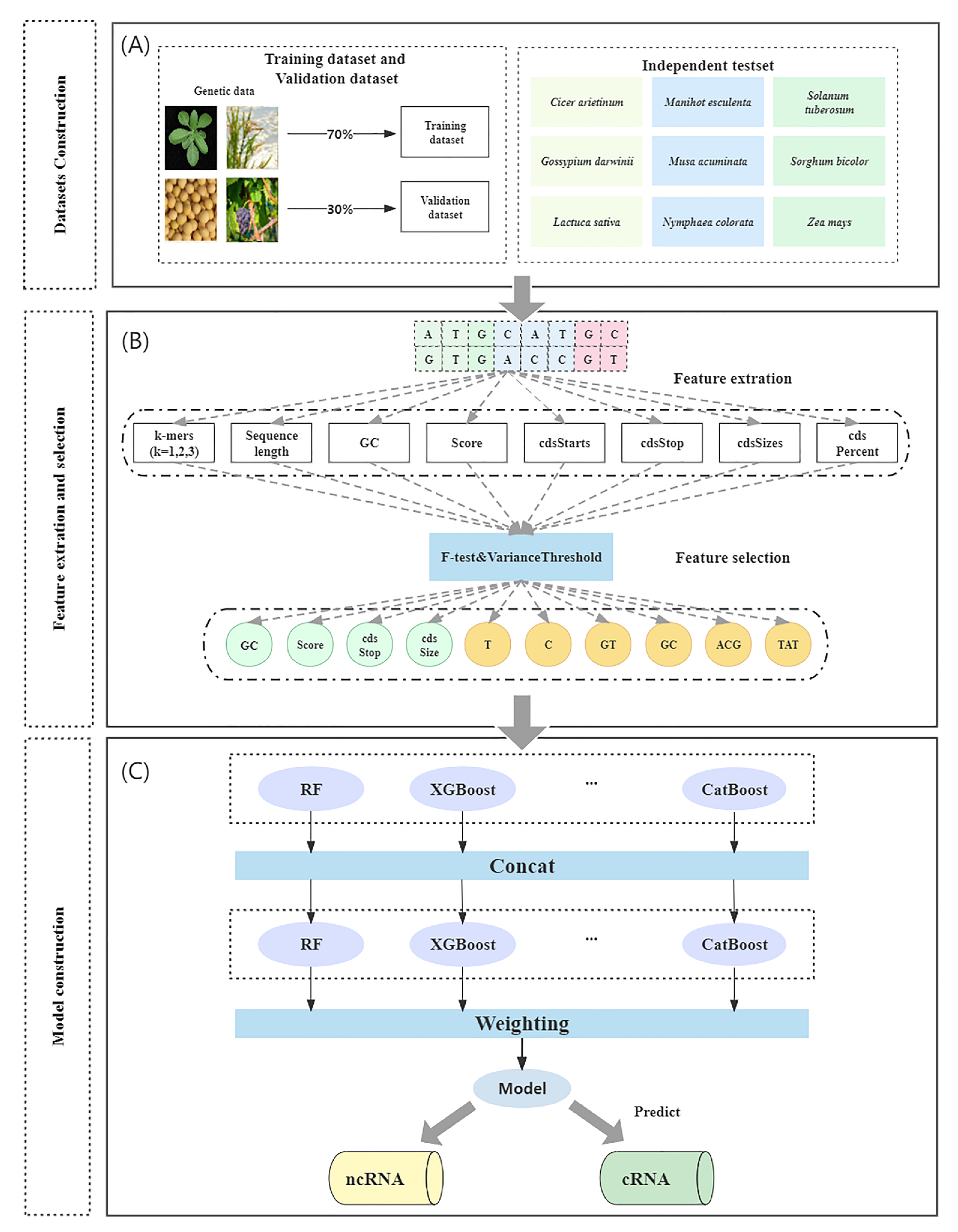
Figure 7 depicts the tool’s overall workflow, which consists of three steps: (1) dataset construction, (2) feature extraction and selection, and (3) model construction.
To create the dataset, RNA sequences were obtained from the GreeNC, CANTATA, RNAcentral, and Phytozome databases. Secondly, feature selection methods were used to extract and filter features. Finally, machine learning models were compared to determine the most effective model for ncRNA identification.
