SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer †
Abstract
1. Introduction
2. Results
2.1. Predicted Cis-Elements of PNPT1 Promoter
2.2. Analysis of Cis-Elements with a PNPT1-Promoter Construct
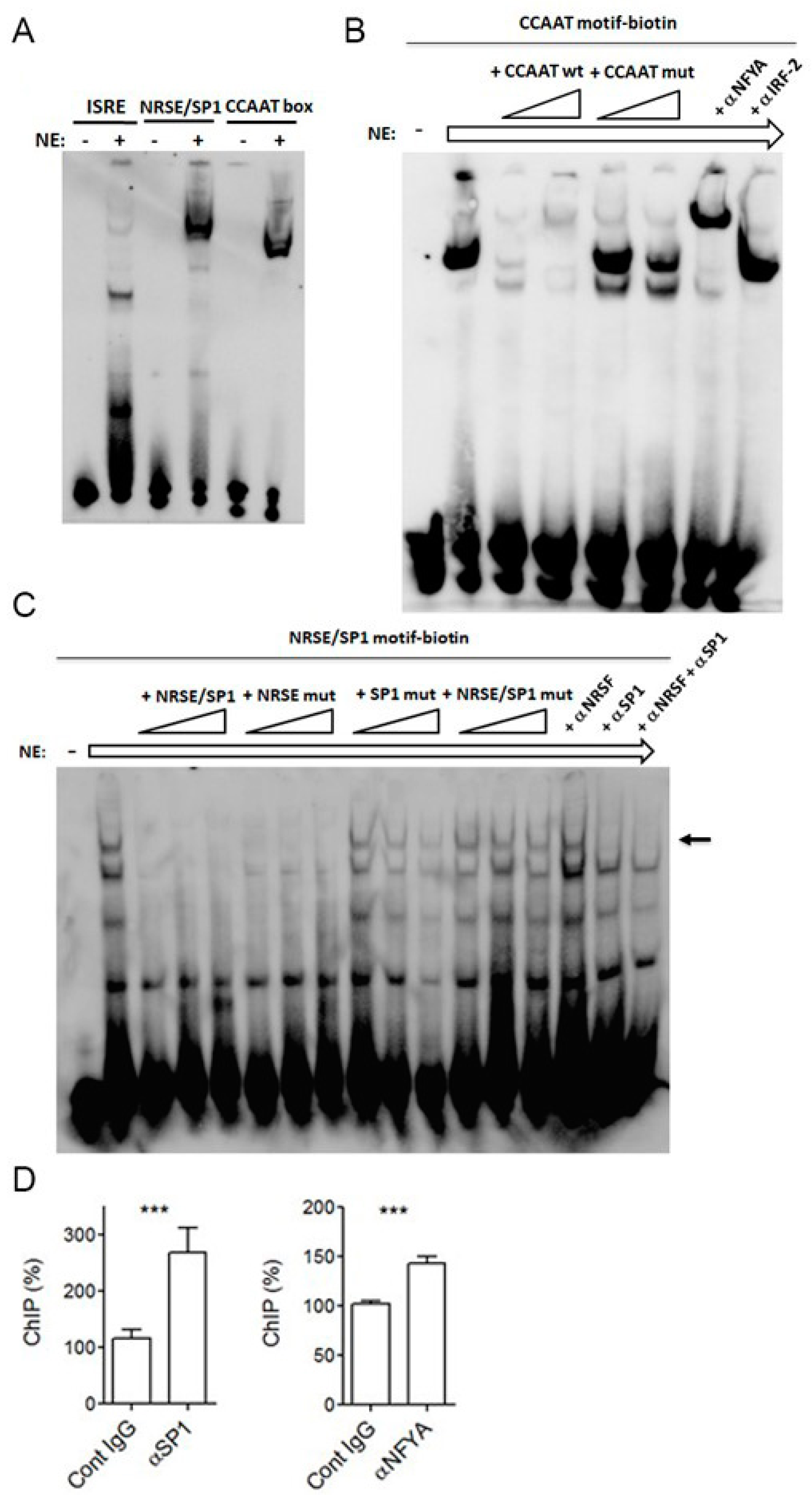
2.3. SP1 and NFYA Bind PNPT1 Promoter
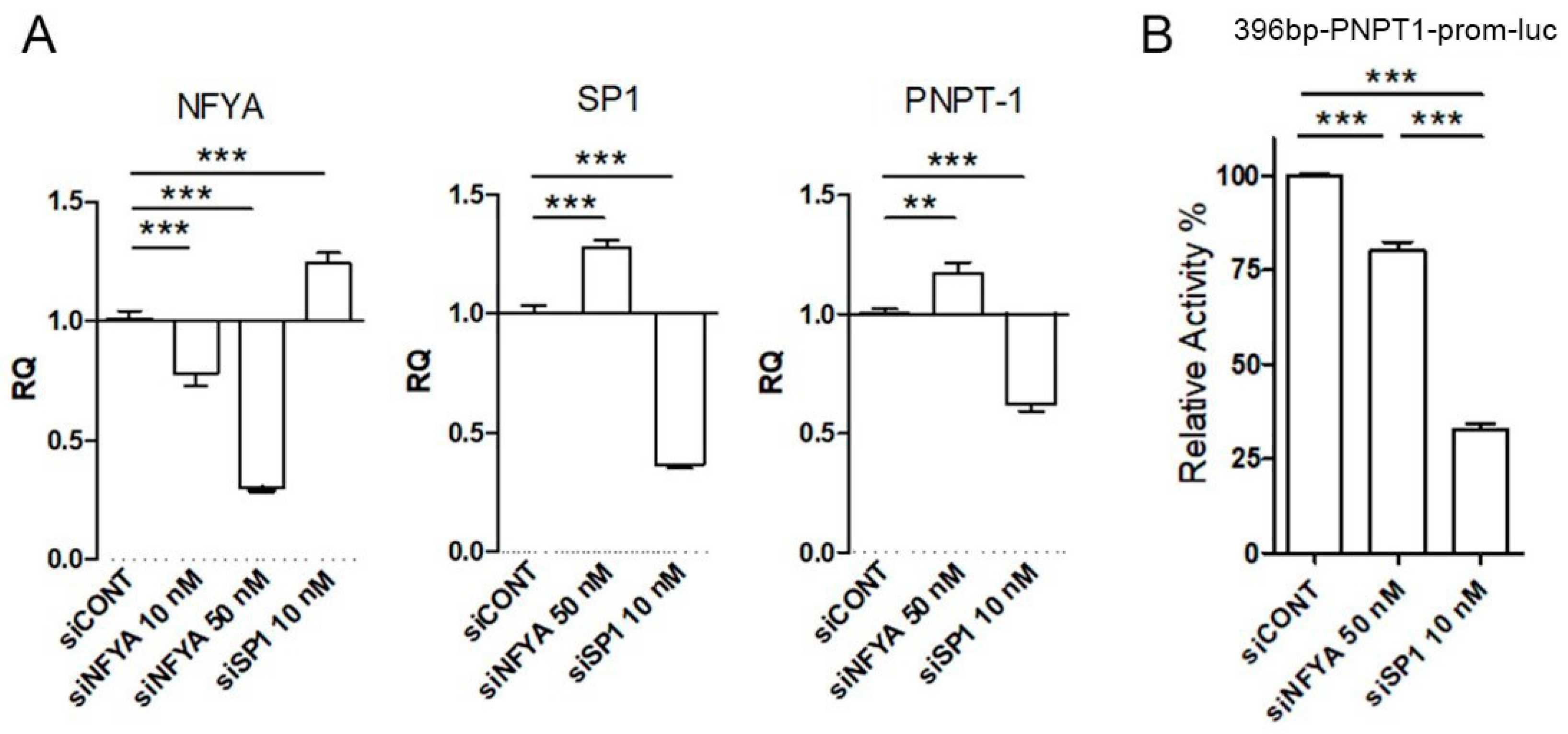
2.4. PNPT1 Promoter Regulation by SP1 and NFYA in HeLa Cells
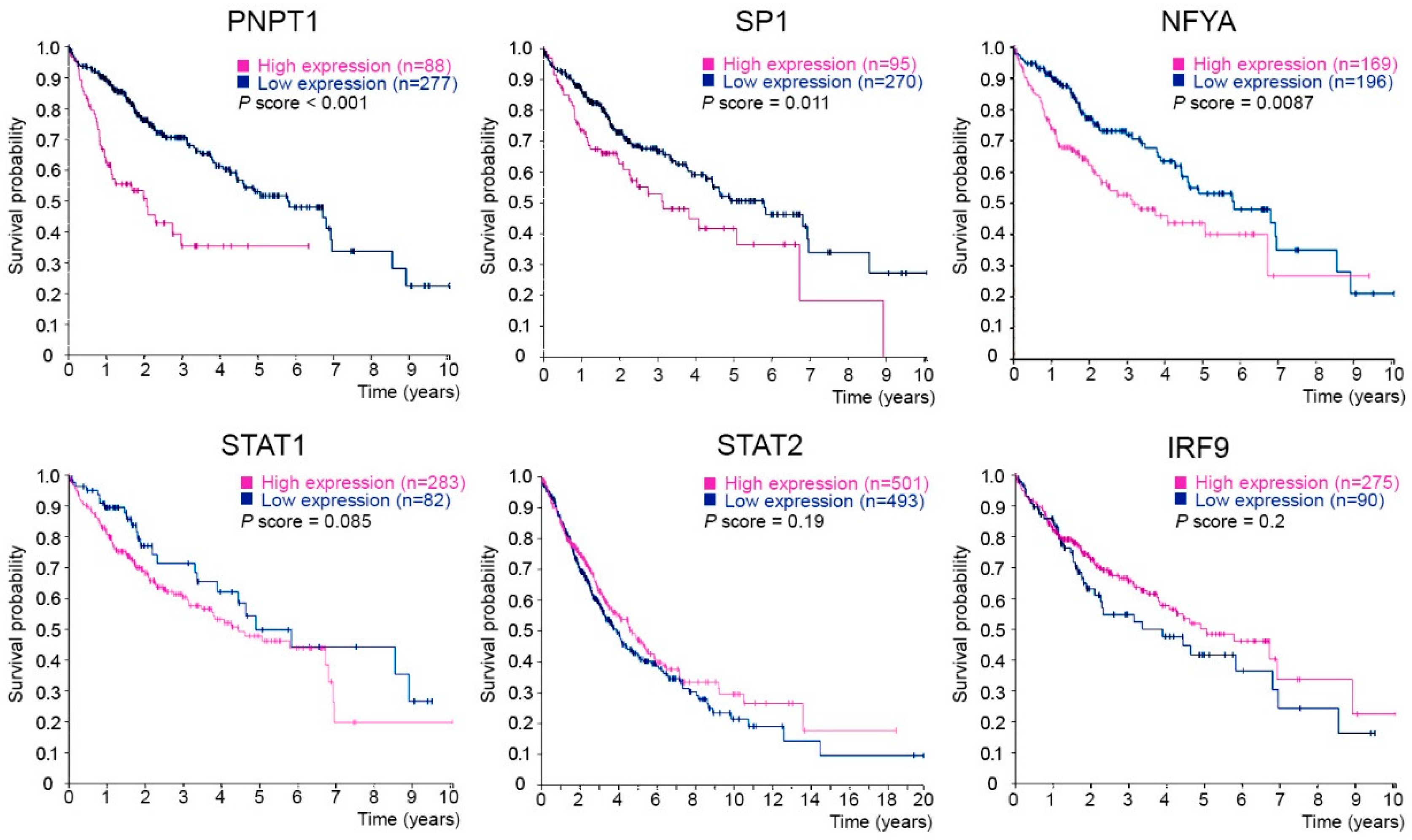
2.5. PNPT1, SP1 and NFYA Overexpression Is Correlated with a Poor Prognosis in Liver Cancer Patients
3. Discussion
4. Materials and Methods
4.1. Analysis of the Promoter Region of the PNPT1 Gene
4.2. Cell Culture, Mitochondrial Activity and Quantification
4.3. DNA Constructs
4.4. Transfection of siRNA Probes
4.5. Purification of RNA, Retrotranscription and qPCR
4.6. Transfection of Reporter Constructs and Luciferase Reporter Assay
4.7. Electromobility Shift Assays
4.8. Chromatin Immunoprecipitation
4.9. Kaplan–Meier Survival Analysis
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
References
- Spinelli, J.; Haigis, M. The multifaceted contributions of mitochondria to cellular metabolism. Nat. Cell Biol. 2018, 20, 745–754. [Google Scholar] [CrossRef]
