Targeted Sequencing of Cytokine-Induced PI3K-Related Genes in Ulcerative Colitis, Colorectal Cancer and Colitis-Associated Cancer
Abstract
1. Introduction
2. Results
2.1. Information on Clinical Samples
2.2. The Technical Performance of the Target Enrichment System Panel
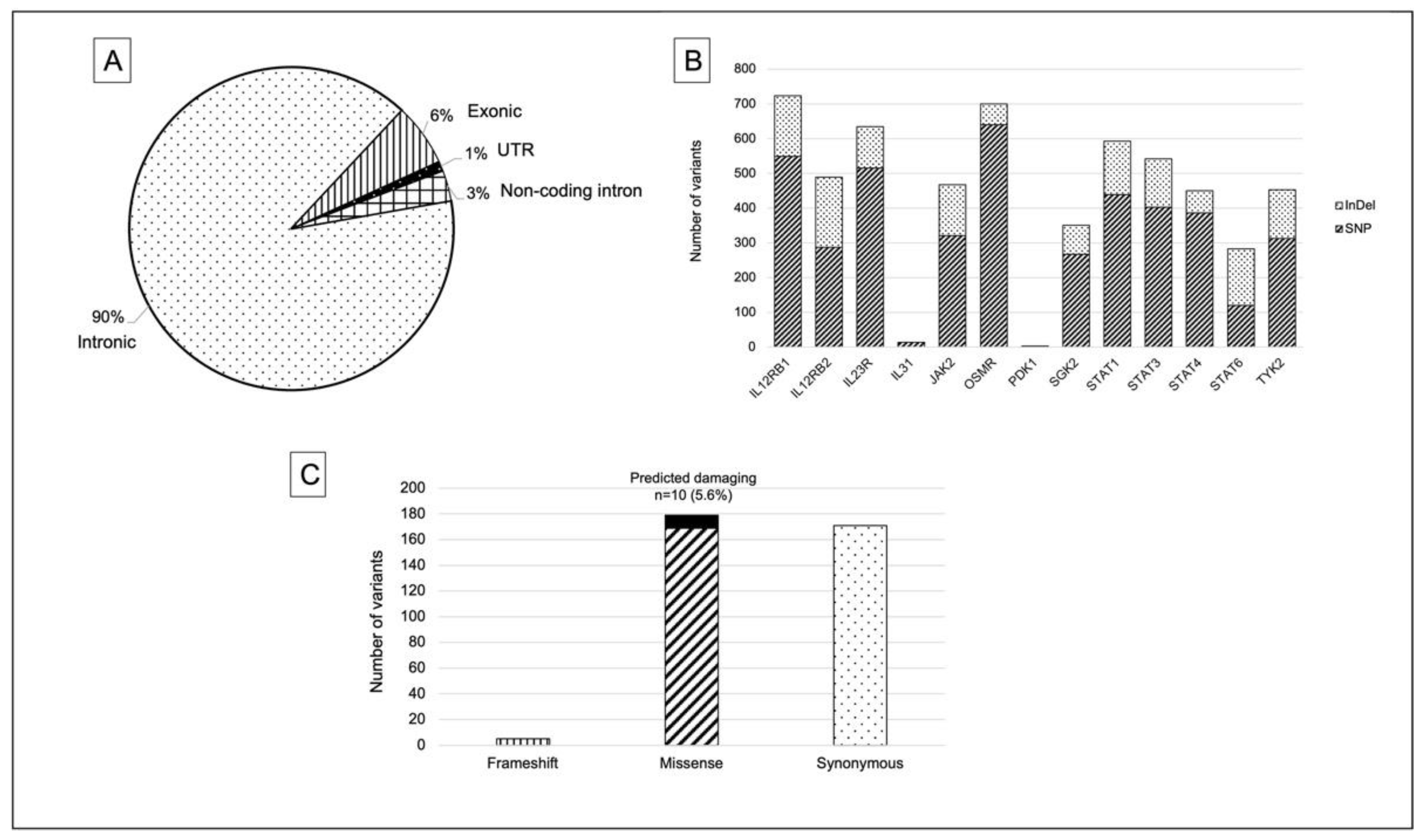
2.3. Summary of Identified Variants in PI3K-Related Genes
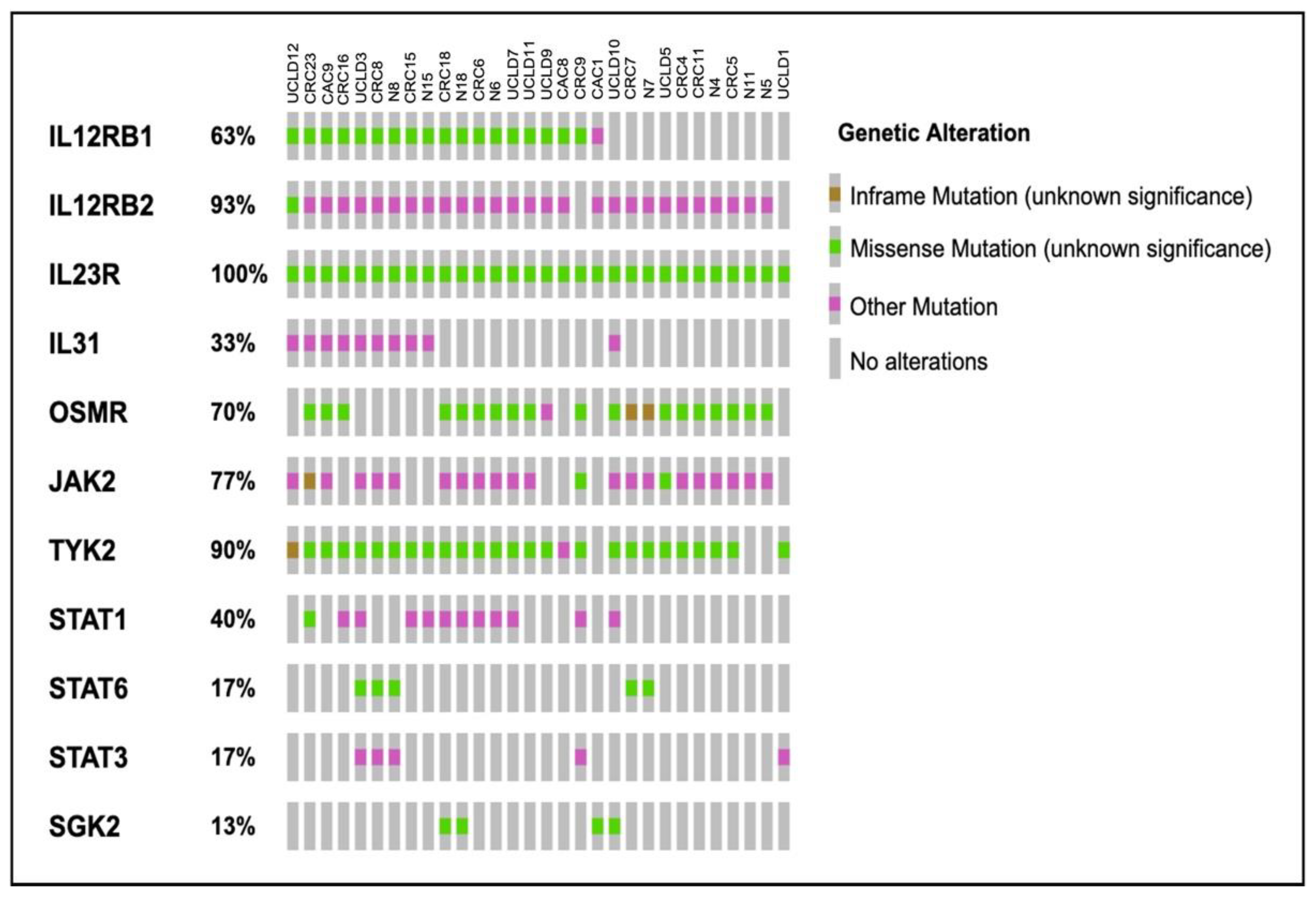
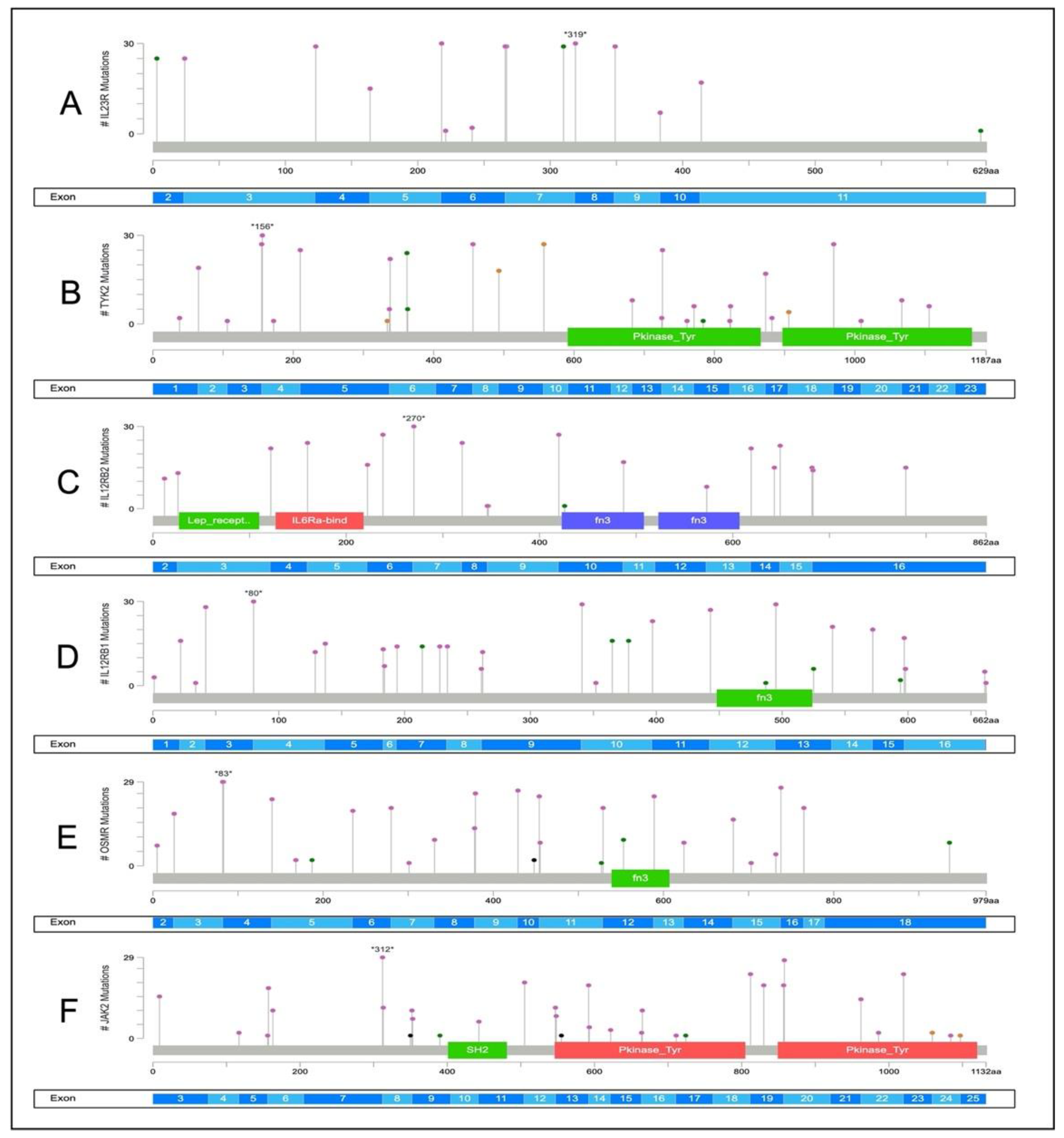
2.4. Somatic Variants Distribution in PI3K-Related Genes among All Samples
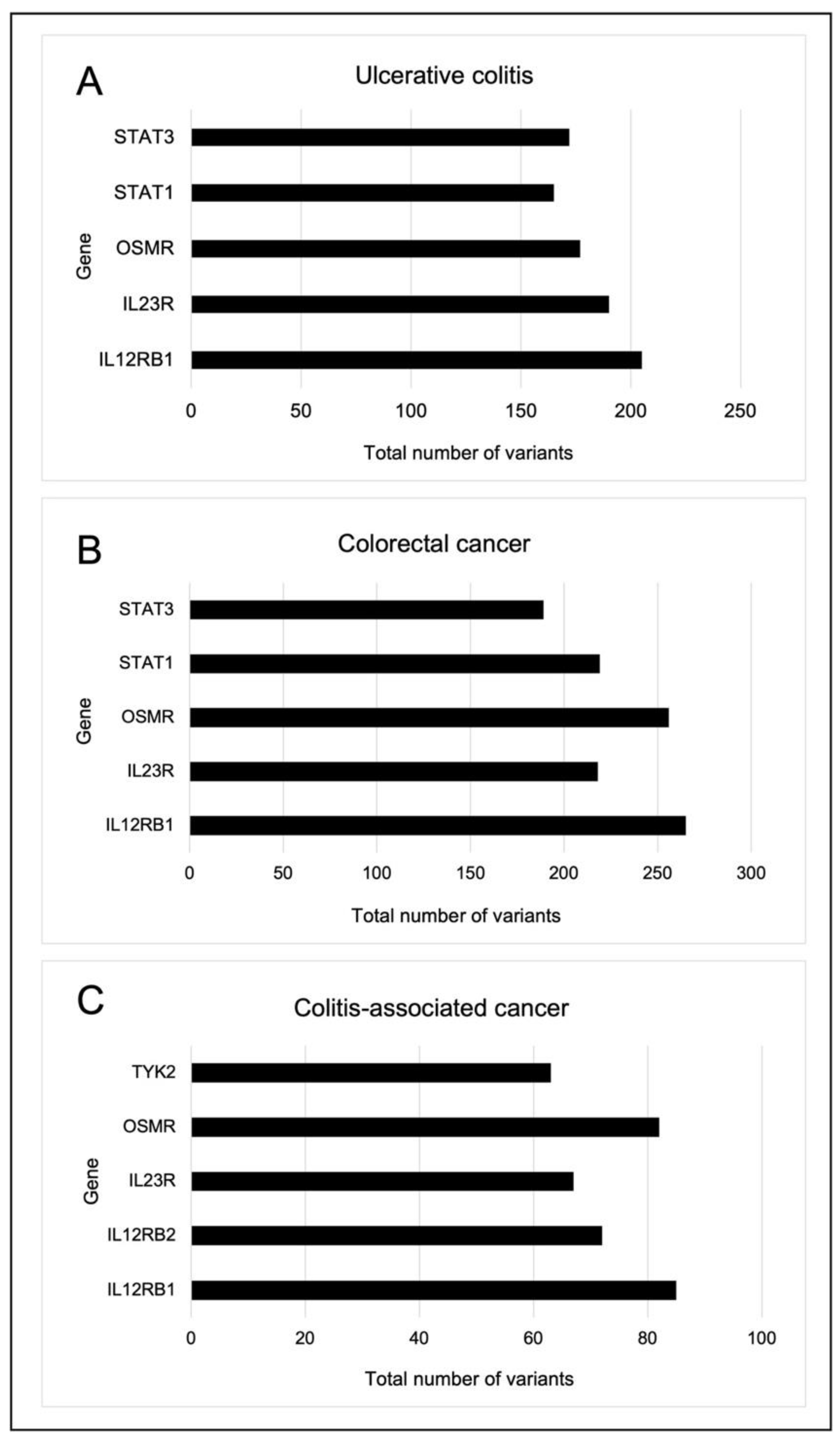
2.5. Somatic Variants Distribution of PI3K-Related Genes per Group
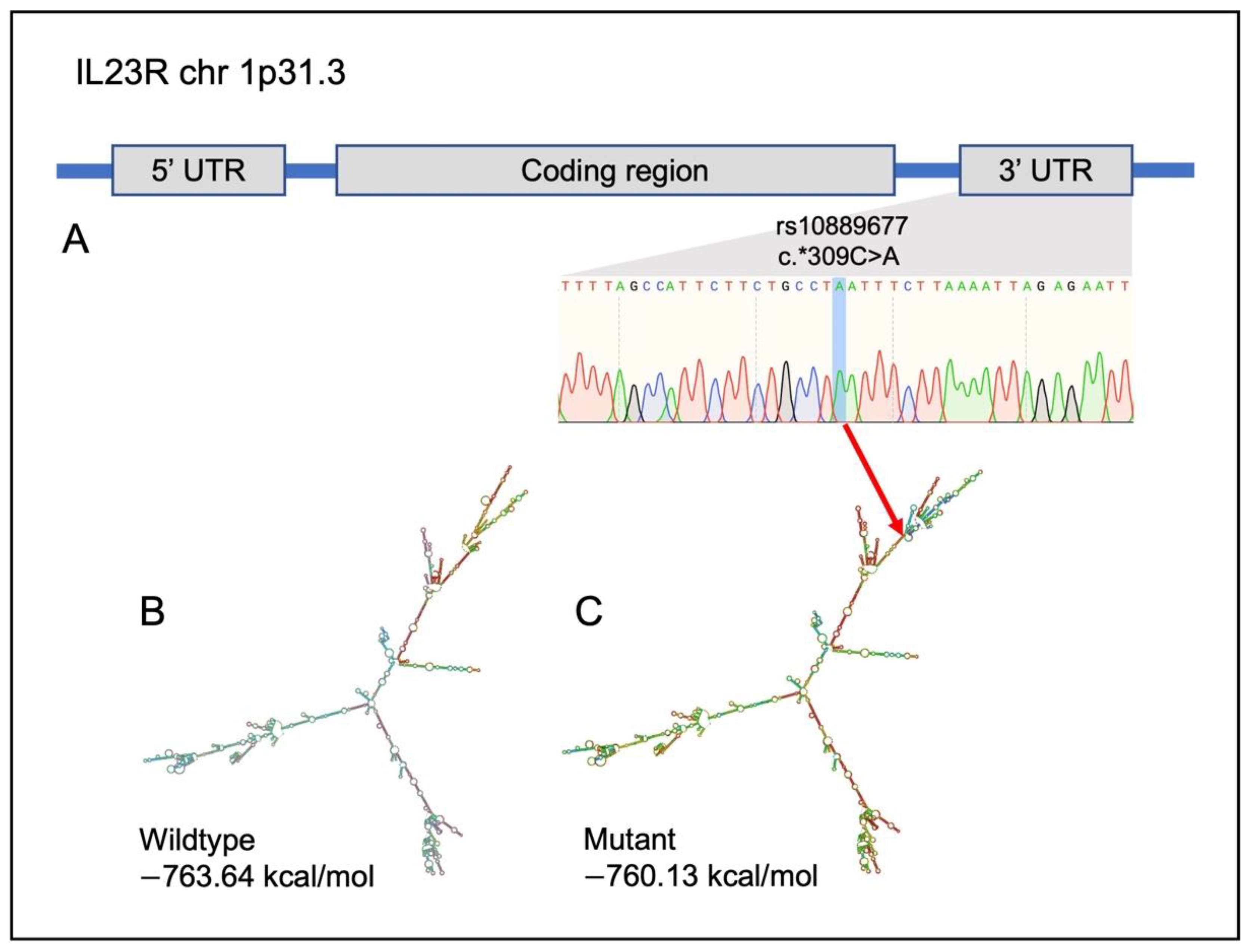
2.6. Identification of Potential Variants That Link UC, CRC and CAC
3. Discussion
4. Materials and Methods
4.1. Sample Collection
4.2. Nucleic Acid Extraction and Quality Assessment
4.3. Targeted Sequencing
4.4. Sequence Alignment and Variant Annotation
4.5. Validation of Genomic Variants
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grivennikov, S.I.; Cominelli, F. Colitis-Associated and Sporadic Colon Cancers: Different Diseases, Different Mutations? Gastroenterology 2016, 150, 808–810. [Google Scholar] [CrossRef]
- Liverani, E.; Scaioli, E.; John Digby, R.; Bellanova, M.; Belluzi, A. How to predict clinical relapse in inflammatory bowel disease patients. World J. Gastroenterol. 2016, 22, 1017–1033. [Google Scholar] [CrossRef] [PubMed]
- Laukoetter, M.G.; Mennigen, R.; Hannig, C.M.; Osada, N.; Rijcken, E.; Vowinkel, T.; Krieglstein, C.F.; Senniger, N.; Anthoni, C.; Bruewer, M. Intestinal Cancer Risk in Crohn’s Disease: A Meta-Analysis. J. Gastrointest. Surg. 2011, 15, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Shen, Z.F.; Wu, B.S.; Xu, C.B.; He, Z.Q.; Chen, T.; Shang, H.T.; Xie, C.F.; Huang, S.Y.; Chen, Y.G.; et al. Risk of Colorectal Cancer in Ulcerative Colitis Patients: A Systematic Review and Meta-Analysis. Gastroenterol. Res. Pract. 2019, 2019, 5363261. [Google Scholar] [CrossRef] [PubMed]
- Zhiqin, W.; Palaniappan, S.; Raja Ali, R.A. Inflammatory Bowel Disease-related Colorectal Cancer in the Asia-Pacific Region: Past, Present, and Future. Intest. Res. 2014, 12, 194. [Google Scholar] [CrossRef]
