Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing
Abstract
1. Introduction
2. Results
2.1. Validation of Custom Multimodal NGS Panel
2.2. DNA SNP Evaluation in Relation to Level of CDDP Resistance
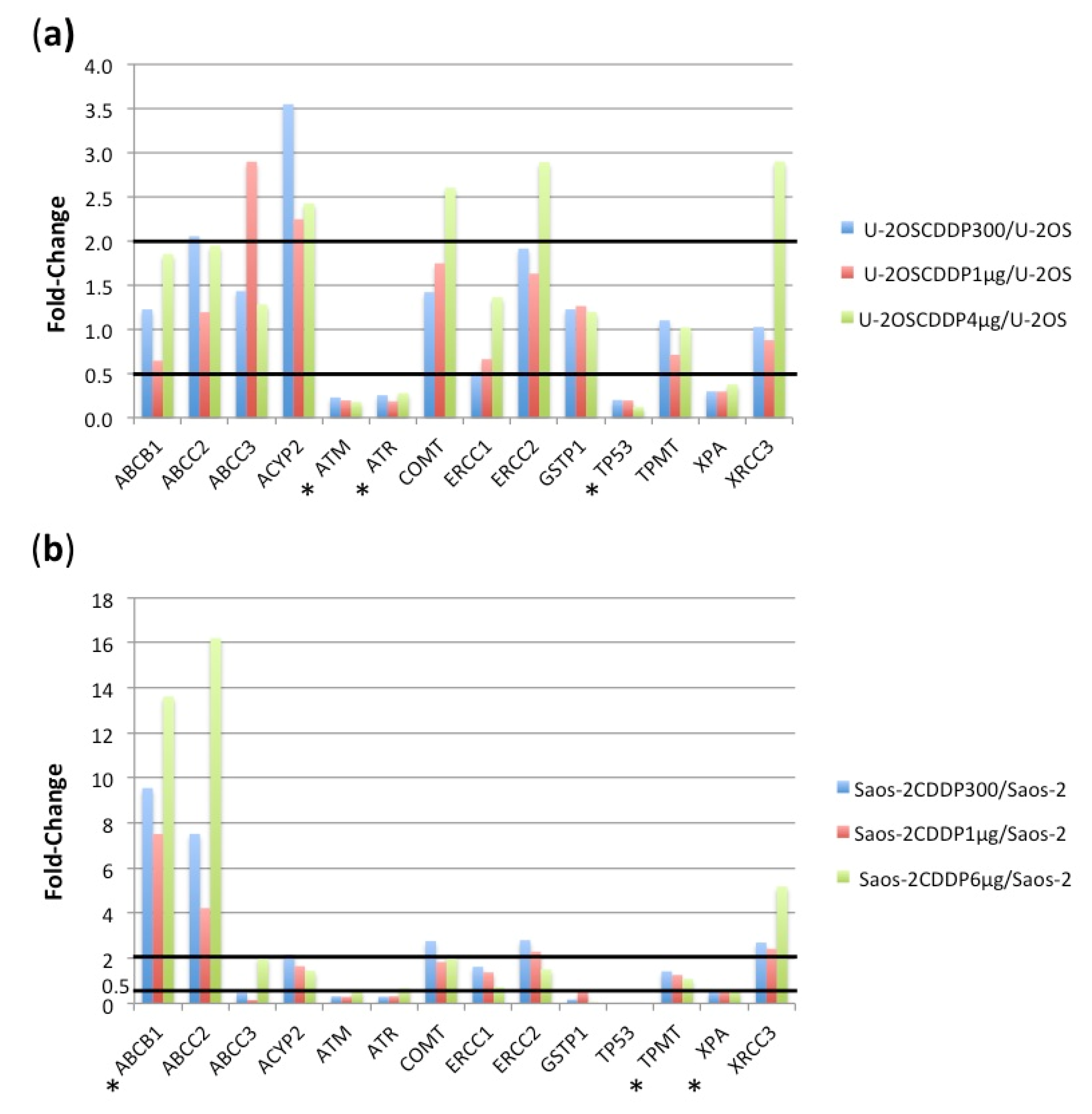
2.2.1. Comparison between U-2OS CDDP-Resistant Variants to Parental U-2OS Cell Line
2.2.2. Comparison between Saos-2 CDDP-Resistant Variants to Parental Saos-2 Cell Line
2.3. RNA SNP Evaluation in Relation to Level of CDDP Resistance
2.3.1. Comparison between U-2OS Cell Line and U-2OS CDDP-Resistant Variants
2.3.2. Comparison between Saos-2 Cell Line and Saos-2 CDDP-Resistant Variants
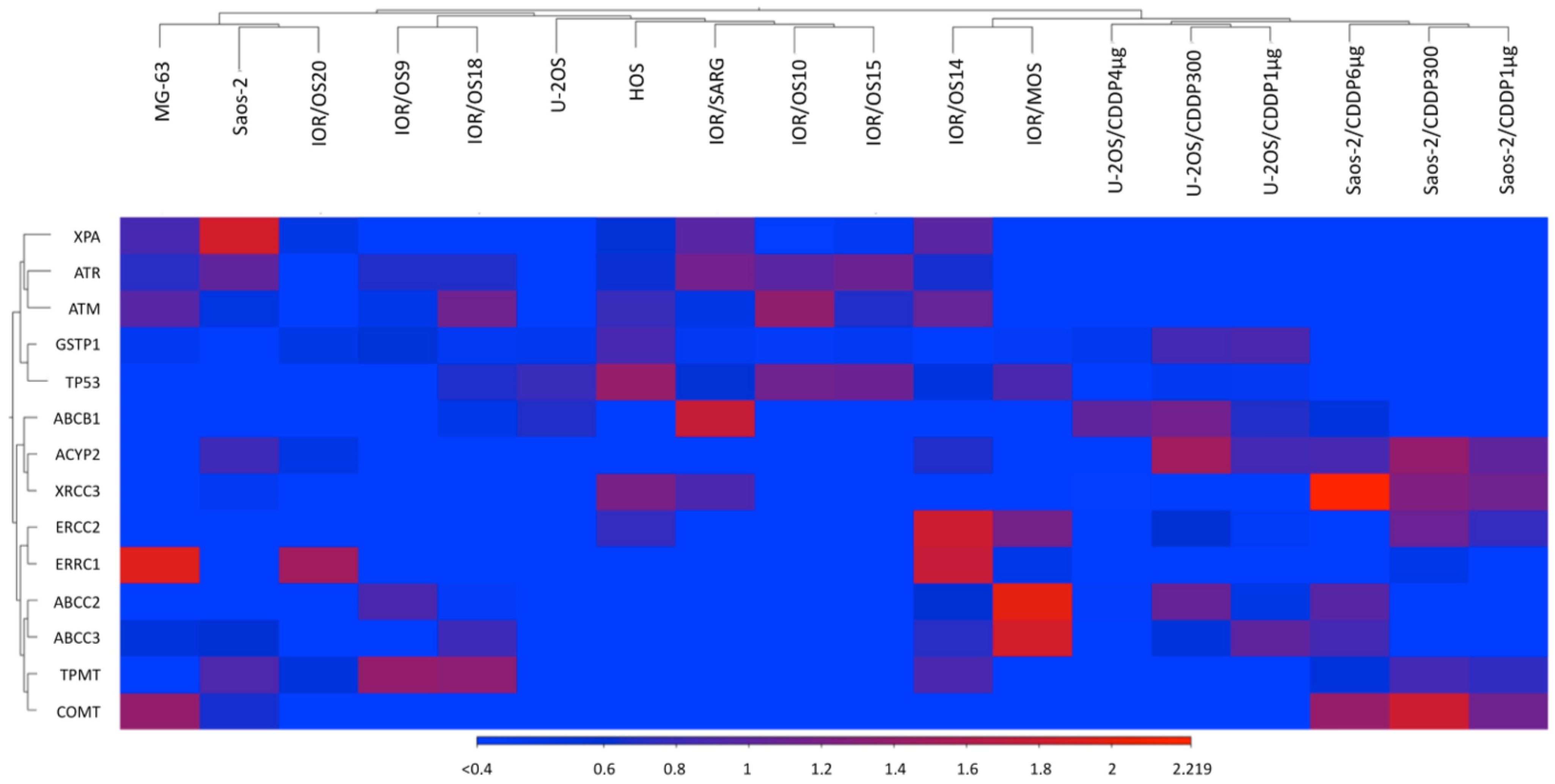
2.4. RNA Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. Extraction of Nucleic Acids
4.3. Custom Multi-Modal Targeted Next Generation Sequencing (mmNGS)
4.4. mmNGS Data Analysis by CLC Genomics Workbench
4.5. SNP Genotyping by Real-Time PCR
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ABCB1 | ATP Binding Cassette Subfamily B Member 1 |
| ABCC2 | ATP Binding Cassette Subfamily C Member 2 |
| ABCC3 | ATP Binding Cassette Subfamily C Member 3 |
| ACYP2 | Acylphosphatase 2 |
| ATM | ATM Serine/Threonine Kinase |
| ATR | ATR Serine/Threonine Kinase |
| CDDP | Cisplatin |
| CLC GWB | CLC Genomics Workbench |
| COMT | Cathechol-O-Methyltransferase |
| ERCC1 | Excision Repair Cross-Complementing 1, Endonuclease Non-Catalytic Subunit |
| ERCC2 | Excision Repair Cross-Complementing 2, TFIIH Core Complex Helicase Subunit |
| GSTP1 | Glutathione S-Transferase Pi 1 |
| HGOS | High-Grade Osteosarcoma |
| IMDM | Iscove’s modified Dulbecco’s medium |
| mmNGS | Multimodal Targeted Next Generation Sequencing |
| MNV | Multi Nucleotide Variant |
| NER | Nucleotide Excision Repair |
| SNP | Single Nucleotide Polymorphism |
| TP53 | Tumor Protein P53 |
| TPM | Transcripts per Million |
| TPMT | Thiopurine S-Methyltransferase |
| UMI | Unique Molecular Indices |
| VAF | Variant Allele Frequency |
| XPA | XPA, DNA Damage Recognition and Repair Factor |
| XRCC3 | X-Ray Repair Cross Complementing 3 |
Appendix A
| Gene Name | Abbreviation | Function |
|---|---|---|
| ATP Binding Cassette Subfamily B Member 1 | ABCB1 | CDDP transport |
| ATP Binding Cassette Subfamily C Member 2 | ABCC2 | CDDP transport |
| ATP Binding Cassette Subfamily C Member 3 | ABCC3 | CDDP-related ototoxicity |
| Acylphosphatase 2 | ACYP2 | CDDP-related ototoxicity |
| ATM Serine/Threonine Kinase | ATM | DNA repair |
| ATR Serine/Threonine Kinase | ATR | DNA repair |
| Cathecol-O-Methyltransferase | COMT | CDDP-related ototoxicity |
