Inflammation-Driven Secretion Potential Is Upregulated in Osteoarthritic Fibroblast-Like Synoviocytes
Abstract
1. Introduction
2. Results
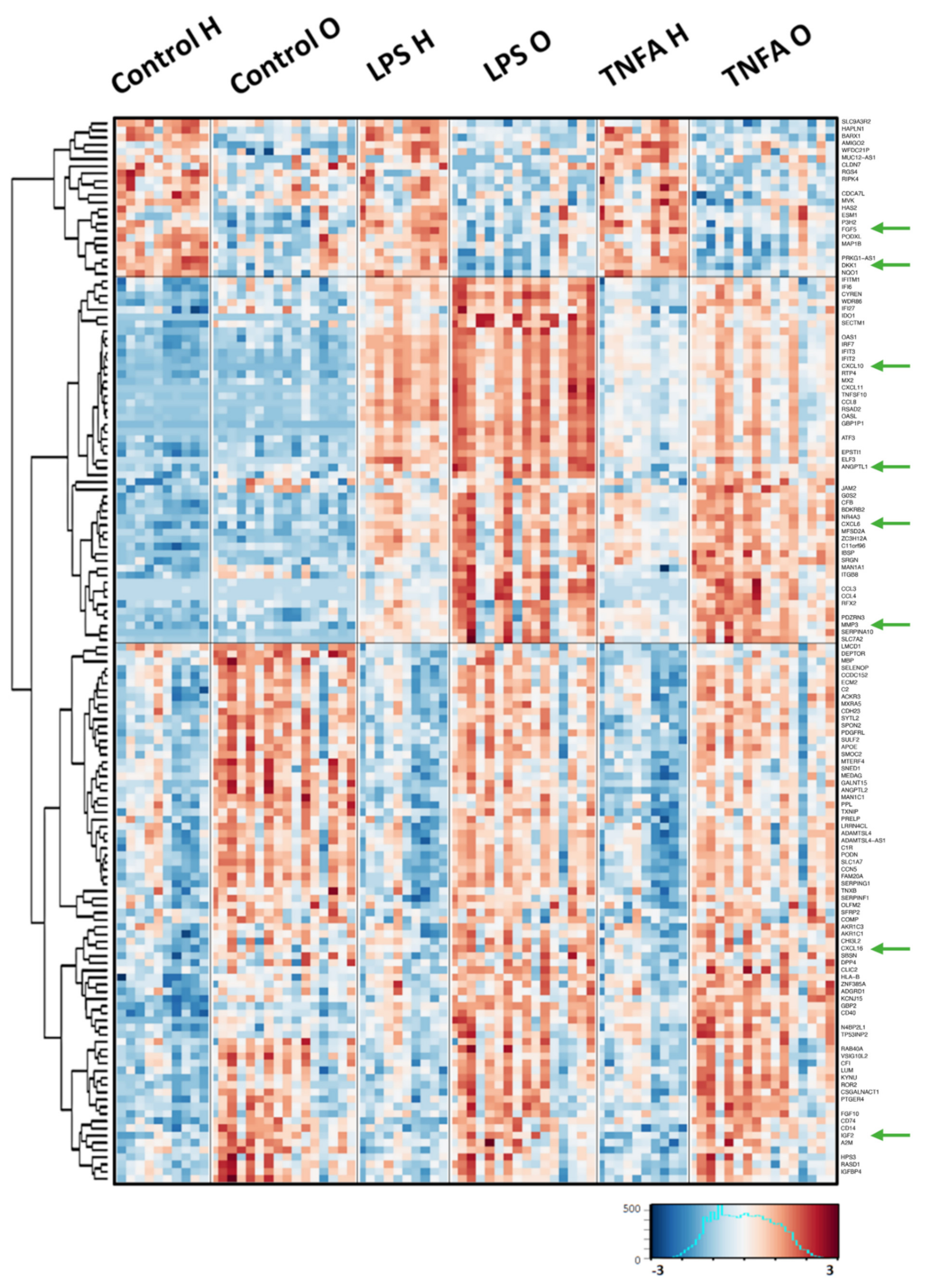
2.1. Comparison of the Gene Expression Response to Pro-Inflammatory Stimulation between OA Patients and Healthy Donors
2.2. Signaling Protein Secretion Analysis Using the ELISA Method
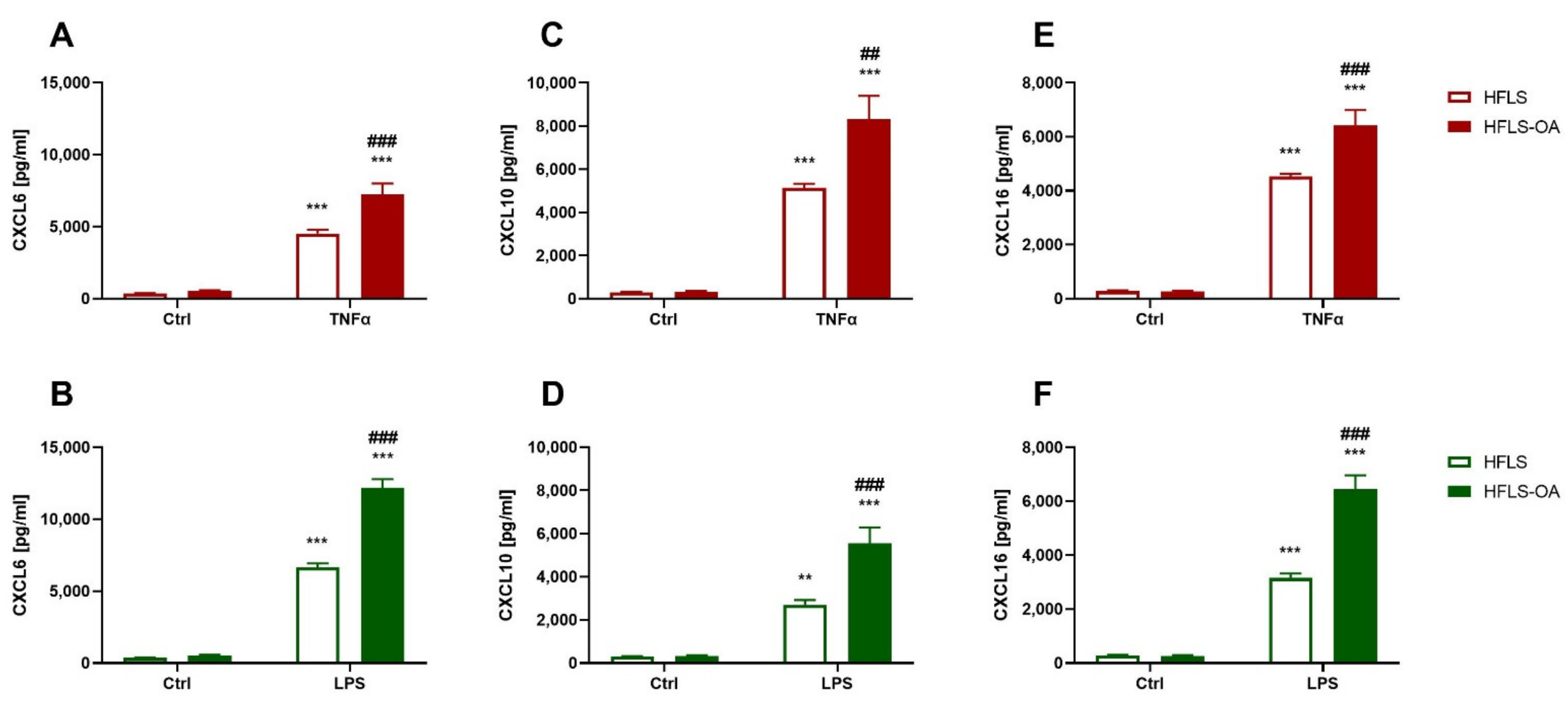
2.2.1. Chemokine Secretion during Inflammation Is Upregulated in HFLS-OA
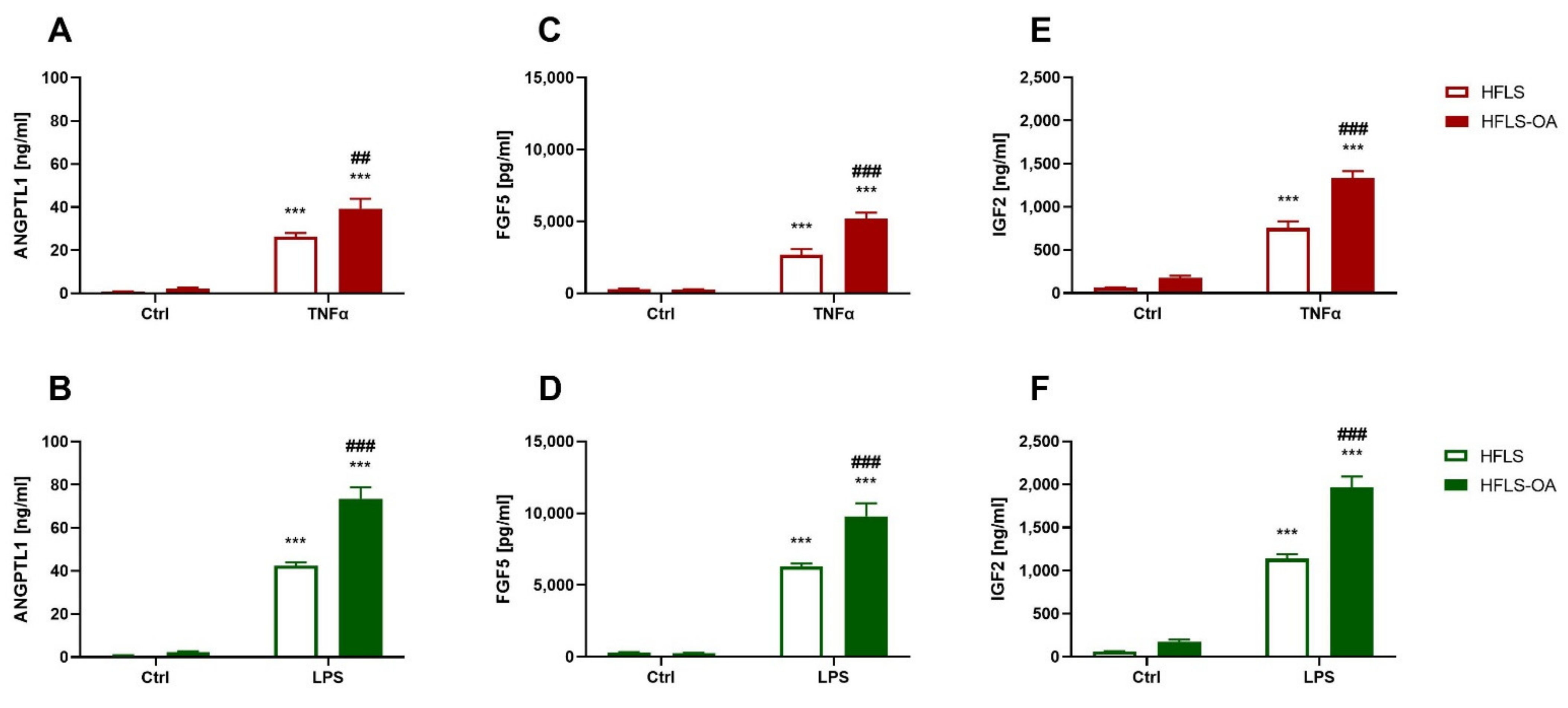
2.2.2. HFLS-OA Released More Growth Factors after Pro-Inflammatory Stimulation
2.2.3. Elevated Proteolytic Enzyme Translation in Pro-Inflammatory-Stimulated HFLS-OA
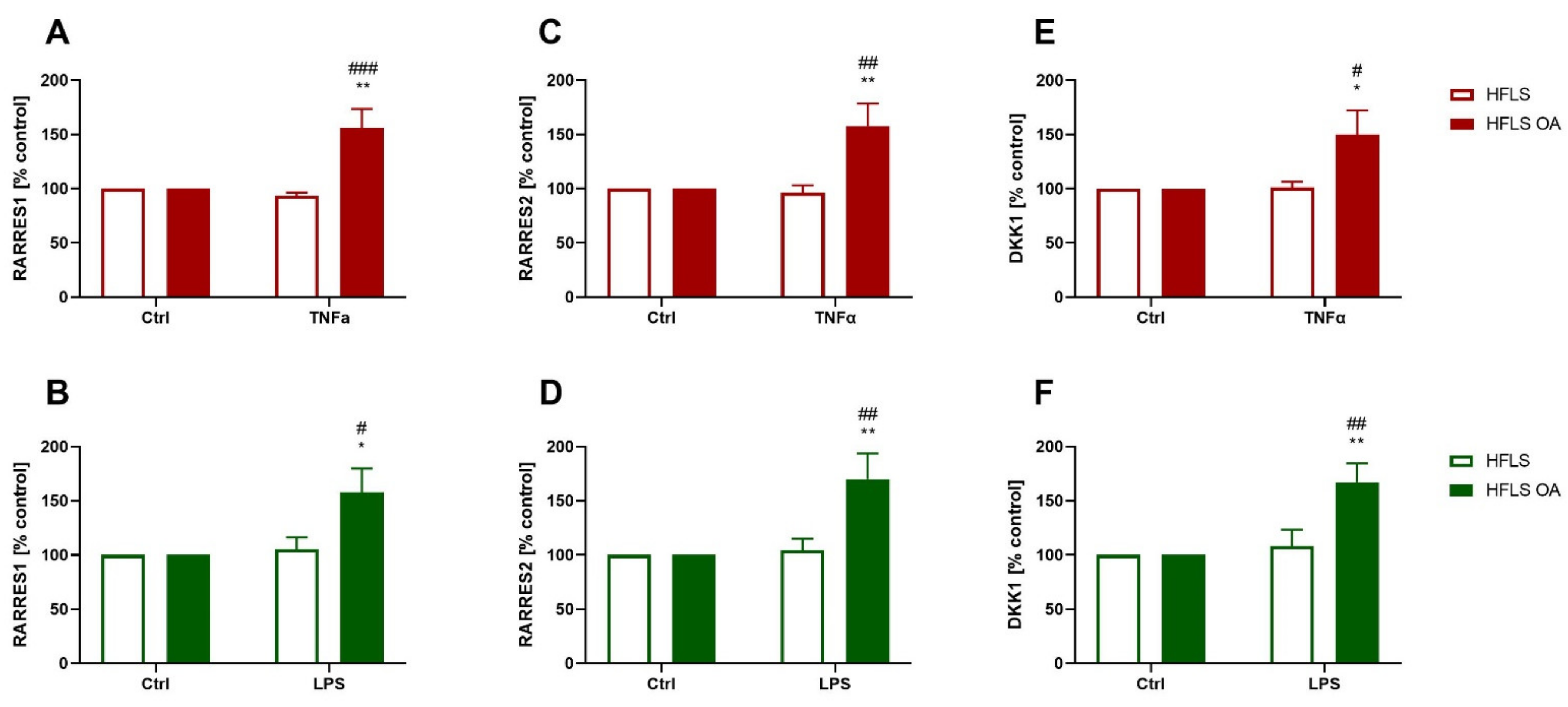
2.2.4. Changes in RARRES1, RARRES2, and DKK1 Expression in HFLS-OA during Inflammation
3. Discussion
4. Materials and Methods
4.1. Human Fibroblast-Like Synoviocytes (HFLS) Preparation
4.2. Cell Culturing and Treatment
4.3. RNA Isolation
4.4. Library Preparation and Transcriptome Sequencing
4.5. RNA-Seq Data Analysis