- Mokhtar, N.M.; Nawawi, K.N.M.; Verasingam, J.; Zhiqin, W.; Sagap, I.; Azman, Z.A.M.; Mazlan, L.; Hamid, H.A.; Yaacob, N.Y.; Rose, I.M.; et al. A four-decade analysis of the incidence trends, sociodemographic and clinical characteristics of inflammatory bowel disease patients at single tertiary centre, Kuala Lumpur, Malaysia. BMC Public Health 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Ananthakrishnan, A.N.; Bernstein, C.N.; Iliopoulos, D.; Macpherson, A.; Neurath, M.F.; Raja Ali, R.A.; Vavricka, S.R.; Fiocchi, C. Environmental triggers in IBD: A review of progress and evidence. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 39–49. [Google Scholar] [CrossRef]
- Kameyama, H.; Nagahashi, M.; Shimada, Y.; Tajima, Y.; Ichikawa, H.; Nakano, M.; Sakata, J.; Kobayashi, T.; Narayanan, S.; Takabe, K.; et al. Genomic characterization of colitis-associated colorectal cancer. World J. Surg. Oncol. 2018, 16, 4–9. [Google Scholar] [CrossRef]
- Watanabe, T.; Konishi, T.; Kishimoto, J.; Kotake, K.; Muto, T.; Sugihara, K. Ulcerative Colitis-associated Colorectal Cancer Shows a Poorer Survival than sporadic colorectal cancer: A Nationwide Japanese Study. Inflamm. Bowel Dis. 2011, 17, 1–7. [Google Scholar] [CrossRef]
- Hartnett, L.; Egan, L.J. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis 2012, 33, 723–731. [Google Scholar] [CrossRef]
- Choi, C.R.; Ignjatovic-Wilson, A.; Askari, A.; Lee, G.H.; Warusavitarne, J.; Moorghen, M.; Thomas-Gibson, S.; Saunders, B.P.; Rutter, M.D.; Graham, T.A.; et al. Low-Grade Dysplasia in Ulcerative Colitis: Risk Factors for Developing High-Grade Dysplasia or Colorectal Cancer. Am. J. Gastroenterol. 2015, 110, 1461–1471. [Google Scholar] [CrossRef] [PubMed]
- Ullman, T.A.; Itzkowitz, S.H. Intestinal inflammation and cancer. Gastroenterology 2011, 140, 1807–1816.e1. [Google Scholar] [CrossRef] [PubMed]
- Claessen, M.M.H.; Schipper, M.E.I.; Oldenburg, B.; Siersema, P.D.; Offerhaus, G.J.A.; Vleggaar, F.P. WNT-pathway activation in IBD-associated colorectal carcinogenesis: Potential biomarkers for colonic surveillance. Cell Oncol. 2010, 32, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Axelrad, J.E.; Lichtiger, S.; Yajnik, V. Inflammatory Bowel Disease: Global view Inflammatory bowel disease and cancer: The role of inflammation, immunosuppression, and cancer treatment. World J. Gastroenterol. 2016, 22, 4794–4801. [Google Scholar] [CrossRef]
- Shrihari, T.G. Dual role of inflammatory mediators in cancer. Ecancermedicalscience 2017, 11, 1–9. [Google Scholar] [CrossRef]
- Galdiero, M.R.; Marone, G.; Mantovani, A. Cancer inflammation and cytokines. Cold Spring Harb. Perspect. Biol. 2018, 10, a028662. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Cominello, F. IL-6 and Stat3 Are Required for Survival of Intestinal Epithelial Cells and Development of Colitis-Associated Cancer. Cancer Cell 2009, 15, 103–113. [Google Scholar] [CrossRef]