| Excision Repair Cross-Complementing 1 | ERCC1 | DNA repair |
| Excision Repair Cross-Complementing 2 | ERCC2 | DNA repair |
| Glutathione S-Transferase P1 | GSTP1 | CDDP detoxification |
| Tumor Protein 53 | TP53 | Genomic stability |
| Thiopurine S-methyltransferase | TPMT | CDDP-related ototoxicity |
| Xeroderma pigmentosum group A | XPA | DNA repair |
| X-Ray Repair Cross Complementing 3 | XRCC3 | DNA repair |
| Gene_Reference Number | Assay ID | Assay Type | Reference Allele |
|---|---|---|---|
| ABCB1_rs1045642 | C___7586657_20 | DME | A |
| ABCB1_rs2032582 | C_11711720C_30 + C_11711720D_40 | DME | A |
| ABCB1_rs1128503 | C___7586662_10 | DME | A |
| ABCC2_rs717620 | C___2814642_10 | DME | T |
| ABCC2_rs2273697 | C__22272980_20 | DME | C |
| ABCC2_rs3740066 | C__11214910_20 | DME | G |
| ABCC2_rs17222723 | C__25591743_30 | DME | T |
| ABCC3_rs4793665 | Not done | Not done | C |
| ABCC3_rs1051640 | Not done | Not done | A |
| ACYP2_rs1872328 | C__11643398_10 | Funct. tested | G |
| ATM_rs664677 | C___2283171_1_ | Validated | C |
| ATM_rs664143 | C___1039783_10 | Validated | A |
| ATR_rs2229032 | C__26021082_10 | Funct. tested | C |
| ATR_rs2227928 | C____157700_10 | Validated | A |
| COMT_rs4646316 | C__29193982_10 | Funct. tested | C |
| COMT_rs9332377 | Not done | Not done | C |
| ERCC1_rs11615 | C___2532959_1_ | Validated | A |
| ERCC1_rs3212986 | Not done | Not done | C |
| ERCC2_rs13181 | C___3145033_10 | Validated | T |
| ERCC2_rs1799793 | C___3145050_10 | Funct. tested | C |
| GSTP1_rs1695 | C___3237198_20 | DME | A |
| TP53_rs1042522 | C___2403545_10 | Funct. tested | G |
| TP53_rs1642785 | C___2880090_10 | Funct. tested | G |
| TPMT_rs12201199 | C__31923406_10 | Funct. tested | A |
| TPMT_rs1142345 | C_____19567_20 | DME | T |
| TPMT_rs1800460 | C__30634116_20 | DME | C |
| XPA_rs1800975 | C____482935_1_ | Validated | T |
| XRCC3_rs861539 | C___8901525_10 | Funct. tested | G |
References
- Harrison, D.J.; Geller, D.S.; Gill, J.D.; Lewis, V.O.; Gorlick, R. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther. 2018, 18, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Lilienthal, I.; Herold, N. Targeting Molecular Mechanisms Underlying Treatment Efficacy and Resistance in Osteosarcoma: A Review of Current and Future Strategies. Int. J. Mol. Sci. 2020, 21, 6885. [Google Scholar] [CrossRef] [PubMed]
- Hattinger, C.M.; Patrizio, M.P.; Luppi, S.; Serra, M. Pharmacogenomics and Pharmacogenetics in Osteosarcoma: Translational Studies and Clinical Impact. Int. J. Mol. Sci. 2020, 21, 4659. [Google Scholar] [CrossRef] [PubMed]
