Acidovorax citrulli Effector AopV Suppresses Plant Immunity and Interacts with Aromatic Dehydratase ADT6 in Watermelon
Abstract
1. Introduction
2. Results
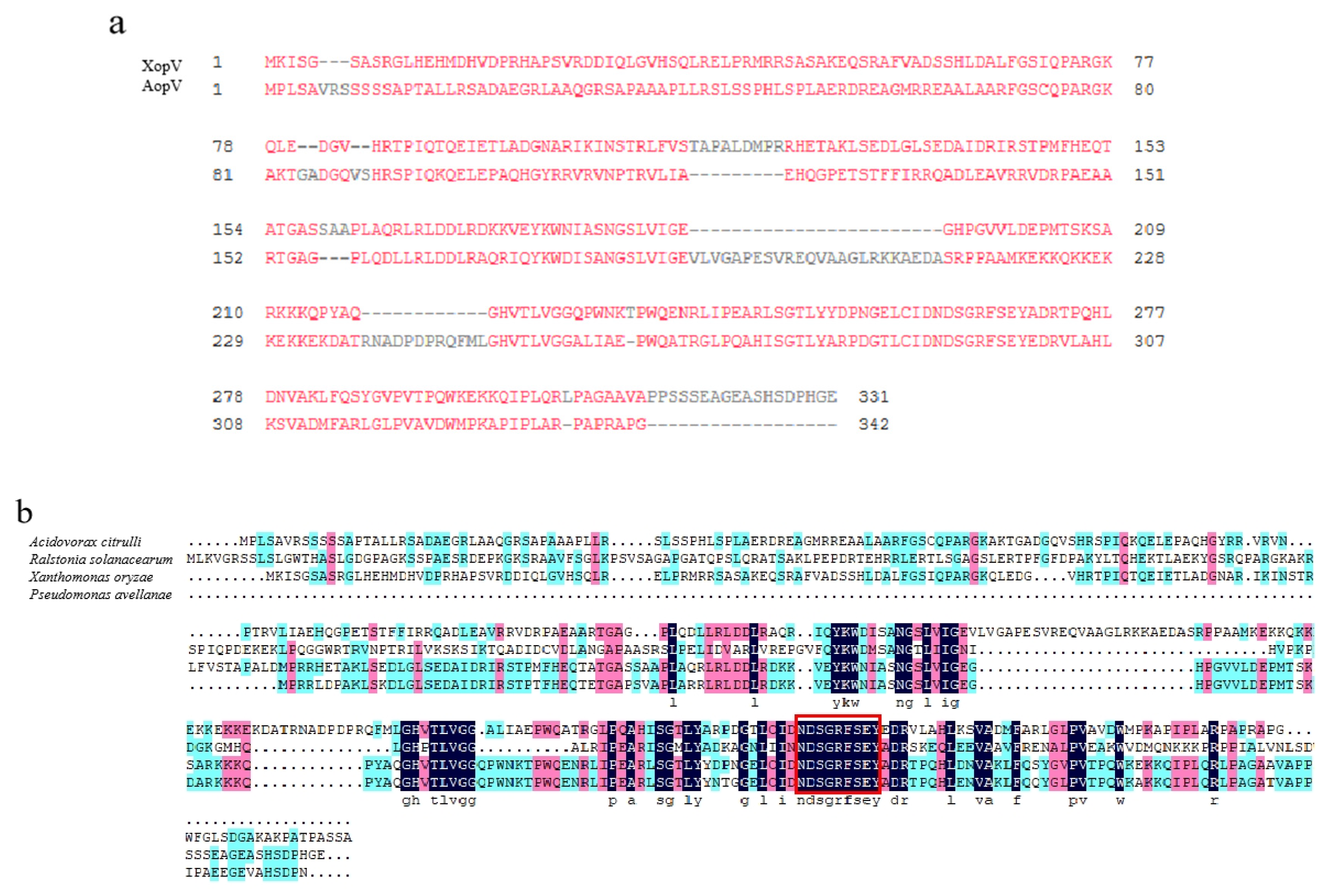
2.1. Sequence Analysis of AopV in A. Citrulli Strain Aac5
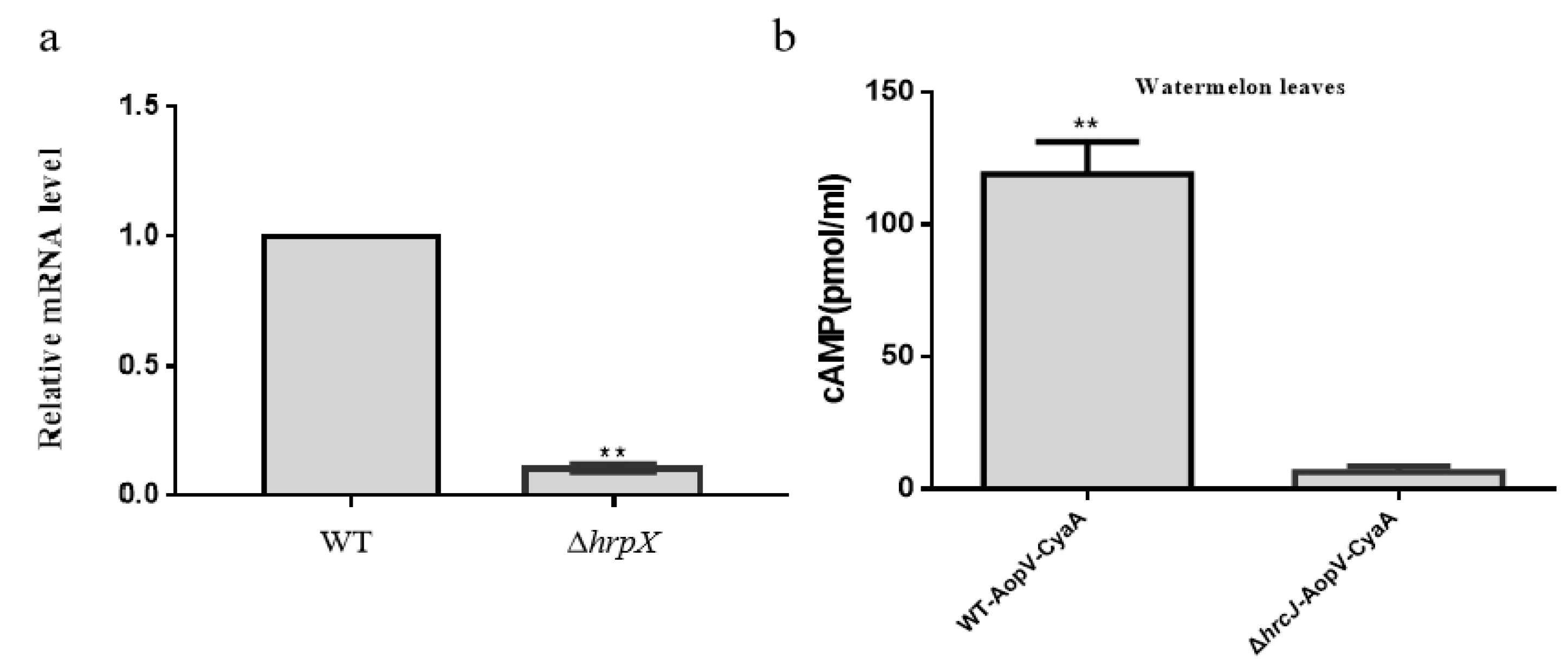
2.2. ApoV Is a Type III Secreted Effector in A. citrulli
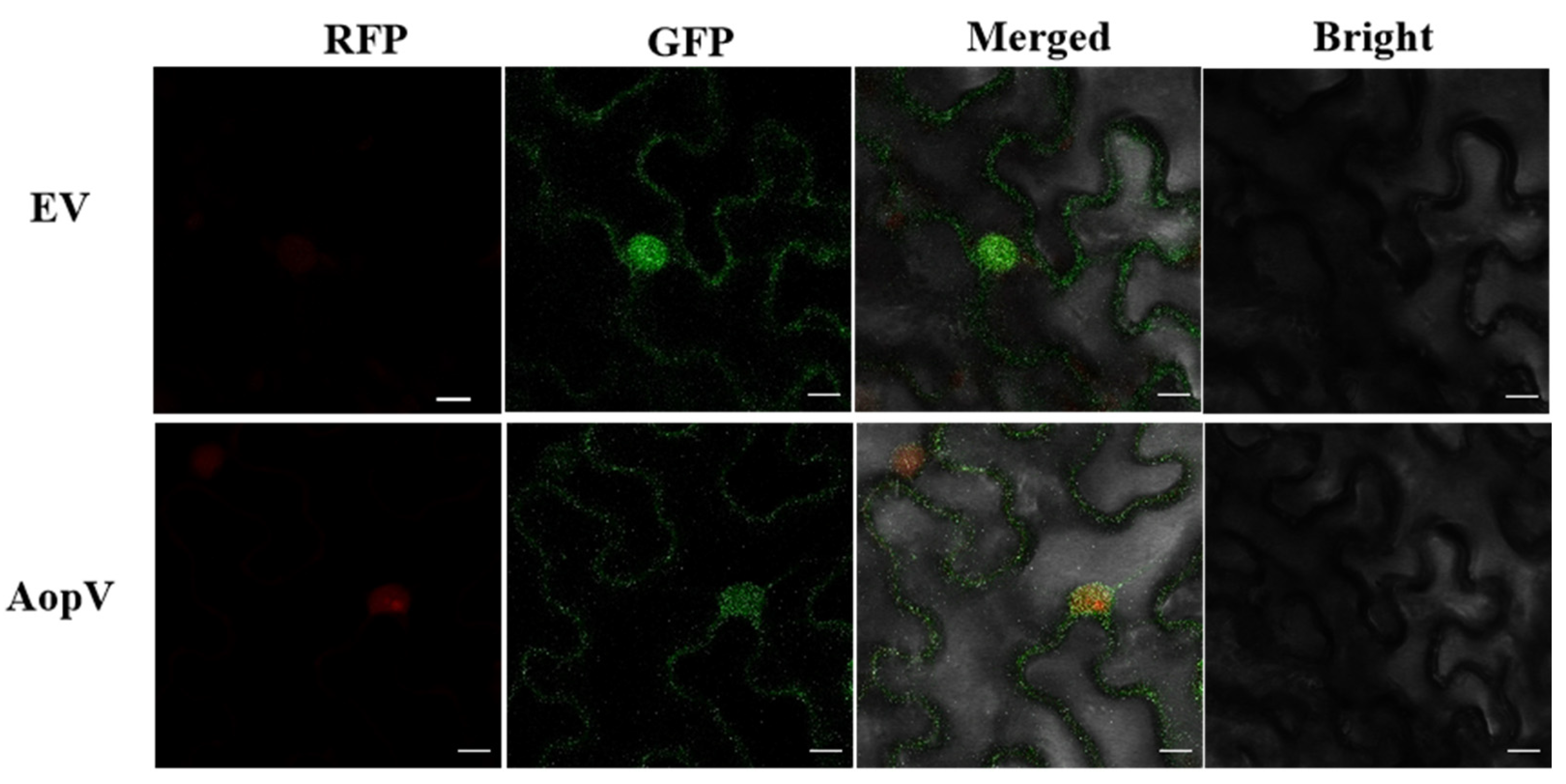
2.3. AopV Is Expressed in the Nucleus and Cell Membrane of N. benthamiana
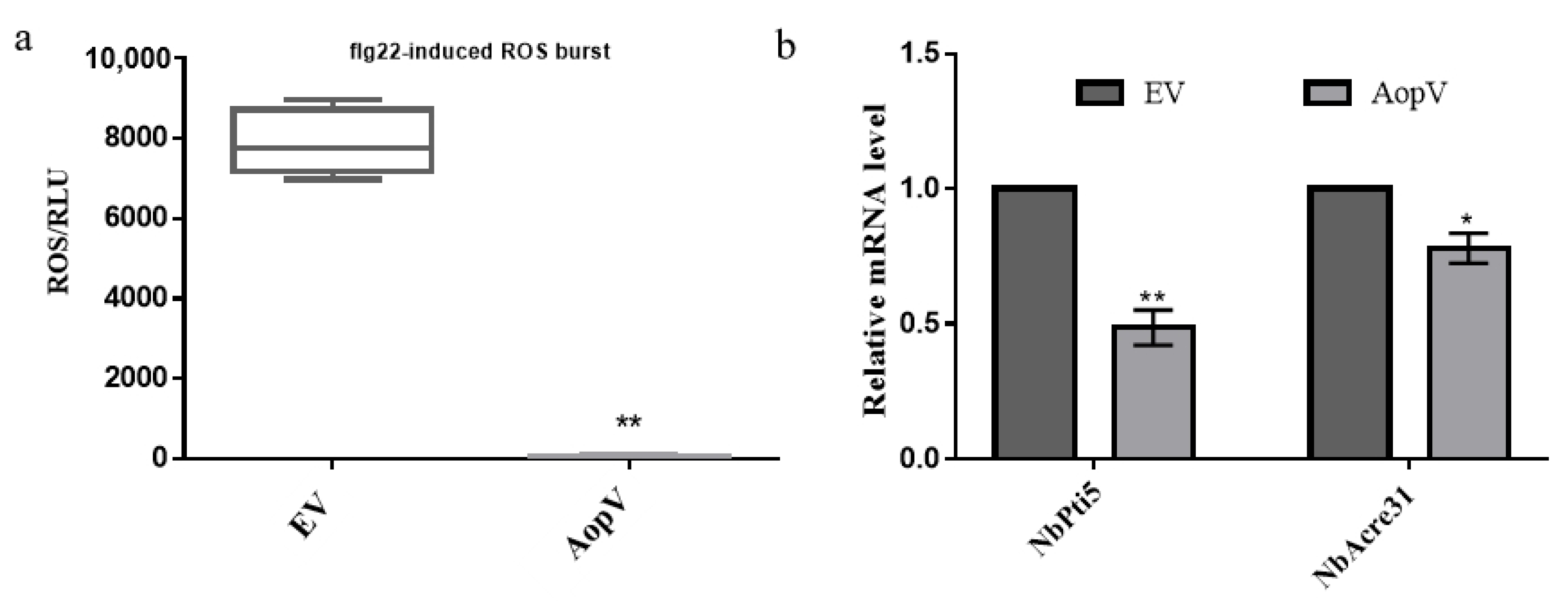
2.4. AopV Inhibits the ROS Burst and Expression Levels of PTI Marker Genes in N. Benthamiana
2.5. AopV Helps the Growth of D36E in N. benthamiana
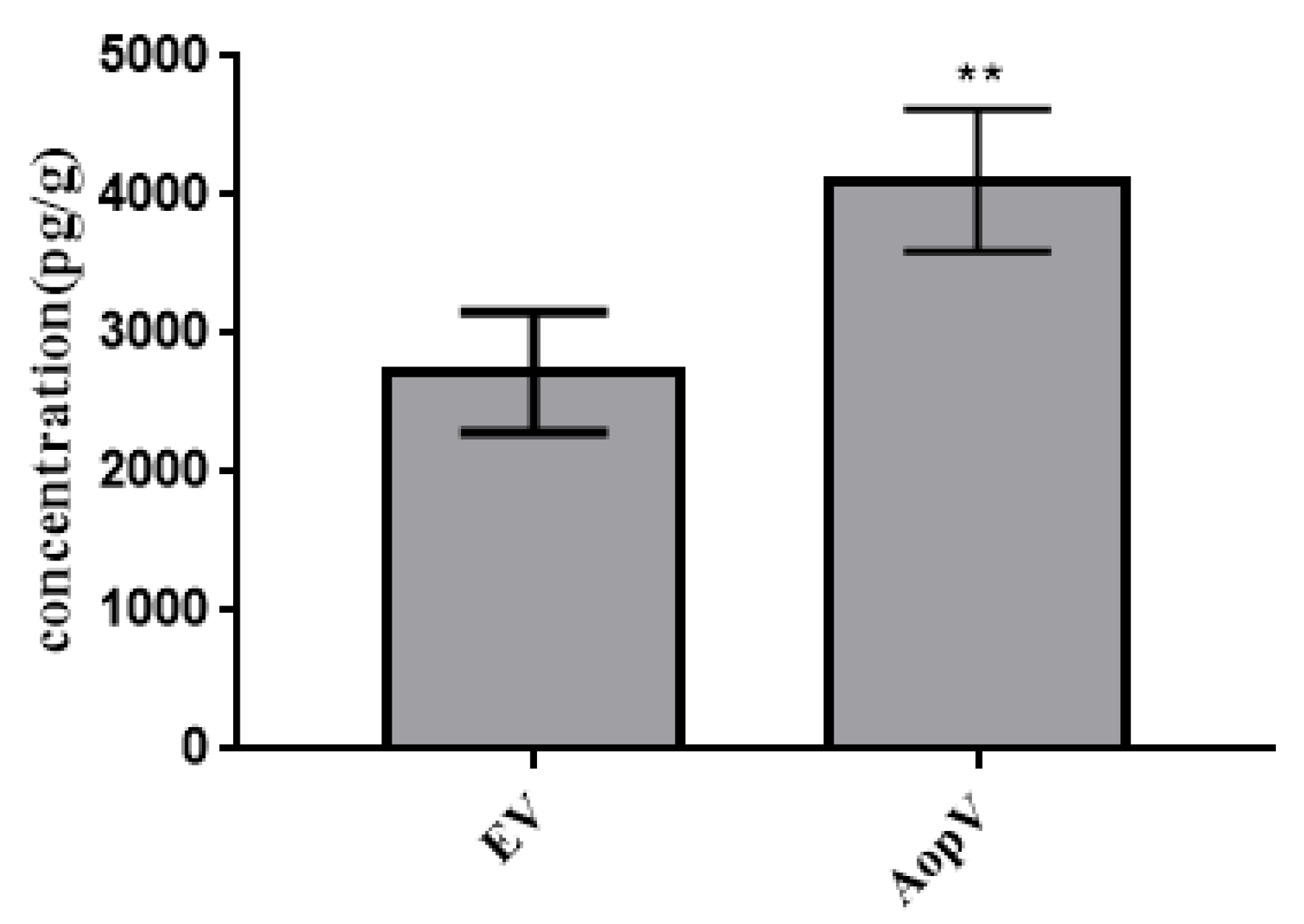
2.6. AopV Stimulates JA Production
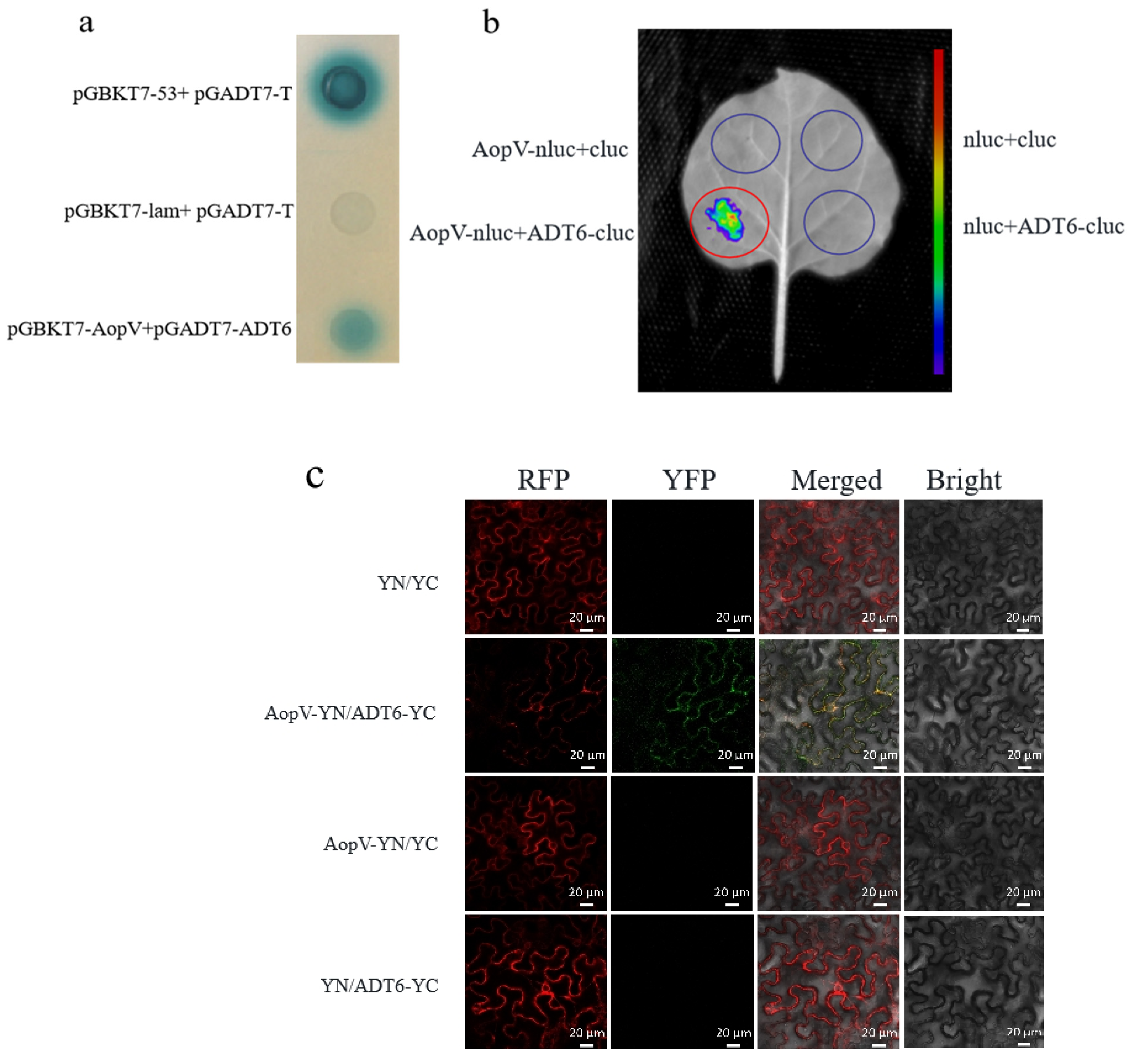
2.7. AopV Interacted with ADT6
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Plant Materials
4.2. AopV Sequence Analysis
4.3. CyaA Translocation Assay
4.4. Agrobacterium Infiltration
4.5. ROS Burst Measurement
4.6. RNA Extraction
4.7. JA Detection
4.8. Subcellular Localization
4.9. Impact of AopV on Pseudomonas Syringae pv. Tomato D36E Growth
4.10. Yeast Two-Hybrid Assay
4.11. Luciferase (LUC) Complementary Imaging (LCI) Assay