- Popov, L. Mitochondrial biogenesis: An update. J. Cell Mol. Med. 2020, 24, 4892–4899. [Google Scholar] [CrossRef]
- Wiedemann, N.; Pfanner, N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Kamenski, P.; Krasheninnikov, I.; Tarassov, I. 40 Years of Studying RNA Import into Mitochondria: From Basic Mechanisms to Gene Therapy Strategies. Mol. Biol. 2019, 53, 813–819. [Google Scholar] [CrossRef]
- Wang, G.; Chen, H.W.; Oktay, Y.; Zhang, J.; Allen, E.L.; Smith, G.M.; Fan, K.C.; Hong, J.S.; French, S.W.; McCaffery, J.M.; et al. PNPASE regulates RNA import into mitochondria. Cell 2010, 142, 456–467. [Google Scholar] [CrossRef]
- Chen, H.W.; Rainey, R.N.; Balatoni, C.E.; Dawson, D.W.; Troke, J.J.; Wasiak, S.; Hong, J.S.; McBride, H.M.; Koehler, C.M.; Teitell, M.A.; et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell Biol. 2006, 26, 8475–8487. [Google Scholar] [CrossRef]
- Shimada, E.; Ahsan, F.M.; Nili, M.; Huang, D.; Atamdede, S.; TeSlaa, T.; Case, D.; Yu, X.; Gregory, B.D.; Perrin, B.J.; et al. PNPase knockout results in mtDNA loss and an altered metabolic gene expression program. PLoS ONE 2018, 13, e0200925. [Google Scholar] [CrossRef]