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef]
- Kager, L.; Tamamyan, G.; Bielack, S. Novel insights and therapeutic interventions for pediatric osteosarcoma. Future Oncol. 2017, 13, 357–368. [Google Scholar] [CrossRef]
- Serra, M.; Hattinger, C.M. The pharmacogenomics of osteosarcoma. Pharmacogenomics J. 2017, 17, 11–20. [Google Scholar] [CrossRef]
- Vos, H.I.; Guchelaar, H.J.; Gelderblom, H.; de Bont, E.S.; Kremer, L.C.; Naber, A.M.; Hakobjan, M.H.; van der Graaf, W.T.; Coenen, M.J.; te Loo, D.M. Replication of a genetic variant in ACYP2 associated with cisplatin-induced hearing loss in patients with osteosarcoma. Pharmacogenet. Genomics 2016, 26, 243–247. [Google Scholar] [CrossRef]
- Windsor, R.E.; Strauss, S.J.; Kallis, C.; Wood, N.E.; Whelan, J.S. Germline genetic polymorphisms may influence chemotherapy response and disease outcome in osteosarcoma: A pilot study. Cancer 2012, 118, 1856–1867. [Google Scholar] [CrossRef]
- Ross, C.J.; Katzov-Eckert, H.; Dube, M.P.; Brooks, B.; Rassekh, S.R.; Barhdadi, A.; Feroz-Zada, Y.; Visscher, H.; Brown, A.M.; Rieder, M.J.; et al. Genetic variants in TPMT and COMT are associated with hearing loss in children receiving cisplatin chemotherapy. Nat. Genet. 2009, 41, 1345–1349. [Google Scholar] [CrossRef]
- Pussegoda, K.; Ross, C.J.; Visscher, H.; Yazdanpanah, M.; Brooks, B.; Rassekh, S.R.; Zada, Y.F.; Dube, M.P.; Carleton, B.C.; Hayden, M.R.; et al. Replication of TPMT and ABCC3 genetic variants highly associated with cisplatin-induced hearing loss in children. Clin. Pharmacol. Ther. 2013, 94, 243–251. [Google Scholar] [CrossRef]
- Hagleitner, M.M.; Coenen, M.J.; Patino-Garcia, A.; de Bont, E.S.; Gonzalez-Neira, A.; Vos, H.I.; van Leeuwen, F.N.; Gelderblom, H.; Hoogerbrugge, P.M.; Guchelaar, H.J.; et al. Influence of genetic variants in TPMT and COMT associated with cisplatin induced hearing loss in patients with cancer: Two new cohorts and a meta-analysis reveal significant heterogeneity between cohorts. PLoS ONE 2014, 9, e115869. [Google Scholar] [CrossRef]
- Pasello, M.; Michelacci, F.; Scionti, I.; Hattinger, C.M.; Zuntini, M.; Caccuri, A.M.; Scotlandi, K.; Picci, P.; Serra, M. Overcoming glutathione S-transferase P1-related cisplatin resistance in osteosarcoma. Cancer Res. 2008, 68, 6661–6668. [Google Scholar] [CrossRef]
- Li, J.Z.; Tian, Z.Q.; Jiang, S.N.; Feng, T. Effect of variation of ABCB1 and GSTP1 on osteosarcoma survival after chemotherapy. Genet. Mol. Res. 2014, 13, 3186–3192. [Google Scholar] [CrossRef]