4.12. BiFC Assays
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jones, J.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef]
- Zipfel, C. Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 2009, 12, 414–420. [Google Scholar] [CrossRef]
- Lee, A.H.; Middleton, M.A.; Guttman, D.S.; Desveaux, D. Phytopathogen type III effectors as probes of biological systems. Microb. Biotechnol. 2013, 6, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Chisholm, S.T.; Coaker, G.; Day, B.; Staskawicz, B.J. Host-Microbe interactions: Shaping the evolution of the plant immune response. Cell 2006, 124, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Alfano, J.R. Plant targets for Pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011, 14, 39–46. [Google Scholar] [CrossRef]
- Zhang, J.; Shao, F.; Li, Y.; Cui, H.T.; Chen, L.J.; Li, H.T.; Zou, Y.; Long, C.Z.; Lan, L.F.; Chai, J.J.; et al. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 2007, 1, 175–185. [Google Scholar] [CrossRef]
- Ma, W.; Xu, X.; Cai, L.; Cao, Y.; Chen, G.A. Xanthomonas oryzae type III effector XopL causes cell death through mediating ferredoxin degradation in N. benthamiana. Phytopathol. Res. 2020, 2, 1–12. [Google Scholar] [CrossRef]
- Shan, L.B.; He, P.; Li, J.M.; Heese, A.; Peck, S.C.; Nürnberger, T.; Martin, G.B.; Sheen, J. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 2008, 4, 17–27. [Google Scholar] [CrossRef]
- Gupta, M.K.; Nathawat, R.; Sinha, D.; Haque, A.S.; Sankaranarayanan, R.; Sont, R.V. Mutations in the Predicted Active Site of Xanthomonas oryzae pv. oryzae XopQ Differentially Affect Virulence, Suppression of Host Innate Immunity, and Induction of the HR in a Nonhost Plant. Mol. Plant-Microbe Interact. 2015, 28, 195. [Google Scholar] [CrossRef]
- Schultink, A.; Qi, T.; Lee, A.; Steinbrenner, A.D.; Staskawicz, B. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. 2017, 92, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Nürnberger, T.; Joosten, M.H.A.J. Of PAMPs and effectors: The blurred PTI-ETI dichotomy. Plant Cell 2011, 23, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef]
- Klessig, D.F.; Durner, J.; Noad, R.; Navarre, D.A.; Wendehenne, D.; Kumar, D.; Zhou, J.M.; Shah, J.; Zhang, S.; Kachroo, P.; et al. Nitric oxide and salicylic acid signaling in plant defense. Proc. Natl. Acad. Sci. USA 2000, 97, 8849–8855. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.V.; Lee, H.I.; Creelman, R.A.; Davis, M.K.R. Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 2000, 12, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef]
- Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; Delaney, T.P.; Uknes, S. A central role of salicylic acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar]
- Chaturvedi, R.; Shah, J. Salicylic Acid in Plant Disease Resistance. Springer: Heidelberg, The Netherlands, 2007. [Google Scholar]
- Zhang, X.; Yang, Y.; Zhao, M.; Yang, L.; Jiang, J.; Walcott, R.; Yang, S.; Zhao, T. Acidovorax citrulli type III effector AopP suppresses plant immunity by targeting the watermelon transcription factor WRKY6. Front. Plant Sci. 2020, 11, 579218. [Google Scholar] [CrossRef]
- Hu, Z.J.; Shao, S.J.; Zheng, C.F.; Sun, Z.H.; Shi, J.Y.; Yu, J.Q.; Qi, Z.Y.; Shi, K. Induction of systemic resistance in tomato against Botrytis cinerea by N-decanoyl-homoserine lactone via jasmonic acid signaling. Planta 2018, 247, 1217–1227. [Google Scholar] [CrossRef]
- Jiang, S.S.; Yao, J.; Ma, K.Y.; Zhou, H.B.; Song, J.K. Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 2013, 9, e1003715. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Teixeira, P.J.P.L.; Biswas, S.; Finkel, O.M.; He, Y.J.; Salas-González, I.; English, M.E.; Epple, P.; Mieczkowski, P.; Dangl, J. Pseudomonas syringae type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 2017, 21, 156–168. [Google Scholar] [CrossRef]
- Burdman, S.; Walcott, R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, T.; Jiménez-Guerrero, I.; Tamir-Ariel, D.; Yarnitzky, T.; Burdman, S. The GDSL-lipolytic enzyme Lip1 is required for full virulence of the cucurbit pathogenic bacterium Acidovorax citrulli. Microorganisms 2022, 5, 1016. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Minsavage, G.V.; Le, T.; Jones, J.B.; Walcott, R.R. Efficacy of a nonpathogenic Acidovorax citrulli strain as a biocontrol seed treatment for bacterial fruit blotch of cucurbits. Plant Dis. 2011, 95, 697–704. [Google Scholar] [CrossRef]
- Bahar, O.; Goffer, T.; Burdman, S. Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol. Plant-Microbe Interact. 2009, 22, 909–920. [Google Scholar] [CrossRef]