- Matilainen, S.; Carroll, C.J.; Richter, U.; Euro, L.; Pohjanpelto, M.; Paetau, A.; Isohanni, P.; Suomalainen, A. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum. Mol. Genet. 2017, 26, 3352–3361. [Google Scholar] [CrossRef]
- Grunberg-Manago, M.; Oritz, P.; Ochoa, S. Enzymatic synthesis of nucleic acidlike polynucleotides. Science 1955, 122, 907–910. [Google Scholar] [CrossRef]
- Cameron, T.; Matz, L.; De Lay, N. Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet. 2018, 14, e1007654. [Google Scholar] [CrossRef]
- Falchi, F.; Pizzoccheri, R.; Briani, F. Activity and Function in Human Cells of the Evolutionary Conserved Exonuclease Polynucleotide Phosphorylase. Int. J. Mol. Sci. 2022, 23, 1652. [Google Scholar] [CrossRef]
- Hung, V.; Zou, P.; Rhee, H.W.; Udeshi, N.D.; Cracan, V.; Svinkina, T.; Carr, S.A.; Mootha, V.K.; Ting, A.Y. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell 2014, 55, 332–341. [Google Scholar] [CrossRef]
- Rainey, R.; Glavin, J.; Chen, H.; French, S.; Teitell, M.; Koehler, C. A new function in translocation for the mitochondrial i-AAA protease Yme1: Import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 2006, 26, 8488–8497. [Google Scholar] [CrossRef]