4.1. Dataset Construction
To construct the experimental dataset, we considered two factors. On the one hand, the diversity of plants and the abundance of annotation data were taken into consideration. On the other hand, considering the balance of the data, we chose four plants as our training and validation datasets (
Table 4), which included two model plants, i.e.,
Arabidopsis thaliana and
Oryza sativa, in addition to two non-model plants, i.e.,
Glycine max and
Vitis vinifera. We used ncRNAs as the positive sample data and coding RNAs as the negative sample data in the dataset. Negative samples were obtained from Phytozome.v13 [
39]. Positive samples were obtained from three public databases, including GreeNC [
40], CANTATA [
41], and RNAcentral [
42]. For all data, first, we used cd-hit-est-2D in the CD-hit tool [
43] to eliminate redundant sequences between the test and training sets at a threshold of 80% [
22,
35,
44,
45]. Second, in order to balance the datasets, random selections of 4000 data were made for each plant, of which 2000 were positive samples and 2000 were negative samples. The positive sample data consisted of 1800 lncRNAs and 200 sncRNAs, and the negative sample data consisted of 2000 mRNAs (
Table 5) [
18,
46]. Thus, the baseline dataset consisted of a total of 16,000 protein sequences from four plants. Meanwhile, we analyzed the length distribution of the positive and negative datasets, as shown in
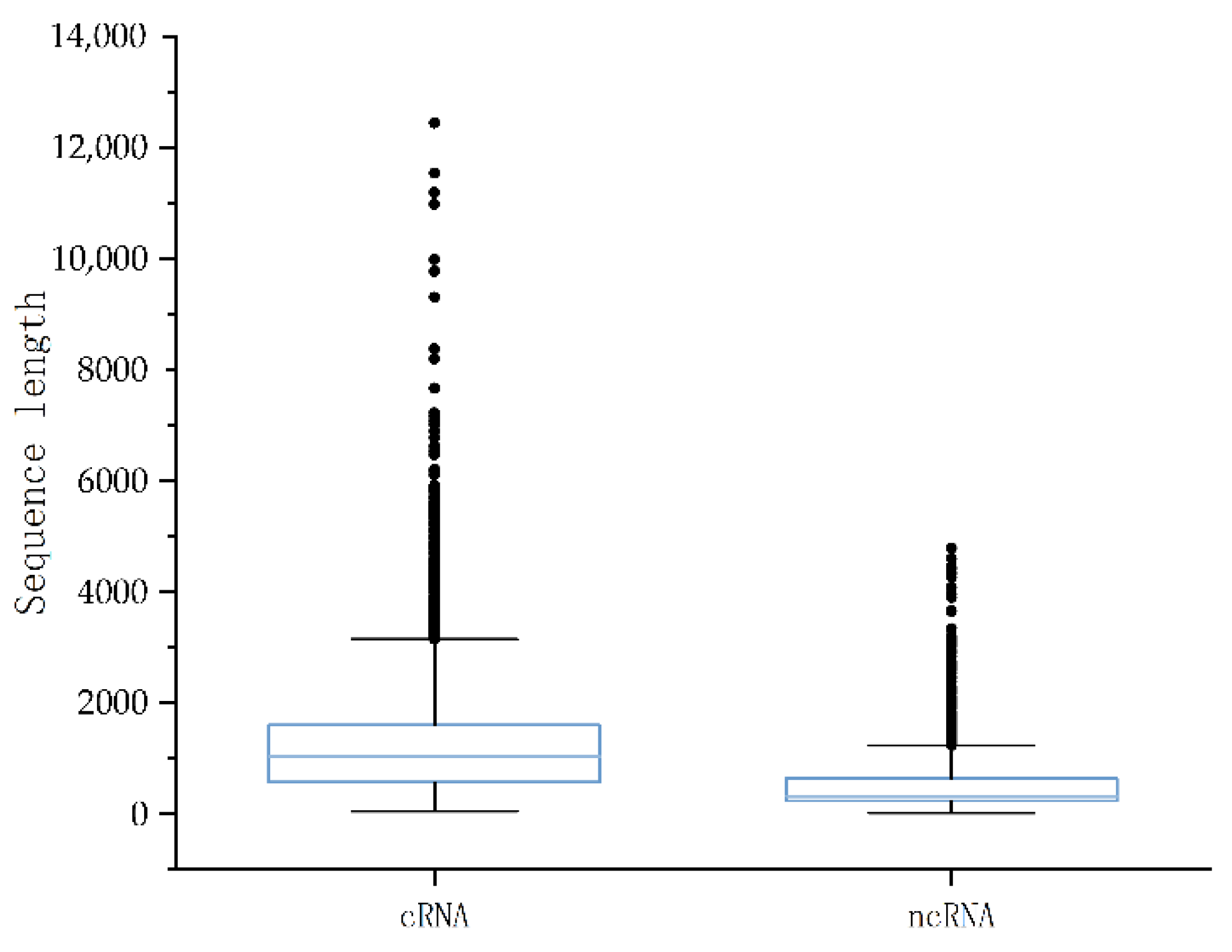
Figure 8. The median length of the coding RNAs data was 1029 and the data were mostly concentrated in the range of 0–2000. The ncRNA data had a median length of 321 and the data were mostly concentrated in the range of 0–1000. Finally, we proportionally divided the dataset into 70% training data and 30% validation data. Additionally, nine independent test sets were created for nine plants. (
Table 6):
Cicer arietinum,
Gossypium darwinii,
Lactuca sativa,
Manihot esculenta,
Musa acuminata,
Nymphaea colorata,
Solanum tuberosum,
Sorghum bicolor, and
Zea mays. To eliminate redundant sequences, the data for these nine independent test sets were taken from the four databases mentioned above and filtered at a threshold of 80%.
4.2. Feature Extraction and Selection
This experiment initially extracted 91 features (
Table 7). The 86 features of k-mer frequency, sequence length, and GC content were obtained using the Python script program (
https://github.com/midisec/PINC, accessed on 22 August 2022); the five features of Score and CDS were obtained using the UCSC Genome txCdsPredict program in the browser (
http://hgdown-load.soe.ucsc.edu/admin/jksrc.zip, accessed on 11 November 2014) [