- Fei, N.Y.; Ji, W.Q.; Yang, L.L.; Yu, C.Y.; Qiao, P.; Yan, J.P.; Guan, W.; Yang, Y.W.; Zhao, T.C. Hcp of the type VI secretion system (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int. J. Mol. Sci. 2022, 23, 9632. [Google Scholar] [CrossRef]
- Ji, W.Q.; Zhao, M.; Fei, N.Y.; Yang, L.L.; Qiao, P.; Walcott, R.; Yang, Y.Y.; Zhao, T.C. Essential Acidovorax citrulli virulence gene hrpE activates host immune response against pathogen. Int. J. Mol. Sci. 2022, 23, 9144. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, M.; Yan, J.; Yang, L.; Yang, Y.; Guan, W.; Walcott, R.; Zhao, T. Involvement of hrpX and hrpG in the virulence of Acidovorax citrulli strain Aac5, Causal Agent of Bacterial Fruit Blotch in Cucurbits. Front. Microbiol. 2018, 9, 507. [Google Scholar] [CrossRef]
- Traore, S.M.; Eckshtain-Levi, N.; Miao, J.; Sparks, C.; Wang, Z.; Wang, K.; Li, Q.; Burdman, S.; Walcott, R.; Welbaum, G.E. Nicotiana species as surrogate host for studying the pathogenicity of Acidovorax citrulli, the causal agent of bacterial fruit blotch of cucurbits. Mol. Plant Pathol. 2019, 20, 800–814. [Google Scholar] [CrossRef]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Da Silva, G.M.; Wagner, N.; Shkedy, D.; Zhao, M.; Pizarro, L.; Bar, M.; Walcott, R.; Sessa, G.; et