- Qu, X.; Tang, Y.; Hua, S. Immunological approaches towards cancer and inflammation: A cross talk. Front. Immunol. 2018, 9, 699. [Google Scholar] [CrossRef]
- Ciraolo, E.; Gulluni, F.; Hirsch, E. Methods to measure the enzymatic activity of PI3Ks. Methods Enzym. 2014, 543, 115–140. [Google Scholar]
- Abdul, S.N.; Ab Mutalib, N.S.; Sean, K.S.; Syafruddin, S.E.; Ishak, M.; Sagap, I.; Mazlan, L.; Rose, I.M.; Abu, N.; Mokhtar, N.M.; et al. Molecular characterization of somatic alterations in Dukes’ B and C colorectal cancers by targeted sequencing. Front. Pharm. 2017, 8, 465. [Google Scholar] [CrossRef]
- Low, E.N.D.; Mokhtar, N.M.; Wong, Z.; Raja Ali, R.A. Colonic mucosal transcriptomic changes in patients with long-duration ulcerative colitis revealed colitis-associated cancer pathways. J. Crohns Colitis 2017, 13, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal Complementary Data Sources and Analysis Options. Sci. Signal. 2014, 6, 1–20. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Du, L.; Kim, J.J.; Shen, J.; Chen, B.; Dai, N. KRAS and TP53 mutations in inflammatory bowel disease associated colorectal cancer: A meta-analysis. Oncotarget 2017, 8, 22175–22186. [Google Scholar] [CrossRef]
- Ong, J.; Giuffre, A.; Joshi, S.; Ravi, H.; Pabon-Pena, C.; Novak, B.; Visitacion, M.; Hamady, M.; Useche, F.; Arezi, B.; et al. Overview of the Agilent Technologies SureSelectTM Target Enrichment System. J. Mol. Tech. 2011, 22, S30–S31. [Google Scholar]
- Duerr, R.H.; Taylor, K.D.; Brant, S.R.; Rioux, J.D.; Silverberg, M.S.; Daly, M.J.; Steinhart, A.H.; Abraham, C.; Regueiro, M.; Griffiths, A.; et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006, 314, 1461–1463. [Google Scholar] [CrossRef]
- Sivanesan, D.; Beauchamp, C.; Quinou, C.; Lee, J.; Lesage, S.; Chemtob, S.; Rioux, J.D.; Michnick, S.W. IL23R (Interleukin 23 Receptor) variants protective against inflammatory bowel diseases (IBD) display loss of function due to impaired protein stability and intracellular trafficking. J. Biol. Chem. 2016, 291, 8673–8685. [Google Scholar] [CrossRef]
- Zhu, Y.; Jiang, H.G.; Chen, Z.H.; Lu, B.H.; Li, J.; Shen, X.N. Genetic association between IL23R rs11209026 and rs10889677 polymorphisms and risk of Crohn’s disease and ulcerative colitis: Evidence from 41 studies. Inflamm. Res. 2020, 69, 87–103. [Google Scholar] [CrossRef]