- Liu, S.; Yi, Z.; Ling, M.; Shi, J.; Qiu, Y.; Yang, S. Predictive potential of ABCB1, ABCC3, and GSTP1 gene polymorphisms on osteosarcoma survival after chemotherapy. Tumour Biol. 2014, 35, 9897–9904. [Google Scholar] [CrossRef]
- Teng, J.W.; Yang, Z.M.; Li, J.; Xu, B. Predictive role of Glutathione S-transferases (GSTs) on the prognosis of osteosarcoma patients treated with chemotherapy. Pak. J. Med. Sci. 2013, 29, 1182–1186. [Google Scholar] [CrossRef]
- Zhang, S.L.; Mao, N.F.; Sun, J.Y.; Shi, Z.C.; Wang, B.; Sun, Y.J. Predictive potential of glutathione S-transferase polymorphisms for prognosis of osteosarcoma patients on chemotherapy. Asian Pac. J. Cancer Prev. 2012, 13, 2705–2709. [Google Scholar] [CrossRef][Green Version]
- Yang, L.M.; Li, X.H.; Bao, C.F. Glutathione S-transferase P1 and DNA polymorphisms influence response to chemotherapy and prognosis of bone tumors. Asian Pac. J. Cancer Prev. 2012, 13, 5883–5886. [Google Scholar] [CrossRef][Green Version]
- Goricar, K.; Kovac, V.; Jazbec, J.; Zakotnik, B.; Lamovec, J.; Dolzan, V. Genetic variability of DNA repair mechanisms and glutathione-S-transferase genes influences treatment outcome in osteosarcoma. Cancer Epidemiol. 2015, 39, 182–188. [Google Scholar] [CrossRef]
- Cao, Z.H.; Yin, H.P.; Jiang, N.; Yu, B. Association between ERCC1 and ERCC2 gene polymorphisms and chemotherapy response and overall survival in osteosarcoma. Genet. Mol. Res. 2015, 14, 10145–10151. [Google Scholar] [CrossRef]
- Ji, W.P.; He, N.B. Investigation on the DNA repaired gene polymorphisms and response to chemotherapy and overall survival of osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 894–899. [Google Scholar]
- Sun, Y.; Wu, Y.; Li, W.; Kong, Z.; Zou, X. Genetic polymorphisms in nucleotide excision repair pathway influences response to chemotherapy and overall survival in osteosarcoma. Int. J. Clin. Exp. Pathol. 2015, 8, 7905–7912. [Google Scholar]
- Zhang, H.; Ge, J.; Hong, H.; Bi, L.; Sun, Z. Genetic polymorphisms in ERCC1 and ERCC2 genes are associated with response to chemotherapy in osteosarcoma patients among Chinese population: A meta-analysis. World J. Surg. Oncol. 2017, 15, 75. [Google Scholar] [CrossRef][Green Version]
- Zhang, Q.; Lv, L.Y.; Li, B.J.; Zhang, J.; Wei, F. Investigation of ERCC1 and ERCC2 gene polymorphisms and response to chemotherapy and overall survival in osteosarcoma. Genet. Mol. Res. 2015, 14, 11235–11241. [Google Scholar] [CrossRef]
- Hao, T.; Feng, W.; Zhang, J.; Sun, Y.J.; Wang, G. Association of four ERCC1 and ERCC2 SNPs with survival of bone tumour patients. Asian Pac. J. Cancer Prev. 2012, 13, 3821–3824. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.J.; Zhu, Y.; Guo, X.J.; Tian, Z.Z. Genetic variability of genes involved in DNA repair influence treatment outcome in osteosarcoma. Genet. Mol. Res. 2015, 14, 11652–11657. [Google Scholar] [CrossRef] [PubMed]