al. Show me your secret(ed) weapons: A multifaceted approach reveals a wide arsenal of type III-secreted effectors in the cucurbit pathogenic bacterium Acidovorax citrulli and novel effectors in the Acidovorax genus. Mol. Plant Pathol. 2020, 21, 12877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, M.; Jiang, J.; Yang, L.; Yang, Y.W.; Yang, S.S.; Walcott, R.; Qiu, D.W.; Zhao, T. Identification and functional analysis of AopN, an Acidovorax citrulli effector that induces programmed cell death in plants. Int. J. Mol. Sci. 2020, 21, 6050. [Google Scholar] [CrossRef] [PubMed]
- Turne, J.G.; Ellis, C.; Devoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, S153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, F.; Melotto, M.; Yao, J.; He, S.Y. Jasmonate signaling and manipulation by pathogens and insects. J. Exp. Bot. 2017, 68, 1371–1385. [Google Scholar] [CrossRef] [PubMed]
- Corea, O.R.A. Arabidopsis Arogenate Dehydratases: Influence on Aromatic Amino Acid and Downstream Phenylpropanoid Biosynthesis. Ph.D. Thesis, Gradworks. Washington State University, Pullman, WA, USA, 2011. Available online: http://hdl.handle.net/2376/3526 (accessed on 25 September 2022).