- Poole, E.M.; Curtin, K.; Hsu, L.; Duggan, D.J.; Makar, K.W.; Xiao, L.; Carlson, C.S.; Caan, B.J.; Potter, J.D.; Slattery, M.L.; et al. Genetic variability in IL23R and risk of colorectal adenoma and colorectal cancer. Cancer Epidemiol. 2012, 36, e104–e110. [Google Scholar] [CrossRef][Green Version]
- Park, H.J.; Kim, S.K.; Park, H.K.; Chung, J.H. Association of IL23R polymorphism (rs7530511) with intracerebral hemorrhage in Korean population. Neurol. Sci. 2016, 37, 983–985. [Google Scholar] [CrossRef] [PubMed]
- El-Gedamy, M.; El-khayat, Z.; Abol-Enein, H.; El-said, A. Rs-1884444 G/T variant in IL-23 receptor is likely to modify risk of bladder urothelial carcinoma by regulating IL-23/IL-17 inflammatory pathway. Cytokine 2021, 138, 155355. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yue, C.; Jin, G.; Guo, H.; Ma, H.; Wang, G.; Huangm, S.; Wu, F.; Zhao, X. Rs1884444 variant in IL23R gene is associated with a decreased risk in esophageal cancer in Chinese population. Mol. Carcinog. 2019, 58, 1822–1831. [Google Scholar] [CrossRef]
- Peng, L.L.; Wang, Y.; Zhu, F.L.; Xu, W.D.; Ji, X.L.; Ni, J. IL-23R mutation is associated with ulcerative colitis: A systemic review and meta-analysis. Oncotarget 2017, 8, 4849–4863. [Google Scholar] [CrossRef] [PubMed]
- Mosallaei, M.; Simonian, M.; Esmaeilzadeh, E.; Bagheri, H.; Miraghajani, M.; Salehi, A.R.; Mehrzad, V.; Salehi, R. Single nucleotide polymorphism rs10889677 in miRNAs Let-7e and Let-7f binding site of IL23R gene is a strong colorectal cancer determinant: Report and meta-analysis. Cancer Genet. 2019, 239, 46–53. [Google Scholar] [CrossRef]
- Cheng, X.; Taranath, R.; Mattheakis, L.; Bhandari, A.; Liu, D. The biomarker profile of PTG-200, an oral peptide antagonist of IL-23 receptor, tracks with efficacy in a preclinical model of IBD. J. Crohn’s Colitis 2017, 11, S502–S540. [Google Scholar] [CrossRef]
- Cheng, X.; Lee, T.Y.; Ledet, G.; Zemade, G.; Tovera, M.; Campbell, R.; Purro, N.; Annamali, T.; Masjedizadeh, M.; Liu, D.; et al. Safety, Tolerability, and Pharmacokinetics of PTG-200, an Oral GI-Restricted Peptide Antagonist of IL-23 Receptor, in Normal Healthy Volunteers. Am. J. Gastroenterol. 2019, 114, S439–S440. [Google Scholar] [CrossRef]
- van de Vosse, E.; Haverkamp, M.H.; Ramirez-Alejo, N.; Martinez-Gallo, M.; Blancas-Galicia, L.; Metin, A.; Garty, B.Z.; Sun-Tan, C.; Broides, A.; de Paus, R.A.; et al. IL-12Rβ1 deficiency: Mutation update and description of the IL12RB1 variation database. Hum. Mutat. 2013, 34, 1329–1339. [Google Scholar] [CrossRef]