4.6. Protein Isolation
4.7. Enzyme-Linked Immunosorbent Assay (ELISA)
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palazzo, C.; Nguyen, C.; Lefevre-Colau, M.M.; Rannou, F.; Poiraudeau, S. Risk Factors and Burden of Osteoarthritis. Ann. Phys. Rehabil. Med. 2016, 59, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Sacitharan, P.K. Part II Clinical Science. Subcellular Biochemistry. In Biochemistry and Cell Biology of Ageing; Springer: Singapore, 2019. [Google Scholar]
- Li, G.; Yin, J.; Gao, J.; Cheng, T.S.; Pavlos, N.J.; Zhang, C.; Zheng, M.H. Subchondral Bone in Osteoarthritis: Insight into Risk Factors and Microstructural Changes. Arthritis Res. Ther. 2013, 15, 223. [Google Scholar] [CrossRef] [PubMed]
- Primorac, D.; Molnar, V.; Rod, E.; Jeleč, Ž.; Čukelj, F.; Matišić, V.; Vrdoljak, T.; Hudetz, D.; Hajsok, H.; Borić, I. Knee Osteoarthritis: A Review of Pathogenesis and State-Of-The-Art Non-Operative Therapeutic Considerations. Genes 2020, 11, 854. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, E.B.; Casado, P.L.; Neto, V.M.; Duarte, M.E.L.; Aguiar, D.P. Synovial Fluid and Synovial Membrane Mesenchymal Stem Cells: Latest Discoveries and Therapeutic Perspectives. Stem Cell Res. Ther. 2014, 5, 112. [Google Scholar] [CrossRef] [PubMed]
- Gait, A.D.; Hodgson, R.; Parkes, M.J.; Hutchinson, C.E.; O’Neill, T.W.; Maricar, N.; Marjanovic, E.J.; Cootes, T.F.; Felson, D.T. Synovial Volume vs Synovial Measurements from Dynamic Contrast Enhanced MRI as Measures of Response in Osteoarthritis. Osteoarthr. Cartil. 2016, 24, 1392. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; He, C. Pro-Inflammatory Cytokines: The Link between Obesity and Osteoarthritis. Cytokine Growth Factor Rev. 2018, 44, 38–50. [Google Scholar] [CrossRef]
- Scanzello, C.R.; Goldring, S.R. The Role of Synovitis in Osteoarthritis Pathogenesis. Bone 2012, 51, 249–257. [Google Scholar] [CrossRef]
- Standarization of Osteoarthritis Definitions. Available online: https://oarsi.org/research/standardization-osteoarthritis-definitions (accessed on 23 September 2022).