- Waadt, R.; Schmidt, L.K.; Lohse, M.; Hashimoto, K.; Bock, R.; Kudla, J. Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008, 56, 505–516. [Google Scholar] [CrossRef]
- Jwa, N.S.; Hwang, B.K. Convergent evolution of pathogen effectors toward reactive oxygen species signaling networks in plants. Front. Plant Sci. 2017, 9, 1687. [Google Scholar] [CrossRef]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef]
- Wei, H.L.; Zhang, W.; Collmer, A. Modular study of the type III effector repertoire in Pseudomonas syringae pv. tomato DC3000 reveals a matrix of effector interplay in pathogenesis. Cell Rep. 2018, 23, 1630–1638. [Google Scholar] [CrossRef]
- Deb, S.; Ghosh, P.; Patel, H.K.; Sonti, R.V. Interaction of the Xanthomonas effectors XopQ and XopX results in induction of rice immune responses. Plant J. 2020, 104, 332–350. [Google Scholar] [CrossRef]
- Jeon, H.; Kim, W.; Kim, B.; Lee, S.; Segonzac, C. Ralstonia solanacearum type III effectors with predicted nuclear localization signal localize to various cell compartments and modulate immune responses in Nicotiana spp. Plant Pathol. J. 2020, 36, 43–53. [Google Scholar] [CrossRef]
- Martinez-Argudo, I.; Blocker, A.J. The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol. Microbiol. 2010, 78, 1365–1378. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Song, C.; Fang, Y.; Zhou, J.; Bing, Y. Non-TAL effectors from Xanthomonas oryzae pv. oryzae suppress peptidoglycan-triggered MAPK activation in rice. Front. Plant Sci. 2018, 9, 1857. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.N.; Chen, L.; Si, N.G.; Jiang, W.J.; Zhou, Z.G.; Liu, J.L.; Zhang, L.Q. Identification of benzyloxy carbonimidoyl dicyanide derivatives as novel type III secretion system inhibitors via high-throughput screening. Front. Plant Sci. 2019, 10, 1059. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Y.; Wang, C.K.; Soong, S.C.; To, K.Y. Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breed. 2003, 11, 287–293. [Google Scholar] [CrossRef]
- Priller, J.P.; Reid, S.; Konein, P.; Dietrich, P.; Sonnewald, S. The Xanthomonas campestris pv. vesicatoria type-3 effector XopB inhibits plant defence responses by interfering with ROS production. PLoS ONE 2016, 11, e0159107. [Google Scholar] [CrossRef]
- Wang, S.; Xu, L.; Li, G.; Chen, P.; Kai, X.; Xie, Z. An ELISA for the determination of salicylic acid in plants using a monoclonal antibody. Plant Sci. 2002, 162, 529–535. [Google Scholar] [CrossRef]
- Nelson, B.K.; Cai, X.; Andreas, N. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2010, 51, 1126–1136. [Google Scholar] [CrossRef]
- Yang, F.; Tian, F.; Li, X.; Fan, S.; Chen, H.; Wu, M.; Yang, C.H.; He, C. The degenerate EAL-GGDEF domain protein filp functions as a cyclic di-GMP receptor and specifically interacts with the PilZ-domain protein PXO_02715 to regulate virulence in Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 2014, 27, 578. [Google Scholar] [CrossRef][Green Version]
- Chen, H.M.; Zou, Y.; Shang, Y.L.; Lin, H.Q.; Wang, Y.J.; Cai, R.; Tang, X.Y.; Zhou, J.M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008, 146, 368–376. [Google Scholar] [CrossRef]
- Yang, S.; Pan, L.; Chen, Y.; Yang, D.; Liu, Q.; Jian, H. Heterodera avenae GLAND5 effector interacts with pyruvate dehydrogenase subunit of plant to promote nematode parasitism. Front. Plant Sci. 2019, 10, 1241. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, J.; Zhao, M.; Zhang, X.; Yang, L.; Fei, N.; Ji, W.; Guan, W.; Walcott, R.; Yang, Y.; Zhao, T. Acidovorax citrulli Effector AopV Suppresses Plant Immunity and Interacts with Aromatic Dehydratase ADT6 in Watermelon. Int. J. Mol. Sci. 2022, 23, 11719. https://doi.org/10.3390/ijms231911719
Jiang J, Zhao M, Zhang X, Yang L, Fei N, Ji W, Guan W, Walcott R, Yang Y, Zhao T. Acidovorax citrulli Effector AopV Suppresses Plant Immunity and Interacts with Aromatic Dehydratase ADT6 in Watermelon. International Journal of Molecular Sciences. 2022; 23(19):11719. https://doi.org/10.3390/ijms231911719
Chicago/Turabian StyleJiang, Jie, Mei Zhao, Xiaoxiao Zhang, Linlin Yang, Nuoya Fei, Weiqin Ji, Wei Guan, Ron Walcott, Yuwen Yang, and Tingchang Zhao. 2022. "Acidovorax citrulli Effector AopV Suppresses Plant Immunity and Interacts with Aromatic Dehydratase ADT6 in Watermelon" International Journal of Molecular Sciences 23, no. 19: 11719. https://doi.org/10.3390/ijms231911719
APA StyleJiang, J., Zhao, M., Zhang, X., Yang, L., Fei, N., Ji, W., Guan, W., Walcott, R., Yang, Y., & Zhao, T. (2022). Acidovorax citrulli Effector AopV Suppresses Plant Immunity and Interacts with Aromatic Dehydratase ADT6 in Watermelon. International Journal of Molecular Sciences, 23(19), 11719. https://doi.org/10.3390/ijms231911719