- Obiedat, H.; Alrabadi, N.; Sultan, E.; Al Shatti, M.; Zihlif, M. The effect of ERCC1 and ERCC2 gene polymorphysims on response to cisplatin based therapy in osteosarcoma patients. BMC Med. Genet. 2018, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Caronia, D.; Patino-Garcia, A.; Milne, R.L.; Zalacain-Diez, M.; Pita, G.; Alonso, M.R.; Moreno, L.T.; Sierrasesumaga-Ariznabarreta, L.; Benitez, J.; Gonzalez-Neira, A. Common variations in ERCC2 are associated with response to cisplatin chemotherapy and clinical outcome in osteosarcoma patients. Pharmacogenomics J. 2009, 9, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Biason, P.; Hattinger, C.M.; Innocenti, F.; Talamini, R.; Alberghini, M.; Scotlandi, K.; Zanusso, C.; Serra, M.; Toffoli, G. Nucleotide excision repair gene variants and association with survival in osteosarcoma patients treated with neoadjuvant chemotherapy. Pharmacogenomics J. 2012, 12, 476–483. [Google Scholar] [CrossRef]
- Li, J.; Liu, S.; Wang, W.; Zhang, K.; Liu, Z.; Zhang, C.; Chen, S.; Wu, S. ERCC polymorphisms and prognosis of patients with osteosarcoma. Tumour Biol. 2014, 35, 10129–10136. [Google Scholar] [CrossRef]
- Pasqui, A.; Boddi, A.; Campanacci, D.A.; Scoccianti, G.; Bernini, A.; Grasso, D.; Gambale, E.; Scolari, F.; Palchetti, I.; Palomba, A.; et al. Alteration of the Nucleotide Excision Repair (NER) Pathway in Soft Tissue Sarcoma. Int. J. Mol. Sci. 2022, 23, 8360. [Google Scholar] [CrossRef]
- Peissert, S.; Sauer, F.; Grabarczyk, D.B.; Braun, C.; Sander, G.; Poterszman, A.; Egly, J.M.; Kuper, J.; Kisker, C. In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair. Nat. Commun. 2020, 11, 1667. [Google Scholar] [CrossRef]
- Kiss, R.C.; Xia, F.; Acklin, S. Targeting DNA Damage Response and Repair to Enhance Therapeutic Index in Cisplatin-Based Cancer Treatment. Int. J. Mol. Sci. 2021, 22, 8199. [Google Scholar] [CrossRef]
- Rocha, C.R.R.; Silva, M.M.; Quinet, A.; Cabral-Neto, J.B.; Menck, C.F.M. DNA repair pathways and cisplatin resistance: An intimate relationship. Clinics 2018, 73, e478s. [Google Scholar] [CrossRef]
- Benini, S.; Baldini, N.; Manara, M.C.; Chano, T.; Serra, M.; Rizzi, S.; Lollini, P.L.; Picci, P.; Scotlandi, K. Redundancy of autocrine loops in human osteosarcoma cells. Int. J. Cancer 1999, 80, 581–588. [Google Scholar] [CrossRef]

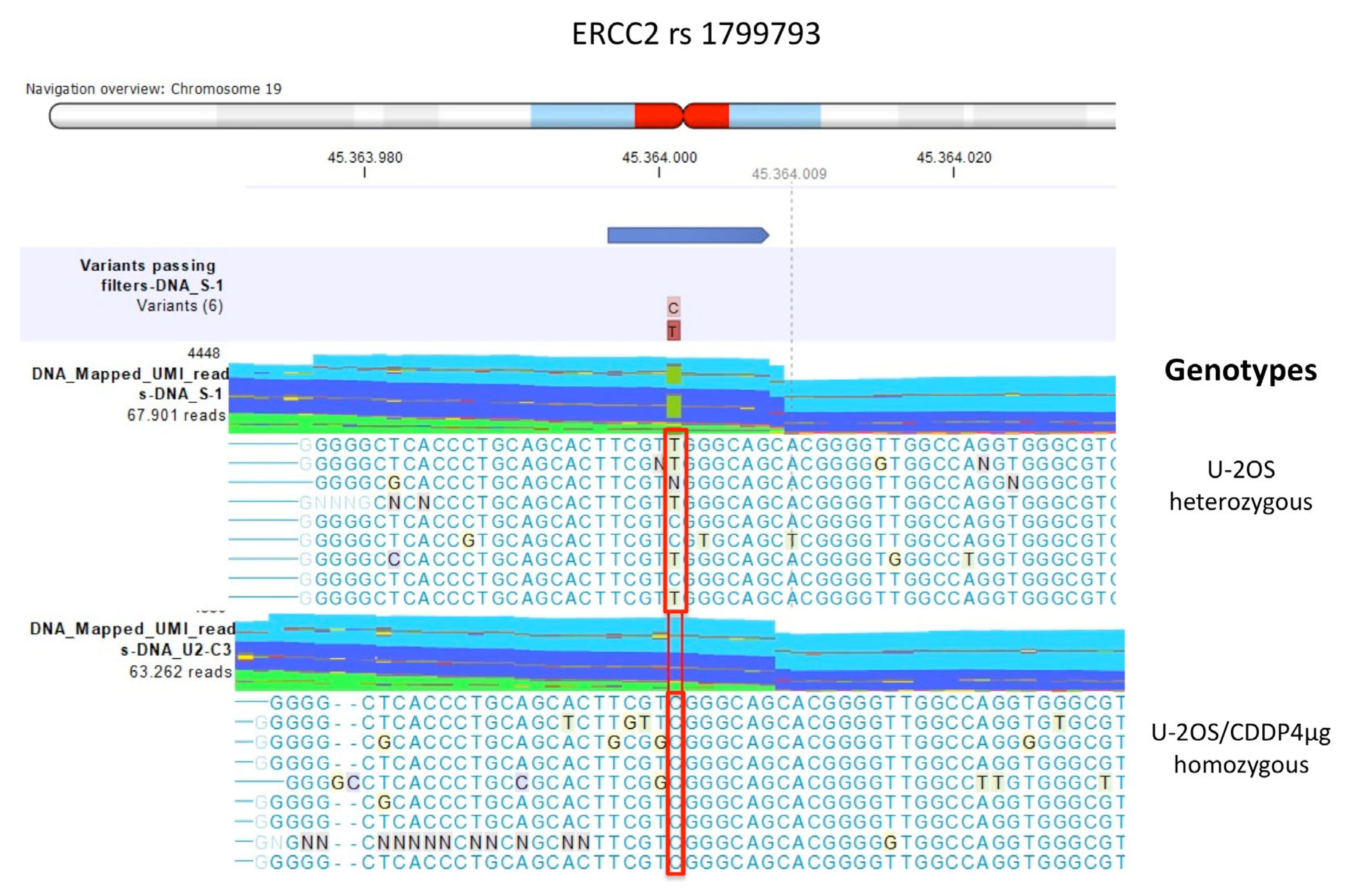
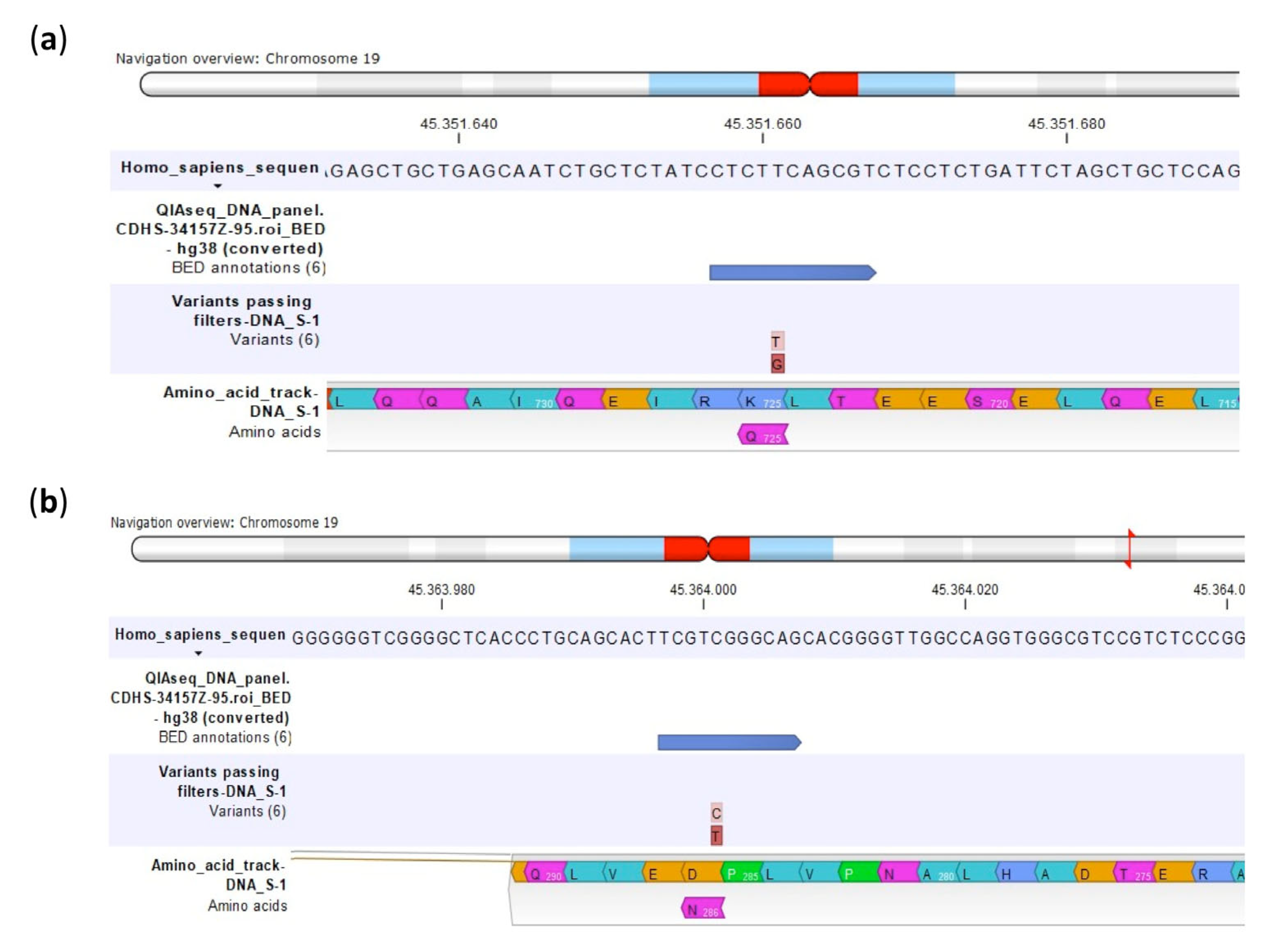