- Lin, Y.J.; Anzaghe, M.; Schülke, S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells 2020, 9, 880. [Google Scholar] [CrossRef]
- Galozzi, P.; Bindoli, S.; Doria, A.; Oliviero, F.; Sfriso, P. Autoinflammatory Features in Gouty Arthritis. J. Clin. Med. 2021, 10, 1880. [Google Scholar] [CrossRef]
- Zaripova, L.N.; Midgley, A.; Christmas, S.E.; Beresford, M.W.; Baildam, E.M.; Oldershaw, R.A. Juvenile Idiopathic Arthritis: From Aetiopathogenesis to Therapeutic Approaches. Pediatr. Rheumatol. 2021, 19, 135. [Google Scholar] [CrossRef]
- Hui, A.Y.; McCarty, W.J.; Masuda, K.; Firestein, G.S.; Sah, R.L. A Systems Biology Approach to Synovial Joint Lubrication in Health, Injury, and Disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 15–37. [Google Scholar] [CrossRef]
- Tchetverikov, I.; Lohmander, L.S.; Verzijl, N.; Huizinga, T.W.J.; TeKoppele, J.M.; Hanemaaijer, R.; DeGroot, J. MMP Protein and Activity Levels in Synovial Fluid from Patients with Joint Injury, Inflammatory Arthritis, and Osteoarthritis. Ann. Rheum. Dis. 2005, 64, 694–698. [Google Scholar] [CrossRef]
- Tamer, T.M. Hyaluronan and Synovial Joint: Function, Distribution and Healing. Interdiscip. Toxicol. 2013, 6, 111–125. [Google Scholar] [CrossRef]
- Daghestani, H.N.; Kraus, V.B. Inflammatory Biomarkers in Osteoarthritis. Osteoarthr. Cartil. 2015, 23, 1890–1896. [Google Scholar] [CrossRef]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The Basic Science of Articular Cartilage: Structure, Composition, and Function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Mehana, E.S.E.; Khafaga, A.F.; El-Blehi, S.S. The Role of Matrix Metalloproteinases in Osteoarthritis Pathogenesis: An Updated Review. Life Sci. 2019, 234, 116786. [Google Scholar] [CrossRef]
- Goshi, N.; Morgan, R.K.; Lein, P.J.; Seker, E. A Primary Neural Cell Culture Model to Study Neuron, Astrocyte, and Microglia Interactions in Neuroinflammation. J. Neuroinflammation 2020, 17, 155. [Google Scholar] [CrossRef]
- Zu, Y.; Mu, Y.; Li, Q.; Zhang, S.T.; Yan, H.J. Icariin Alleviates Osteoarthritis by Inhibiting NLRP3-Mediated Pyroptosis. J. Orthop. Surg. Res. 2019, 14, 307. [Google Scholar] [CrossRef]
- Feng, Y.; Mei, L.; Wang, M.; Huang, Q.; Huang, R. Anti-Inflammatory and Pro-Apoptotic Effects of 18beta-Glycyrrhetinic Acid In Vitro and In Vivo Models of Rheumatoid Arthritis. Front. Pharmacol. 2021, 12, 681525. [Google Scholar] [CrossRef]
- Ai, M.; Lin, S.; Zhang, M.; Wu, T.; Yang, N.; Li, Y.; Li, L. Cirsilineol Attenuates LPS-Induced Inflammation in Both in Vivo and in Vitro Models via Inhibiting TLR-4/NFkB/IKK Signaling Pathway. J. Biochem. Mol. Toxicol. 2021, 35, e22799. [Google Scholar] [CrossRef]
- Xie, C.; Chen, Q. Adipokines: New Therapeutic Target for Osteoarthritis? Curr. Rheumatol. Rep. 2019, 21, 71. [Google Scholar] [CrossRef]
- Li, Z.M.; Li, M. Improvement in Orthopedic Outcome Score and Reduction in IL-1β, CXCL13, and TNF-α in Synovial Fluid of Osteoarthritis Patients Following Arthroscopic Knee Surgery. Genet. Mol. Res. 2017, 16, 1–9. [Google Scholar] [CrossRef]
- Boffa, A.; Merli, G.; Andriolo, L.; Lattermann, C.; Salzmann, G.M.; Filardo, G. Synovial Fluid Biomarkers in Knee Osteoarthritis: A Systematic Reviewand Quantitative Evaluation Using BIPEDs Criteria. Cartilage 2021, 13, 82S. [Google Scholar] [CrossRef]
- Eymard, F.; Pigenet, A.; Citadelle, D.; Flouzat-Lachaniette, C.H.; Poignard, A.; Benelli, C.; Berenbaum, F.; Chevalier, X.; Houard, X. Induction of an Inflammatory and Prodegradative Phenotype in Autologous Fibroblast-like Synoviocytes by the Infrapatellar Fat Pad from Patients with Knee Osteoarthritis. Arthritis Rheumatol. 2014, 66, 2165–2174. [Google Scholar] [CrossRef]
- Qiao, L.; Li, Y.; Sun, S. Insulin Exacerbates Inflammation in Fibroblast-Like Synoviocytes. Inflammation 2020, 43, 916. [Google Scholar] [CrossRef]
- Sanchez-Lopez, E.; Coras, R.; Torres, A.; Lane, N.E.; Guma, M. Synovial Inflammation in Osteoarthritis Progression HHS Public Access. Nat. Rev. Rheumatol. 2022, 18, 258–275. [Google Scholar] [CrossRef]
- Ma, J.; Niu, D.S.; Wan, N.J.; Qin, Y.; Guo, C.J. Elevated Chemerin Levels in Synovial Fluid and Synovial Membrane from Patients with Knee Osteoarthritis. Int. J. Clin. Exp. Pathol. 2015, 8, 13393. [Google Scholar]
- Shepherd, C.; Zhu, D.; Skelton, A.J.; Combe, J.; Threadgold, H.; Zhu, L.; Vincent, T.L.; Stuart, P.; Reynard, L.N.; Loughlin, J. Functional Characterization of the Osteoarthritis Genetic Risk Residing at ALDH1A2 Identifies Rs12915901 as a Key Target Variant. Arthritis Rheumatol. 2018, 70, 1577–1587. [Google Scholar] [CrossRef]