- Danese, S.; Peyrin-Biroulet, L. Selective tyrosine kinase 2 inhibition for treatment of inflammatory bowel disease: New hope on the rise. Inflamm. Bowel Dis. 2021, 27, 2023–2030. [Google Scholar] [CrossRef]
- Villanueva, M.T. TYK2 inhibition shows promise. Nature 2019, 18, 668–669. [Google Scholar] [CrossRef]
- Tao, J.H.; Zou, Y.F.; Feng, X.L.; Li, J.; Wang, F.; Pan, F.M.; Ye, D.Q. Meta-analysis of TYK2 gene polymorphisms association with susceptibility to autoimmune and inflammatory diseases. Mol. Biol. Rep. 2010, 38, 4663–4672. [Google Scholar] [CrossRef] [PubMed]
- Can, G.; Tezel, A.; Gürkan, H.; Can, H.; Yilmaz, B.; Unsal, G.; Soylu, A.R.; Ummit, H.C. Tyrosine kinase-2 gene polymorphisms are associated with ulcerative colitis and Crohn’s disease in Turkish Population. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rotival, M.; Patin, E.; Michel, F.; Pellegrini, S. Two common disease-associated TYK2 variants impact exon splicing and TYK2 dosage. PLoS ONE 2020, 15, e0225289. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Hegazy, A.N.; Owens, B.M.J.; Bullers, S.J.; Linggi, B.; Buonocore, S.; Coccia, M.; Gortz, D.; This, S.; Stockenhuber, K.; et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor–neutralizing therapy in patients with inflammatory bowel disease. Nat. Med. 2017, 23, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Hong, I.K.; Eun, Y.G.; Chung, D.H.; Kwon, K.H.; Kim, D.Y. Association of the oncostatin M receptor gene polymorphisms with papillary thyroid cancer in the Korean population. Clin. Exp. Otorhinolaryngol. 2011, 4, 193–198. [Google Scholar] [CrossRef]
- Deng, S.; He, S.Y.; Zhao, P.; Zhang, P. The role of oncostatin M receptor gene polymorphisms in bladder cancer. World J. Surg. Oncol 2019, 17, 1–9. [Google Scholar] [CrossRef]
- Gaspar, P.; Moura, G.; Santos, M.A.S.; Oliveira, J.L. mRNA secondary structure optimization using a correlated stem-loop prediction. Nucleic Acids Res. 2012, 41, e73. [Google Scholar] [CrossRef]
- Chen, M.H.; Fang, C.; Wu, N.Y.; Xia, Y.H.; Zeng, Y.J.; Ouyang, W. Genetic variation of rs12918566 affects GRIN2A expression and is associated with spontaneous movement response during sevoflurane anesthesia induction. Brain Behav. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Zwiers, A.; Kraal, L.; van de Pouw Kraan, T.C.; Wurdinger, T.; Bouma, G.; Kraal, G. Cutting edge: A variant of the IL-23R gene associated with inflammatory bowel disease induces loss of microRNA regulation and enhanced protein production. J. Immunol. 2012, 188, 1573–1577. [Google Scholar] [CrossRef]