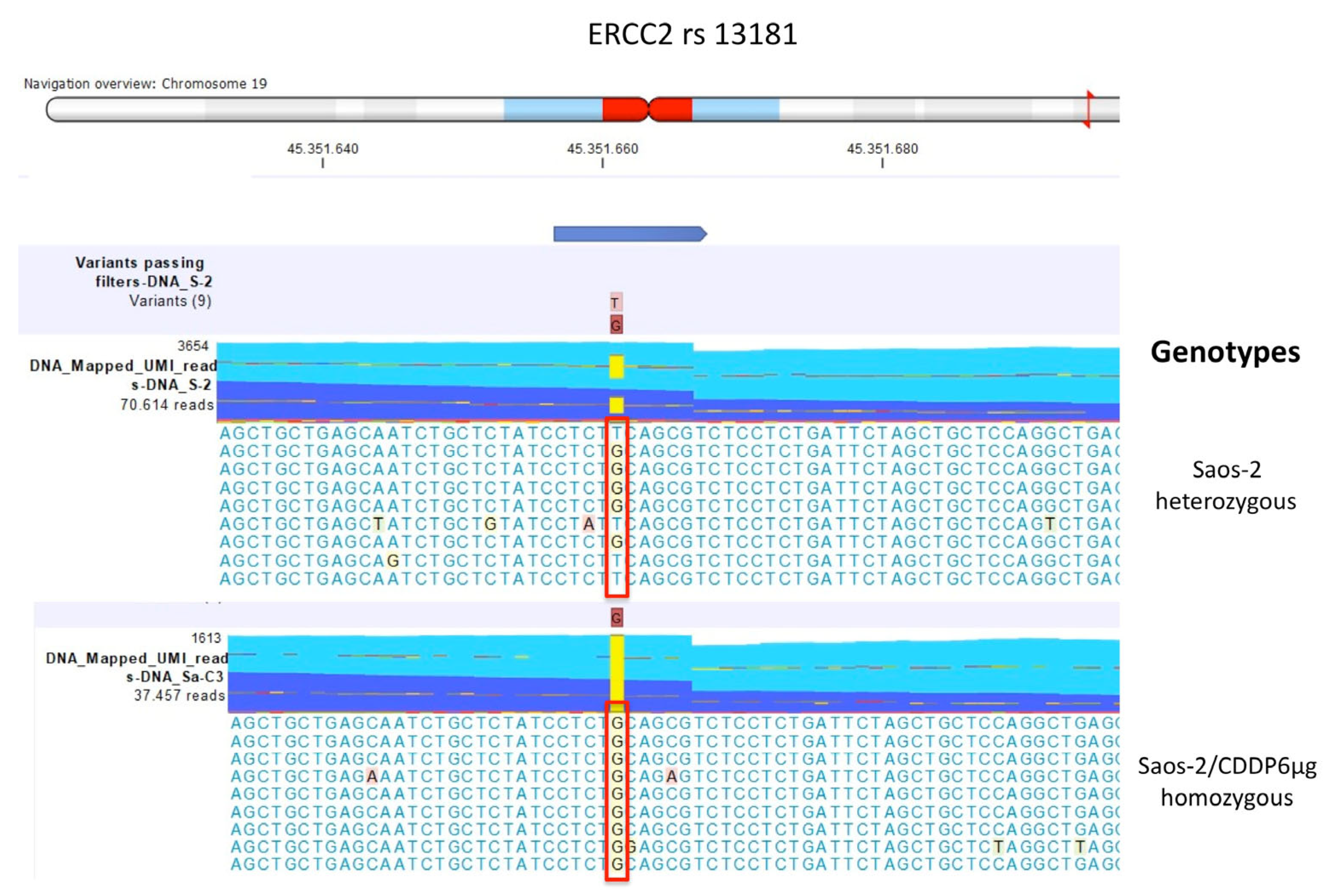
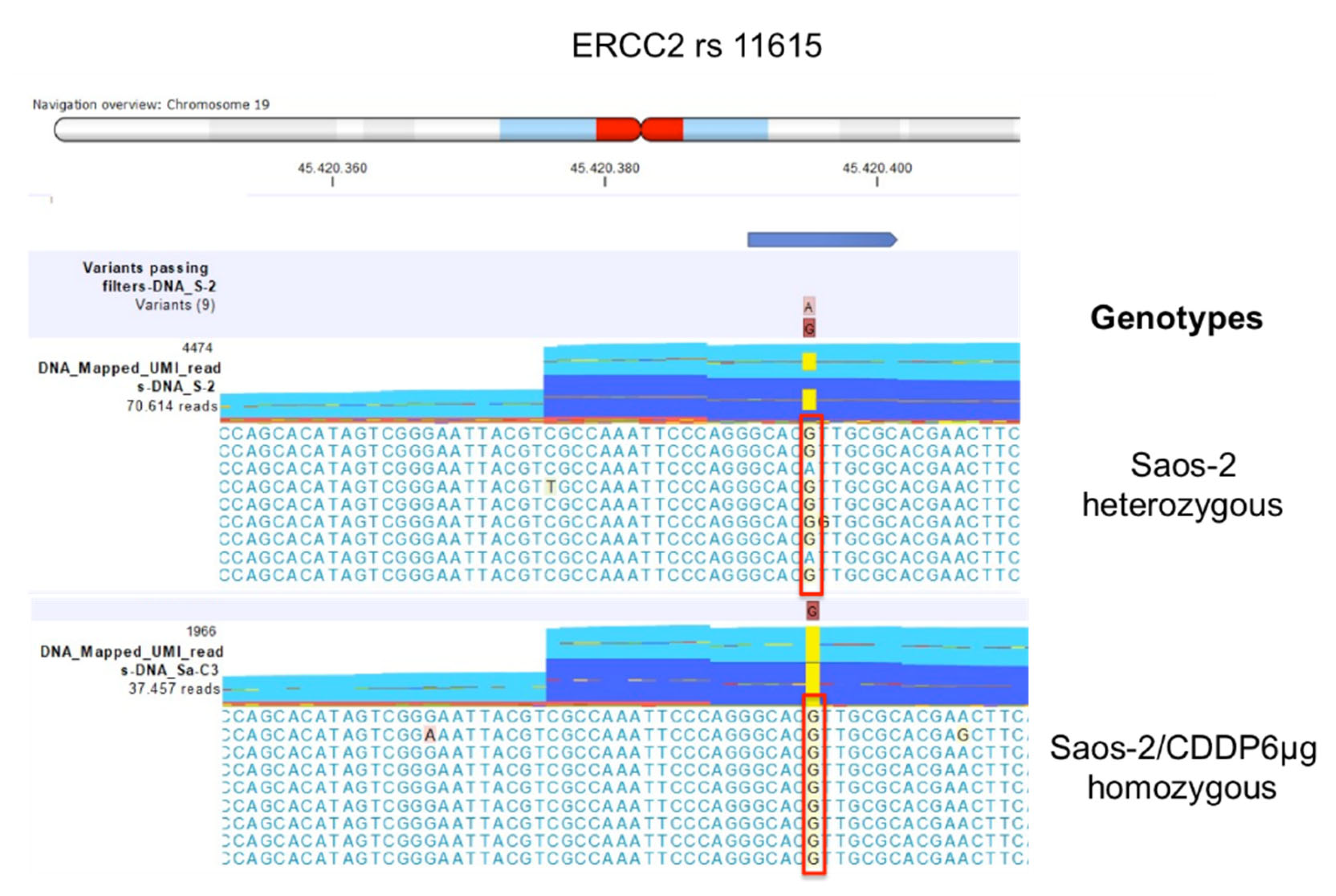
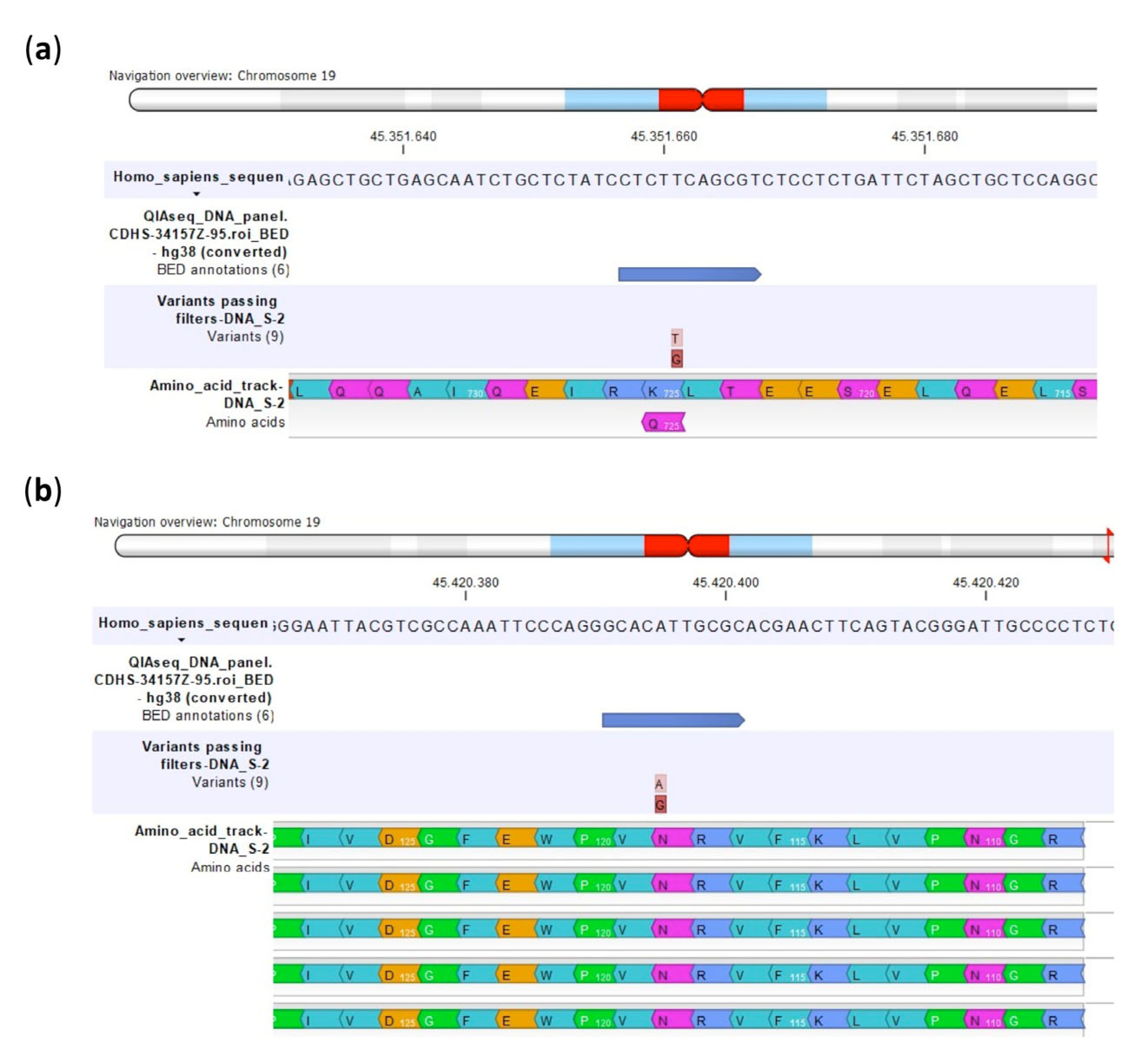
| Polymorphism | U-2OS | Saos-2 | IOR/OS9 | IOR/OS10 | IOR/OS14 | IOR/OS15 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | |
| ABCB1 rs1045642 | GA | GA | GA | GA | GA | GA | GG | GG | GG | GG | ||
| ABCB1 rs2032582 | CA | CA | CA | CA | CA | CA | CA | CA | CC | CC | CC | CC |
| ABCB1 rs1128503 | GG | GG | GA | GA | GA | GA | GA | GA | GG | GG | GG | GG |
| ABCC2 rs717620 | CT | CT | ||||||||||
| ABCC2 rs2273697 | GA | GA | AA | AA | ||||||||
| ABCC2 rs3740066 | CT | CT | TT | TT | TT | TT | ||||||
| ABCC2 rs17222723 | ||||||||||||
| ABCC3 rs4793665 | / | CT | / | TT | / | TC | ||||||
| ABCC3 rs1051640 | ||||||||||||
| ACYP2 rs1872328 | ||||||||||||
| ATM rs664677 | TT | TT | CT | CT | TT | TT | TT | TT | TT | TT | ||
| ATM C11orf65 rs664143 | GG | GG | GA | GA | GG | GG | GG | GG | GG | GG | ||
| ATR rs2229032 | TT | TT | ||||||||||
| ATR rs2227928 | GA | GA | GG | GG | GG | GG | GG/GA | GA | ||||
| COMT rs4646316 | TT | TT | TT | TT | CT | CT | ||||||
| COMT rs9332377 | ||||||||||||
| ERCC1 rs11615 | GA | GA | GA | GA | GA | GA | GG | GA | ||||
| ERCC1 rs3212986 | / | AC | / | AC | ||||||||
| ERCC2 rs13181 | GT | GT | GT | GT | GT | GT | GT | GT | GT | GT | GG | GG |
| ERCC2 rs1799793 | CT | CT | TT | TT | CT | CT | CC | TT | ||||
| GSTP1 rs1695 | AG | AG | AG | AG | AG | AG | AG | AG | ||||
| TP53 rs1042522 | CC | CC | CC | CC | ||||||||
| TP53 rs1642785 | CC | CC | ||||||||||
| TPMT rs12201199 | ||||||||||||
| TPMT rs1142345 | ||||||||||||
| TPMT rs1800460 | ||||||||||||
| XPA rs1800975 | CT | CC | CT | CT | CC | CC | CC | CC | ||||
| XRCC3;KLC1 rs861539 | AG | AG | AA | AA | AA | AA | GG | AG | AA | AA | ||
| Polymorphism | IOR/OS18 | IOR/OS20 | IOR/MOS | IOR/SARG | HOS | MG-63 | ||||||
| TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | |
| ABCB1 rs1045642 | GA | GA | GG | GG | GG | GA | GG | GG | ||||
| ABCB1 rs2032582 | CA | CA | CC | CC | CC | CC | CC | CC | ||||
| ABCB1 rs1128503 | GA | GA | GG | GG | GG | GG | GG | GG | ||||
| ABCC2 rs717620 | CT | CT | CT | CT | ||||||||
| ABCC2 rs2273697 | AA | AA | GA | GA | ||||||||
| ABCC2 rs3740066 | CT | CT | CT | CT | CT | CT | ||||||
| ABCC2 rs17222723 | ||||||||||||
| ABCC3 rs4793665 | / | TC | / | TC | / | CT | / | CT | / | TT | / | CT |
| ABCC3 rs1051640 | / | GA | / | GA | ||||||||
| ACYP2 rs1872328 | ||||||||||||
| ATM rs664677 | CT | CT | TT | TT | CT | CT | TT | TT | TT | TT | TT | TT |
| ATM;C11orf65 rs664143 | GA | GA | GG | GG | GA | GA | GG | GG | GG | GG | GG | GG |