- Chen, A.; Lee, K.; He, J.C. Autocrine and Paracrine Effects of a Novel Podocyte Gene, RARRES1. Kidney Int. 2021, 100, 745–747. [Google Scholar] [CrossRef]
- Helfer, G.; Wu, Q.F. Chemerin: A Multifaceted Adipokine Involved in Metabolic Disorders. J. Endocrinol. 2018, 238, R79–R94. [Google Scholar] [CrossRef]
- Mathiessen, A.; Conaghan, P.G. Synovitis in Osteoarthritis: Current Understanding with Therapeutic Implications. Arthritis Res. Ther. 2017, 19, 18. [Google Scholar] [CrossRef] [PubMed]
- Sokolove, J.; Lepus, C.M. Role of Inflammation in the Pathogenesis of Osteoarthritis: Latest Findings and Interpretations. Ther. Adv. Musculoskelet. Dis. 2013, 5, 77–94. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, S.; Chen, D. TGF-β Signaling and the Development of Osteoarthritis. Bone Res. 2014, 2, 14002. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.A.; Rolauffs, B. Onset and Progression of Human Osteoarthritis—Can Growth Factors, Inflammatory Cytokines, or Differential MiRNA Expression Concomitantly Induce Proliferation, ECM Degradation, and Inflammation in Articular Cartilage? Int. J. Mol. Sci. 2018, 19, 2282. [Google Scholar] [CrossRef]
- Heinhuis, B.; Koenders, M.I.; Van Riel, P.L.; Van De Loo, F.A.; Dinarello, C.A.; Netea, M.G.; Van Den Berg, W.B.; Joosten, L.A.B. Tumour Necrosis Factor Alpha-Driven IL-32 Expression in Rheumatoid Arthritis Synovial Tissue Amplifies an Inflammatory Cascade. Ann. Rheum. Dis. 2011, 70, 660–667. [Google Scholar] [CrossRef]
- Scanzello, C.R. Chemokines and Inflammation in Osteoarthritis: Insights From Patients and Animal Models. J. Orhop. Res. 2017, 35, 735–739. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Yang, Z.B.; Zhang, Z.J.; Zhang, Z.Q.; Kang, Y.; Huang, G.X.; Wang, S.W.; Huang, H.; Liao, W.M. CCL3 Serves as a Potential Plasma Biomarker in Knee Degeneration (Osteoarthritis). Osteoarthr. Cartil. 2015, 23, 1405–1411. [Google Scholar] [CrossRef]
- Sandell, L.J.; Xing, X.; Franz, C.; Davies, S.; Chang, L.W.; Patra, D. Exuberant Expression of Chemokine Genes by Adult Human Articular Chondrocytes in Response to IL-1β. Osteoarthr. Cartil. 2008, 16, 1560–1571. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Muraoka, S.; Kusunoki, N.; Masuoka, S.; Yamada, S.; Ogasawara, H.; Imai, T.; Akasaka, Y.; Tochigi, N.; Takahashi, H.; et al. Resistin Upregulates Chemokine Production by Fibroblast-like Synoviocytes from Patients with Rheumatoid Arthritis. Arthritis Res. Ther. 2017, 19, 6–11. [Google Scholar] [CrossRef]
- Siegel, R.J.; Singh, A.K.; Panipinto, P.M.; Shaikh, F.S.; Vinh, J.; Han, S.U.; Kenney, H.M.; Schwarz, E.M.; Crowson, C.S.; Khuder, S.A.; et al. Extracellular Sulfatase-2 Is Overexpressed in Rheumatoid Arthritis and Mediates the TNF-α-Induced Inflammatory Activation of Synovial Fibroblasts. Cell. Mol. Immunol. 2022, 19, 1185–1195. [Google Scholar] [CrossRef]
- Saruga, T.; Imaizumi, T.; Kawaguchi, S.; Seya, K.; Matsumiya, T.; Sasaki, E.; Sasaki, N.; Uesato, R.; Ishibashi, Y. Role of MDA5 in Regulating CXCL10 Expression Induced by TLR3 Signaling in Human Rheumatoid Fibroblast-like Synoviocytes. Mol. Biol. Rep. 2021, 48, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-H.; Xu, L.-L.; Zhao, J.-X.; Sun, L.; Yao, Z.-Q.; Deng, X.-L.; Liu, R.; Yang, L.; Xing, R.; Liu, X.-Y. CXCL16 Upregulates RANKL Expression in Rheumatoid Arthritis Synovial Fibroblasts through the JAK2/STAT3 and P38/MAPK Signaling Pathway. Inflamm. Res. 2015, 65, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, N.; Haqqi, T.M. Epigallocatechin-3-Gallate Suppresses the Global Interleukin-1beta-Induced Inflammatory Response in Human Chondrocytes. Arthritis Res. Ther. 2011, 13, R93. [Google Scholar] [CrossRef]
- Pierer, M.; Rethage, J.; Seibl, R.; Lauener, R.; Brentano, F.; Wagner, U.; Hantzschel, H.; Michel, B.A.; Gay, R.E.; Gay, S.; et al. Chemokine Secretion of Rheumatoid Arthritis Synovial Fibroblasts Stimulated by Toll-Like Receptor 2 Ligands. J. Immunol. 2004, 172, 1256–1265. [Google Scholar] [CrossRef]
- Song, G.; Lu, Q.; Fan, H.; Zhang, X.; Ge, L.; Tian, R.; Wang, S.; Feng, T.; Pan, J.; Feng, J.; et al. Inhibition of Hexokinases Holds Potential as Treatment Strategy for Rheumatoid Arthritis. Arthritis Res. Ther. 2019, 21, 87. [Google Scholar] [CrossRef]
- Zou, Y.; Zeng, S.; Huang, M.; Qiu, Q.; Xiao, Y.; Shi, M.; Zhan, Z.; Liang, L.; Yang, X.; Xu, H. Inhibition of 6-Phosphofructo-2-Kinase Suppresses Fibroblast-like Synoviocytes-Mediated Synovial Inflammation and Joint Destruction in Rheumatoid Arthritis. Br. J. Pharmacol. 2017, 174, 893–908. [Google Scholar] [CrossRef]