- Zheng, J.; Jiang, L.; Zhang, L.; Yang, L.; Deng, J.; You, Y.; Li, N.; Wu, H.; Li, W.; Lu, J.; et al. Functional genetic variations in the IL-23 receptor gene are associated with risk of breast, lung and nasopharyngeal cancer in Chinese populations. Carcinogenesis 2012, 33, 2409–2416. [Google Scholar] [CrossRef]
- Xie, Y.H.; Chen, Y.X.; Fang, J.Y. Comprehensive review of targeted therapy for colorectal cancer. Signal. Transduct. Target 2020, 5, 22. [Google Scholar] [CrossRef] [PubMed]
- Quiniou, C.; Domínguez-Punaro, M.; Cloutier, F.; Erfani, A.; Ennaciri, J.; Sivanesan, D.; Sanchez, M.; Chognard, G.; Hou, X.; Rivera, J.C.; et al. Specific targeting of the IL-23 receptor, using a novel small peptide noncompetitive antagonist, decreases the inflammatory response. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R1216–R1230. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using polyphen-2. In Current Protocols in Human Genetics; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, 37–43. [Google Scholar] [CrossRef]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Bamford, S.; Dawson, E.; Forbes, S.; Clements, J.; Pettett, R.; Dogan, A.; Flanagan, A.; Teague, J.; Futreal, P.A.; Stratton, M.R.; et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br. J. Cancer 2004, 91, 355–358. [Google Scholar] [CrossRef]
- McKusick, V.A. Mendelian Inheritance in Man: A catalog of Human Genes and Genetic Disorders; JHU Press: Baltimor, MA, USA, 1998. [Google Scholar]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Stenson, P.D.; Ball, E.V.; Mort, M.; Phillips, A.D.; Shiel, J.A.; Thomas, N.S.; Abeysinghe, S.; Krawczak, M.; Cooper, D.N. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003, 21, 577–581. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Haw, R. Functional Interaction Network Construction and Analysis for Disease Discovery. Methods Mol. Biol. 2017, 1558, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef]
| UC (n = 8) | CRC (n = 11) | Normal (n = 8) | CAC (n = 3) | |
|---|---|---|---|---|
| Median age (range) | 65.5 (60–69) | 60 (36–74) | 64 (51–74) | 59 (20–69) |
| Race | ||||
| Malay | 3 | 10 | 7 | 1 |
| Chinese | 3 | 1 | 1 | - |
| Indian | 2 | - | - | 2 |
| Gender | ||||
| Male | 4 | 5 | 3 | - |
| Female | 4 | 6 | 5 | 3 |
| Smoking status | ||||
| Ex-smoker | - | 1 | 1 | - |
| Non-smoker | 8 | 10 | 7 | 3 |