| ATR rs2229032 | CT | CT | CT | CT | TT | TT | ||||||
| ATR rs2227928 | GG | GG | GG | GG | GG | GG | GG | GG | GG | GG | ||
| COMT rs4646316 | CT | CT | ||||||||||
| COMT rs9332377 | / | TT | ||||||||||
| ERCC1 rs11615 | GG | GG | GG | GG | GG | GG | GA | GA | ||||
| ERCC1 rs3212986 | / | AA | / | AC | / | AC | ||||||
| ERCC2 rs13181 | GT | GT | GG | GG | ||||||||
| ERCC2 rs1799793 | TT | TT | CC | TT | TT | TT | ||||||
| GSTP1 rs1695 | AG | AG | ||||||||||
| TP53 rs1042522 | CC | CC | CC | CC | CC | CC | CC | CC | CC | CC | ||
| TP53 rs1642785 | ||||||||||||
| TPMT rs12201199 | ||||||||||||
| TPMT rs1142345 | ||||||||||||
| TPMT rs1800460 | ||||||||||||
| XPA rs1800975 | CC | CC | CC | CC | CT | CC | ||||||
| XRCC3;KLC1 rs861539 | AA | AA | AG | AG | AG | AG | ||||||
| Polymorphism | U-2OS/ CDDP300 | U-2OS/ CDDP1μg | U-2OS/ CDDP4μg | Saos-2/ CDDP300 | Saos-2/ CDDP1μg | Saos-2/ CDDP6μg | ||||||
| TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | TaqM | NGS | |
| ABCB1 rs1045642 | GA | GA | GA | GA | GA | GA | ||||||
| ABCB1 rs2032582 | CA | CA | CA | CA | CA | CA | CA | CA | CA | CA | CA | CA |
| ABCB1 rs1128503 | GG | GG | GG | GG | GG | GG | GA | GA | GA | GA | GA | GA |
| ABCC2 rs717620 | ||||||||||||
| ABCC2 rs2273697 | ||||||||||||
| ABCC2 rs3740066 | CT | CT | CT | CT | CT | CT | TT | TT | TT | TT | TT | TT |
| ABCC2 rs17222723 | ||||||||||||
| ABCC3 rs4793665 | / | TT | / | TT | ||||||||
| ABCC3 rs1051640 | ||||||||||||
| ACYP2 rs1872328 | ||||||||||||
| ATM rs664677 | TT | TT | TT | TT | TT | TT | CT | CT | CT | CT | CT | CT |
| ATM;C11orf65 rs664143 | GG | GG | GG | GG | GG | GG | GA | GA | GA | GA | GA | GA |
| ATR rs2229032 | ||||||||||||
| ATR rs2227928 | GA | GA | GA | GA | GA | GA | ||||||
| COMT rs4646316 | TT | TT | TT | TT | TT | TT | TT | TT | TT | TT | TT | TT |
| COMT rs9332377 | ||||||||||||
| ERCC1 rs11615 | GA | GA | GA | GA | GG | GG | ||||||
| ERCC1 rs3212986 | ||||||||||||
| ERCC2 rs13181 | GT | GT | TT | GT | GT | GT | GT | GT | GG | GG | ||
| ERCC2 rs1799793 | CT | CT | CT | CT | TT | TT | TT | TT | TT | TT | ||
| GSTP1 rs1695 | AG | AG | AG | AG | AG | AG | AG | AG | AG | AG | AG | AG |
| TP53 rs1042522 | CC | CC | CC | CC | CC | CC | ||||||
| TP53 rs1642785 | ||||||||||||
| TPMT rs12201199 | ||||||||||||
| TPMT rs1142345 | ||||||||||||
| TPMT rs1800460 | ||||||||||||
| XPA rs1800975 | CC | CC | CC | CC | CC | CC | CT | CT | CT | CT | CT | CT |
| XRCC3;KLC1 rs861539 | AG | AG | AG | AG | GG | AG | AA | AA | AA | AA | AA | AA |
| Cell Line | Identified Variants | |
|---|---|---|
| ERCC2 rs13181 | ERCC2 rs1799793 | |