- Furman, B.D.; Kent, C.L.; Huebner, J.L.; Kraus, V.B.; McNulty, A.L.; Guilak, F.; Olson, S.A. CXCL10 Is Upregulated in Synovium and Cartilage Following Articular Fracture. J. Orthop. Res. 2018, 36, 1220–1227. [Google Scholar] [CrossRef]
- Kim, M.S.; Song, H.J.; Lee, S.H.; Lee, C.K. Comparative Study of Various Growth Factors and Cytokines on Type I Collagen and Hyaluronan Production in Human Dermal Fibroblasts. J. Cosmet. Dermatol. 2014, 13, 44–51. [Google Scholar] [CrossRef]
- Endres, M.; Andreas, K.; Kalwitz, G.; Freymann, U.; Neumann, K.; Ringe, J.; Sittinger, M.; Häupl, T.; Kaps, C. Chemokine Profile of Synovial Fluid from Normal, Osteoarthritis and Rheumatoid Arthritis Patients: CCL25, CXCL10 and XCL1 Recruit Human Subchondral Mesenchymal Progenitor Cells. Osteoarthr. Cartil. 2010, 18, 1458–1466. [Google Scholar] [CrossRef]
- Livingstone, C.; Borai, A. Insulin-like Growth Factor-II: Its Role in Metabolic and Endocrine Disease. Clin. Endocrinol. 2014, 80, 773–781. [Google Scholar] [CrossRef]
- Choi, H.M.; Lee, Y.A.; Lee, S.H.; Hong, S.J.; Hahm, D.H.; Choi, S.Y.; Yang, H.I.; Yoo, M.C.; Kim, K.S. Adiponectin May Contribute to Synovitis and Joint Destruction in Rheumatoid Arthritis by Stimulating Vascular Endothelial Growth Factor, Matrix Metalloproteinase-1, and Matrix Metalloproteinase-13 Expression in Fibroblast-like Synoviocytes More than Proinflammatory Mediators. Arthritis Res. Ther. 2009, 11, R161. [Google Scholar] [CrossRef]
- Hamilton, J.L.; Nagao, M.; Levine, B.R.; Chen, D.; Olsen, B.R.; Im, H.J. Targeting VEGF and Its Receptors for the Treatment of Osteoarthritis and Associated Pain. J. Bone Miner. Res. 2016, 31, 911–924. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, W.; Huang, J.; He, B.C.; Zuo, G.W.; Zhang, W.; Luo, Q.; Shi, Q.; Zhang, B.Q.; Wagner, E.R.; et al. Insulin-like Growth Factor 2 (IGF-2) Potentiates BMP-9-Induced Osteogenic Differentiation and Bone Formation. J. Bone Miner. Res. 2010, 25, 2447–2459. [Google Scholar] [CrossRef]
- Timur, U.T.; Jahr, H.; Anderson, J.; Green, D.C.; Emans, P.J.; Smagul, A.; van Rhijn, L.W.; Peffers, M.J.; Welting, T.J.M. Identification of Tissue-Dependent Proteins in Knee OA Synovial Fluid. Osteoarthr. Cartil. 2021, 29, 124–133. [Google Scholar] [CrossRef]
- Uchimura, T.; Foote, A.T.; Smith, E.L.; Matzkin, E.G.; Zeng, L. Insulin-Like Growth Factor II (IGF-II) Inhibits IL-1β-Induced Cartilage Matrix Loss and Promotes Cartilage Integrity in Experimental Osteoarthritis. Physiol. Behav. 2015, 116, 2858–2869. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, R.; Zhang, Q.; Xu, Z.; Xu, F.; Li, D.; Li, Y. Microtia Patients: Auricular Chondrocyte ECM Is Promoted by CGF through IGF-1 Activation of the IGF-1R/PI3K/AKT Pathway. J. Cell. Physiol. 2019, 234, 21817–21824. [Google Scholar] [CrossRef] [PubMed]
- Ishida, M.; Kawao, N.; Mizukami, Y.; Takafuji, Y.; Kaji, H. Influence of Angptl1 on Osteoclast Formation and Osteoblastic Phenotype in Mouse Cells. BMC Musculoskelet. Disord. 2021, 22, 398. [Google Scholar] [CrossRef]
- Santulli, G. Angiopoietin-like Proteins: A Comprehensive Look. Front. Endocrinol. 2014, 5, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Kuo, T.; Tseng, C.; Ma, J. ANGPTL1 Antagonizes MET Receptor Activity to Repress Sorafenib Resistance and Cancer Stemness in Hepatocellular Carcinoma Cells. Hepatology 2016, 64, 1637–1651. [Google Scholar] [CrossRef]
- Jiang, K.; Chen, H.; Fang, Y.; Chen, L.; Zhong, C.; Bu, T.; Dai, S.; Pan, X.; Fu, D.; Qian, Y.; et al. Exosomal ANGPTL1 Attenuates Colorectal Cancer Liver Metastasis by Regulating Kupffer Cell Secretion Pattern and Impeding MMP9 Induced Vascular Leakiness. J. Exp. Clin. Cancer Res. 2021, 40, 21. [Google Scholar] [CrossRef]
- Zeng, G.Q.; Chen, A.B.; Li, W.; Song, J.H.; Gao, C.Y. High MMP-1, MMP-2, and MMP-9 Protein Levels in Osteoarthritis. Genet. Mol. Res. 2015, 14, 14811–14822. [Google Scholar] [CrossRef] [PubMed]
- Hébert, J.M.; Rosenquist, T.; Götz, J.; Martin, G.R. FGF5 as a Regulator of the Hair Growth Cycle: Evidence from Targeted and Spontaneous Mutations. Cell 1994, 78, 1017–1025. [Google Scholar] [CrossRef]
- Allerstorfer, S.; Sonvilla, G.; Fischer, H.; Spiegl-Kreinecker, S.; Gauglhofer, C.; Setinek, U.; Czech, T.; Marosi, C.; Buchroithner, J.; Pichler, J.; et al. FGF5 as an Oncogenic Factor in Human Glioblastoma Multiforme: Autocrine and Paracrine Activities. Oncogene 2008, 27, 4180–4190. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Wang, M.; Yu, Z.; Yin, L.; Liu, C.; Wang, J.; Liu, Y.; Jiang, S.; Ren, Z.; Yin, J. FGF5 Promotes Osteosarcoma Cells Proliferation via Activating MAPK Signaling Pathway. Cancer Manag. Res. 2019, 11, 6457–6466. [Google Scholar] [CrossRef]