| Stage | Not applicable | Not applicable | ||
| I | 4 | - | ||
| II | 3 | - | ||
| III | 4 | 3 | ||
| Adenocarcinoma types | Not applicable | Not applicable | ||
| Poorly differentiated | 1 | 1 | ||
| Moderately differentiated | 9 | - | ||
| Well differentiated | 1 | 2 | ||
| Mayo score (range) | 1–3 | Not applicable | Not applicable | Data unavailable |
| Geboes score (range) | 2A.1–2A.2 | Not applicable | Not applicable | Data unavailable |
| Group | Gene | Location | dbSNP | Changes | Prediction |
|---|---|---|---|---|---|
| UC | IL12Rß1 | Intronic | rs201422056 | g.18174947_18174948del | Not applicable |
| IL12Rß2 | Intronic | rs17838042 | g.67792801G>C | Not applicable | |
| Intronic | rs17129778 | g.67787691A>T | Not applicable | ||
| Intronic | rs17129794 | g.67794918A>C | Not applicable | ||
| Intronic | rs147756804 | g.67796641_67796646del | Not applicable | ||
| IL23R | Intronic | rs41313260 | g.67706309C>T | Not applicable | |
| SGK2 | Intronic | rs73620603 | g.42195665C>T | Not applicable | |
| OSMR | Intronic | rs367864552 | g.38881296_38881299del | Not applicable | |
| Intronic | rs55964556 | g.38931022_38931024del | Not applicable | ||
| Intronic | rs757333768 | g.38881299ins | Not applicable | ||
| STAT4 | Intronic | rs370820216 | g.191898876T>C | Not applicable | |
| STAT6 | Intronic | - | g.57494483_57494499del | Not applicable | |
| Intronic | - | g.57494421ins | Not applicable | ||
| CRC | IL12Rß1 | Exonic | rs370238890 | c.1781G>A; p.G594E | Benign |
| IL12Rß2 | Intronic | - | g.67860953_67860954del | Not applicable | |
| IL23R | Intronic | rs767258696 | g.67699612_67699616del | Not applicable | |
| OSMR | Intronic | rs113727379 | g.38885647C>T | Not applicable | |
| Exonic | rs34675408 | c.561T>G; p.H187Q | Benign | ||
| JAK2 | Intronic | rs3780378 | g.5112288C>T | Not applicable | |
| STAT4 | Intronic | rs35593987 | g.191916526_191916527del | Not applicable | |
| Intronic | rs11272763 | g.191992821ins | Not applicable | ||
| STAT6 | UTR5 | rs71802646 | g.57505072_57505076del | Not applicable | |
| CAC | IL12Rß2 | Intronic | Not available | g.67795960ins | Not applicable |
| TYK2 | Intronic | Not available | g.10477409_10477412del | Not applicable |
| Group | Gene | Location | dbSNP | Change |
|---|---|---|---|---|
| CAC with UC | IL12Rß1 | Intronic | rs372889 | g.18173603T>C |
| Intronic | rs439409 | g.18193613A>G | ||
| Intronic | rs382634 | g.18187562G>A | ||
| Intronic | rs17878594 | g.18173513C>T | ||
| Intronic | Not available | g.18179560_18179562del | ||