| U-2OS | GT | CT |
| U-2OS/CDDP300 | GT | CT |
| U-2OS/CDDP1μg | GT (a)/TT (b) | CT |
| U-2OS/CDDP4μg | TT | CC |
| Amino acid change | ||
| U-2OS | Lys725Gln | Asp286Asn |
| U-2OS/CDDP300 | Lys725Gln | Asp286Asn |
| U-2OS/CDDP1μg | Lys725Gln | Asp286Asn |
| U-2OS/CDDP4μg | / | / |
| Cell Line | Identified Variants | |
|---|---|---|
| ERCC2 rs13181 | ERCC1 rs11615 | |
| Saos-2 | GT | GA |
| Saos-2/CDDP300 | GT | GA |
| Saos-2/CDDP1μg | GT | GA |
| Saos-2/CDDP6μg | GG | GG |
| Amino Acid Change | ||
| Saos-2 | Lys725Gln | Synonymous |
| Saos-2/CDDP300 | Lys725Gln | Synonymous |
| Saos-2/CDDP1μg | Lys725Gln | Synonymous |
| Saos-2/CDDP6μg | Lys725Gln | Synonymous |
| Cell Line | Identified Variants | |
|---|---|---|
| GSTP1 rs1695 | ||
| DNA | RNA | |
| U-2OS | AG | AA |
| U-2OS/CDDP300 | AG | AG |
| U-2OS/CDDP1μg | AG | GAT |
| U-2OS/CDDP4μg | AG | AG |
| Amino acid change | ||
| U-2OS | / | |
| U-2OS/CDDP300 | Ile105Val | |
| U-2OS/CDDP1μg | Ile105Val | |
| U-2OS/CDDP4μg | Ile105Val | |
| Cell Line | Identified Variants | |
|---|---|---|
| GSTP1 rs1695 | ||
| DNA | RNA | |
| Saos-2 | AG | AA |
| Saos-2/CDDP300 | AG | AG |
| Saos-2/CDDP1μg | AG | AG |
| Saos-2/CDDP6μg | AG | AG |
| Amino acid change | ||
| Saos-2 | / | |
| Saos-2/CDDP300 | Ile105Val | |
| Saos-2/CDDP1μg | Ile105Val | |
| Saos-2/CDDP6μg | Ile105Val | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hattinger, C.M.; Casotti, C.; Patrizio, M.P.; Luppi, S.; Fantoni, L.; Scotlandi, K.; Ibrahim, T.; Serra, M. Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing. Int. J. Mol. Sci. 2022, 23, 11787. https://doi.org/10.3390/ijms231911787
Hattinger CM, Casotti C, Patrizio MP, Luppi S, Fantoni L, Scotlandi K, Ibrahim T, Serra M. Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing. International Journal of Molecular Sciences. 2022; 23(19):11787. https://doi.org/10.3390/ijms231911787
Chicago/Turabian StyleHattinger, Claudia Maria, Chiara Casotti, Maria Pia Patrizio, Silvia Luppi, Leonardo Fantoni, Katia Scotlandi, Toni Ibrahim, and Massimo Serra. 2022. "Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing" International Journal of Molecular Sciences 23, no. 19: 11787. https://doi.org/10.3390/ijms231911787
APA StyleHattinger, C. M., Casotti, C., Patrizio, M. P., Luppi, S., Fantoni, L., Scotlandi, K., Ibrahim, T., & Serra, M. (2022). Pharmacogenomic Profiling of Cisplatin-Resistant and -Sensitive Human Osteosarcoma Cell Lines by Multimodal Targeted Next Generation Sequencing. International Journal of Molecular Sciences, 23(19), 11787. https://doi.org/10.3390/ijms231911787