- Ghassemi, S.; Vejdovszky, K.; Sahin, E.; Ratzinger, L.; Schelch, K.; Mohr, T.; Peter-Vörösmarty, B.; Brankovic, J.; Lackner, A.; Leopoldi, A.; et al. FGF5 Is Expressed in Melanoma and Enhances Malignancy in Vitro and in Vivo. Oncotarget 2017, 8, 87750–87762. [Google Scholar] [CrossRef][Green Version]
- Hanaka, H.; Hamada, T.; Ito, M.; Nakashima, H.; Tomita, K.; Seki, S.; Kobayashi, Y.; Imaki, J. Fibroblast Growth Factor-5 Participates in the Progression of Hepatic Fibrosis. Exp. Anim. 2014, 63, 85–92. [Google Scholar] [CrossRef]
- Remst, D.F.G.; Davidson, E.N.B.; van der Kraan, P.M. Unravelling Osteoarthritis-Related Synovial Fibrosis: A Step Closer to Solving Joint Stiffness. Rheumatology 2015, 54, 1954–1963. [Google Scholar] [CrossRef]
- Carballo, C.B.; Nakagawa, Y.; Sekiya, I.; Rodeo, S.A. Basic Science of Articular Cartilage. Clin. Sports Med. 2017, 36, 413–425. [Google Scholar] [CrossRef]
- Chow, Y.; Chin, K. The Role of Inflammation in the Pathogenesis of Osteoarthritis. Mediat. Inflamm. 2020, 2020, 8293921. [Google Scholar] [CrossRef]
- Capsoni, F.; Ongari, A.M.; Lonati, C.; Accetta, R.; Gatti, S.; Catania, A. α-Melanocyte-Stimulating-Hormone (α-MSH) Modulates Human Chondrocyte Activation Induced by Proinflammatory Cytokines. BMC Musculoskelet. Disord. 2015, 16, 154. [Google Scholar] [CrossRef]
- Favero, M.; Belluzzi, E.; Trisolino, G.; Goldring, M.B.; Goldring, S.R.; Cigolotti, A.; Pozzuoli, A.; Ruggieri, P.; Ramonda, R.; Grigolo, B.; et al. Inflammatory Molecules Produced by Meniscus and Synovium in Early and End-Stage Osteoarthritis: A Coculture Study. J. Cell. Physiol. 2019, 234, 11176–11187. [Google Scholar] [CrossRef] [PubMed]
- Haraden, C.A.; Huebner, J.L.; Hsueh, M.F.; Li, Y.J.; Kraus, V.B. Synovial Fluid Biomarkers Associated with Osteoarthritis Severity Reflect Macrophage and Neutrophil Related Inflammation. Arthritis Res. Ther. 2019, 21, 146. [Google Scholar] [CrossRef] [PubMed]
- Pengas, I.; Eldridge, S.; Assiotis, A.; McNicholas, M.; Mendes, J.E.; Laver, L. MMP-3 in the Peripheral Serum as a Biomarker of Knee Osteoarthritis, 40 Years after Open Total Knee Meniscectomy. J. Exp. Orthop. 2018, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Kozawa, E.; Cheng, X.W.; Urakawa, H.; Arai, E.; Yamada, Y.; Kitamura, S.; Sato, K.; Kuzuya, M.; Ishiguro, N.; Nishida, Y. Increased Expression and Activation of Cathepsin K in Human Osteoarthritic Cartilage and Synovial Tissues. J. Orthop. Res. 2016, 34, 127–134. [Google Scholar] [CrossRef]
- Rezus, E.; Burlui, A.; Cardoneanu, A.; Macovei, L.A.; Tamba, B.I.; Rezus, C. From Pathogenesis to Therapy in Knee Osteoarthritis: Bench-to-Bedside. Int. J. Mol. Sci. 2021, 22, 2697. [Google Scholar] [CrossRef]
- Kozawa, E.; Nishida, Y.; Cheng, X.W.; Urakawa, H.; Arai, E.; Futamura, N.; Shi, G.P.; Kuzuya, M.; Hu, L.; Sasaki, T.; et al. Osteoarthritic Change Is Delayed in a Ctsk-Knockout Mouse Model of Osteoarthritis. Arthritis Rheum. 2012, 64, 454–464. [Google Scholar] [CrossRef]
- Yasuda, Y.; Kaleta, J.; Brömme, D. The Role of Cathepsins in Osteoporosis and Arthritis: Rationale for the Design of New Therapeutics. Adv. Drug Deliv. Rev. 2005, 57, 973–993. [Google Scholar] [CrossRef]
- Lambert, C.; Dubuc, J.E.; Montell, E.; Vergés, J.; Munaut, C.; Noël, A.; Henrotin, Y. Gene Expression Pattern of Cells from Inflamed and Normal Areas of Osteoarthritis Synovial Membrane. Arthritis Rheumatol. 2014, 66, 960–968. [Google Scholar] [CrossRef]
- Weitoft, T.; Larsson, A.; Manivel, V.A.; Lysholm, J.; Knight, A.; Rönnelid, J. Cathepsin S and Cathepsin L in Serum and Synovial Fluid in Rheumatoid Arthritis with and without Autoantibodies. Rheumatology 2015, 54, 1923–1928. [Google Scholar] [CrossRef]
- Yao, X.; Cheng, F.; Yu, W.; Rao, T.; Li, W.; Zhao, S.; Zhou, X.; Ning, J. Cathepsin S Regulates Renal Fibrosis in Mouse Models of Mild and Severe Hydronephrosis. Mol. Med. Rep. 2019, 20, 141–150. [Google Scholar] [CrossRef]
- Zhang, L.; Xing, R.; Huang, Z.; Ding, L.; Zhang, L.; Li, M.; Li, X.; Wang, P.; Mao, J. Synovial Fibrosis Involvement in Osteoarthritis. Front. Med. 2021, 8, 684389. [Google Scholar] [CrossRef]
- Daoussis, D.; Andonopoulos, A.P. The Emerging Role of Dickkopf-1 in Bone Biology: Is It the Main Switch Controlling Bone and Joint Remodeling? Semin. Arthritis Rheum. 2011, 41, 170–177. [Google Scholar] [CrossRef]