47]. These features can be classified into three categories: k-mer frequency features, CDS-related features, and other features. The k-mer frequency describes all possible frequencies for the presence of k nucleotides in a sequence, based on methods that have initially been implemented in whole genome shotgun assemblers. When k = 1, each nucleotide can contain a maximum of four A, C, G, or T. When k equals 2, the calculation involves the dinucleotide frequency (i.e., AA, AT, AG, AC, …, TT) and a total of
= 16 species. When k = 3, the calculated three-nucleotide frequencies (i.e., AAA, AAT, AAG, AAC, …, TTT) are computed for a total of
= 64 species. By combining 1–3-mer frequencies for a total of 84 features, k-mer frequencies can capture rich statistical information about negative profiles in plant genomes, according to some research [
48]. CDS is the result of encoded proteins that are interchangeable with ORF in some ways, but differ slightly [
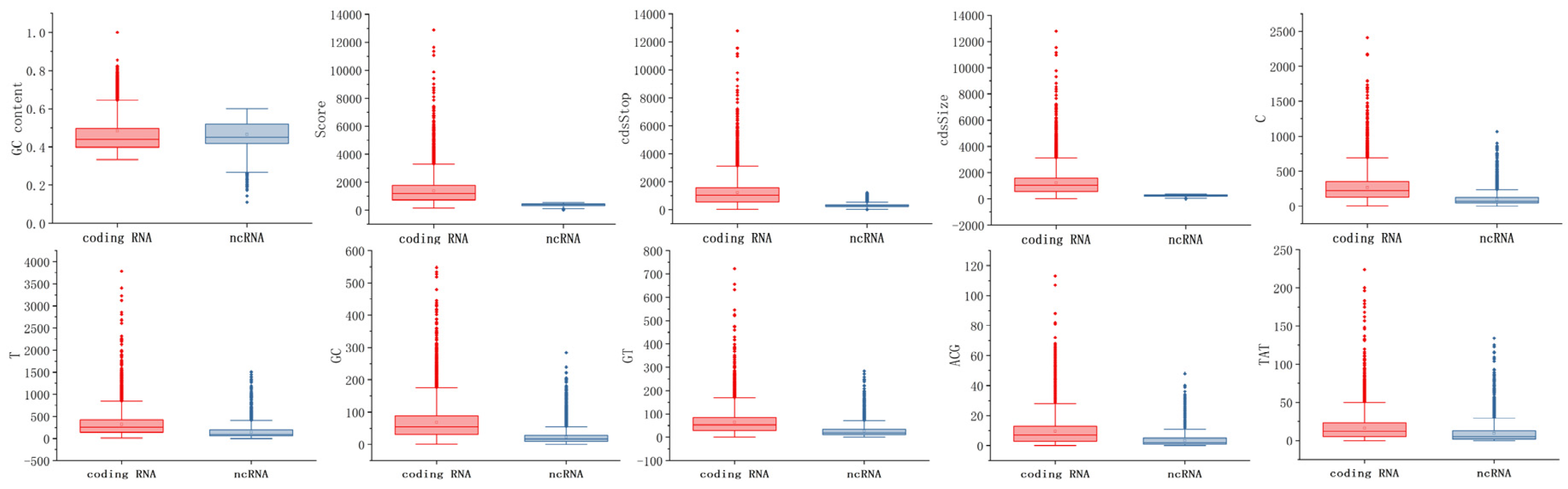
49]. The features Score, cdsStarts, cdsStop, cdsSize, and cdsPercent comprise the second major category of features. Score is the predicted protein score; if it is >800, there is a 90% chance that it is a protein, and if it is >1000, it is virtually certain that it is a protein. cdsStop is the end of the coding region in the transcript, cdsSize is cdsStop minus cdsStart, and cdsPercent is the ratio of cdsSize to the total sequence length. Other features include sequence length and GC content, which are widely used for ncRNA identification. Sequence length indicates the total length of the sequence. GC content is the ratio of guanine and cytosine to the other four DNA bases.
There may be redundant features among the 91 features listed above; therefore, we employed feature selection to filter them. For the feature selection method, redundant features were filtered out using a combination of variance threshold filtering and the F-test. Variance threshold filtering is used to filter features based on their own variance. The smaller a feature’s variance, the less significant its variation, and these insignificant features are eliminated. F-test is a method to determine the relationship between each feature and label. The GC content, Score, cdsStop, cdsSize, and T, C, GT, GC, ACG, and TAT frequencies were among the 91 features identified by this combined feature selection method. Finally, these 10 features were used as the model input.
4.3. Model Construction
Machine learning (ML) is currently utilized in a variety of fields to solve numerous difficult problems. Nevertheless, model construction for machine learning requires human intervention. Manual intervention is required in the feature extraction, model selection, and parameter adjustment processes, which require professionals to optimize and can waste a significant amount of time and resources if errors occur. To reduce these repetitive development costs, the concept of automating the entire machine learning process, automatic machine learning, has been conceived (AutoML). The definition of AutoML is that it is a combination of automation and ML [