| IL12Rß2 | Intronic | rs12410480 | g.67803994G>T | |
| Intronic | rs145598332 | g.67833145ins | ||
| Intronic | rs66726768 | g.67795956_67795960del | ||
| IL23R | Intronic | Not available | g.67672567_67672569del | |
| OSMR | Intronic | rs79215370 | g.38882285C>T | |
| Intronic | rs137968159 | g.38919267_38919270del | ||
| JAK2 | Intronic | rs10283730 | g.5073289G>A | |
| Intronic | rs7865719 | g.5082333A>G | ||
| Intronic | rs138377711 | g.5111358_5111359del | ||
| TYK2 | Intronic | rs12720294 | g.10469699A>G | |
| Intronic | rs12720293 | g.10470293A>G | ||
| Intronic | rs143429818 | g.10469743ins | ||
| STAT1 | Intronic | rs2066803 | g.191839459C>A | |
| Intronic | rs41371944 | g.191844745T>C | ||
| Intronic | rs376961322 | g.191844269_191844270 | ||
| STAT3 | Intronic | rs9909659 | g.40473835G>A | |
| Intronic | rs8081037 | g.40499158C>T | ||
| STAT6 | Intronic | Not available | g.57494483ins | |
| Intronic | Not available | g.57494421ins | ||
| Intronic | rs398019756 | g.57494880_57494881del | ||
| CAC with CRC | JAK2 | Intronic | rs9987451 | g.5113452C>T |
| STAT4 | Intronic | Not available | g.191940749_191940750del |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razali, N.N.; Raja Ali, R.A.; Muhammad Nawawi, K.N.; Yahaya, A.; Mokhtar, N.M. Targeted Sequencing of Cytokine-Induced PI3K-Related Genes in Ulcerative Colitis, Colorectal Cancer and Colitis-Associated Cancer. Int. J. Mol. Sci. 2022, 23, 11472. https://doi.org/10.3390/ijms231911472
Razali NN, Raja Ali RA, Muhammad Nawawi KN, Yahaya A, Mokhtar NM. Targeted Sequencing of Cytokine-Induced PI3K-Related Genes in Ulcerative Colitis, Colorectal Cancer and Colitis-Associated Cancer. International Journal of Molecular Sciences. 2022; 23(19):11472. https://doi.org/10.3390/ijms231911472
Chicago/Turabian StyleRazali, Nurul Nadirah, Raja Affendi Raja Ali, Khairul Najmi Muhammad Nawawi, Azyani Yahaya, and Norfilza M. Mokhtar. 2022. "Targeted Sequencing of Cytokine-Induced PI3K-Related Genes in Ulcerative Colitis, Colorectal Cancer and Colitis-Associated Cancer" International Journal of Molecular Sciences 23, no. 19: 11472. https://doi.org/10.3390/ijms231911472
APA StyleRazali, N. N., Raja Ali, R. A., Muhammad Nawawi, K. N., Yahaya, A., & Mokhtar, N. M. (2022). Targeted Sequencing of Cytokine-Induced PI3K-Related Genes in Ulcerative Colitis, Colorectal Cancer and Colitis-Associated Cancer. International Journal of Molecular Sciences, 23(19), 11472. https://doi.org/10.3390/ijms231911472