- Thomas, T.; Nikolaos, E.; Vasileios, N.; Ioannis, C.; Ioannis, P.; Panayiotis, K.; Athanasios, P.; Narjes, N.A.; Eva, K. Association between Serum and Synovial Fluid Dickkopf-1 Levels with Radiographic Severity in Primary Knee Osteoarthritis Patients. Clin. Rheumatol. 2017, 36, 1865–1872. [Google Scholar] [CrossRef]
- Zheng, L.; Hu, F.; Bian, W.; Li, Y.; Zhang, L.; Shi, L.; Ma, X.; Liu, Y.; Zhang, X.; Li, Z. Dickkopf-1 Perpetuated Synovial Fibroblast Activation and Synovial Angiogenesis in Rheumatoid Arthritis. Clin. Rheumatol. 2021, 40, 4279–4288. [Google Scholar] [CrossRef]
- Weng, L.H.; Wang, C.J.; Ko, J.Y.; Sun, Y.C.; Su, Y.S.; Wang, F.S. Inflammation Induction of Dickkopf-1 Mediates Chondrocyte Apoptosis in Osteoarthritic Joint. Osteoarthr. Cartil. 2009, 17, 933–943. [Google Scholar] [CrossRef]
- Diarra, D.; Stolina, M.; Polzer, K.; Zwerina, J.; Ominsky, M.S.; Dwyer, D.; Korb, A.; Smolen, J.; Hoffmann, M.; Scheinecker, C.; et al. Dickkopf-1 Is a Master Regulator of Joint Remodeling. Nat. Med. 2007, 13, 156–163. [Google Scholar] [CrossRef]
- Möller-Hackbarth, K.; Dabaghie, D.; Charrin, E.; Zambrano, S.; Genové, G.; Li, X.; Wernerson, A.; Lal, M.; Patrakka, J. Retinoic Acid Receptor Responder1 Promotes Development of Glomerular Diseases via the Nuclear Factor-ΚB Signaling Pathway. Kidney Int. 2021, 100, 809–823. [Google Scholar] [CrossRef]
- Santana, L.J.C.; Herrera, F.R.; Rojas, A.P.; Lozano, D.J.M.; Prieto, N.; Castañeda, M.B. Serum Chemerin in a Cohort of Colombian Patients with Primary Osteoarthritis. Reumatol. Clínica 2021, 17, 530–535. [Google Scholar] [CrossRef]
- Gonzalez-Ponce, F.; Gamez-Nava, J.I.; Perez-Guerrero, E.E.; Saldaña-Cruz, A.M.; Vazquez-Villegas, M.L.; Ponce-Guarneros, J.M.; Huerta, M.; Trujillo, X.; Contreras-Haro, B.; Rocha-Muñoz, A.D.; et al. Serum Chemerin Levels: A Potential Biomarker of Joint Inflammation in Women with Rheumatoid Arthritis. PLoS ONE 2021, 16, e0255854. [Google Scholar] [CrossRef]
- Iannone, F.; Lapadula, G. Chemerin/ChemR23 Pathway: A System beyond Chemokines. Arthritis Res. Ther. 2011, 13, 104. [Google Scholar] [CrossRef][Green Version]
- Eisinger, K.; Bauer, S.; Schäffler, A.; Walter, R.; Neumann, E.; Buechler, C.; Müller-Ladner, U.; Frommer, K.W. Chemerin Induces CCL2 and TLR4 in Synovial Fibroblasts of Patients with Rheumatoid Arthritis and Osteoarthritis. Exp. Mol. Pathol. 2012, 92, 90–96. [Google Scholar] [CrossRef]
- Ma, J.; Ren, L.; Guo, C.J.; Wan, N.J.; Niu, D.S. Chemerin Affects the Metabolic and Proliferative Capabilities of Chondrocytes by Increasing the Phosphorylation of AKT/ERK. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3656–3662. [Google Scholar] [CrossRef]
- Lowin, T.; Zhu, W.; Dettmer-Wilde, K.; Straub, R.H. Cortisol-Mediated Adhesion of Synovial Fibroblasts Is Dependent on the Degradation of Anandamide and Activation of the Endocannabinoid System. Arthritis Rheum. 2012, 64, 3867–3876. [Google Scholar] [CrossRef]
- Gawlińska, K.; Gawliński, D.; Korostyński, M.; Borczyk, M.; Frankowska, M.; Piechota, M.; Filip, M.; Przegaliński, E. Maternal Dietary Patterns Are Associated with Susceptibility to a Depressive-like Phenotype in Rat Offspring. Dev. Cogn. Neurosci. 2021, 47, 100879. [Google Scholar] [CrossRef]
- Babraham Bioinformatics. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 23 September 2022).
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2012, 31, 46–53. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chwastek, J.; Kędziora, M.; Borczyk, M.; Korostyński, M.; Starowicz, K. Inflammation-Driven Secretion Potential Is Upregulated in Osteoarthritic Fibroblast-Like Synoviocytes. Int. J. Mol. Sci. 2022, 23, 11817. https://doi.org/10.3390/ijms231911817
Chwastek J, Kędziora M, Borczyk M, Korostyński M, Starowicz K. Inflammation-Driven Secretion Potential Is Upregulated in Osteoarthritic Fibroblast-Like Synoviocytes. International Journal of Molecular Sciences. 2022; 23(19):11817. https://doi.org/10.3390/ijms231911817
Chicago/Turabian StyleChwastek, Jakub, Marta Kędziora, Małgorzata Borczyk, Michał Korostyński, and Katarzyna Starowicz. 2022. "Inflammation-Driven Secretion Potential Is Upregulated in Osteoarthritic Fibroblast-Like Synoviocytes" International Journal of Molecular Sciences 23, no. 19: 11817. https://doi.org/10.3390/ijms231911817
APA StyleChwastek, J., Kędziora, M., Borczyk, M., Korostyński, M., & Starowicz, K. (2022). Inflammation-Driven Secretion Potential Is Upregulated in Osteoarthritic Fibroblast-Like Synoviocytes. International Journal of Molecular Sciences, 23(19), 11817. https://doi.org/10.3390/ijms231911817