50]. From an automation standpoint, AutoML can be viewed as the design of a framework to automate the entire machine learning process, allowing models to automatically learn the correct parameters and configurations without manual intervention. From the standpoint of machine learning, AutoML is a system that is highly capable of learning and generalizing given data and tasks. Recent research on AutoML has focused on the neural network architecture search (NAS) method, which employs a search strategy to test and evaluate a large number of architectures in a search space, and then selects the one that best meets the objectives of a given problem by maximizing the adaptation function. However, the NAS faces two obstacles to the method: first, the amount of computation is excessive, resulting in increased resource consumption. Second, instability may vary each time and the search structure is altered, resulting in varying precision. In our experiments, we compared four automatic machine learning frameworks, AutoGluon, H2O, TPOT, and Autokeras, with three conventional machine learning models, SVM, RF, and Naive Bayes. We determined that AutoGluon was the superior framework, and therefore it was used as the classifier. AutoGluon contains 26 base models including random forest, XGBoost, and a neural network, and in our experiments, we used all the base models for training the model [
51]. AutoGluon is an open-source machine learning training framework for tabular data. It is a framework that attempts to avoid a hyperparametric search as much as possible, training multiple models concurrently and weighting them using a multi-layer stacking strategy to obtain the final output.
4.4. Performance Evaluation
Several widely used performance metrics were evaluated in the experiments, including sensitivity (SE), specificity (SPC), accuracy (ACC), F1-score, positive predictive value (PPV), negative predictive value (NPV), and the Matthews correlation coefficient (MCC). To evaluate the performance of the classifier numerically and visually, the area under the curve (AUC) and ROC curves were also used. These definitions are as follows:
TP represents true positives, the number of correctly identified positive samples, while FN, TN, and FP represent false negatives, true negatives, and false positives, the number of incorrectly identified positive samples, correctly identified negative samples, and incorrectly identified negative samples, respectively.