- Wu, J.; Jiang, Z.; Liu, M.; Gong, X.; Wu, S.; Burns, C.M.; Li, Z. Polynucleotide phosphorylase protects Escherichia coli against oxidative stress. Biochemistry 2009, 48, 2012–2020. [Google Scholar] [CrossRef]
- Liu, X.; Fu, R.; Pan, Y.; Meza-Sosa, K.; Zhang, Z.; Lieberman, J. PNPT1 Release from Mitochondria during Apoptosis Triggers Decay of Poly(A) RNAs. Cell 2018, 174, 187–201.e12. [Google Scholar] [CrossRef]
- Sarkar, D.; Leszczyniecka, M.; Kang, D.; Lebedeva, I.V.; Valerie, K.; Dhar, S.; Pandita, T.K.; Fisher, P.B. Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 2003, 278, 24542–24551. [Google Scholar] [CrossRef]
- Khaw, S.; Wen, C.M.; Koh, C.; Lim, B.; Chang, N.S. Oocyte Factors Suppress Mitochondrial Polynucleotide Phosphorylase to Remodel the Metabolome and Enhance Reprogramming. Cell Rep. 2015, 12, 1080–1088. [Google Scholar] [CrossRef]
- Shyh-Chang, N.; Daley, G.; Cantley, L. Stem cell metabolism in tissue development and aging. Development 2013, 140, 2535–2547. [Google Scholar] [CrossRef]
- Heiden, M.; Cantley, L.; Thompson, C. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Chujo, T.; Ohira, T.; Sakaguchi, Y.; Goshima, N.; Nomura, N.; Nagao, A.; Suzuki, T. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012, 40, 8033–8047. [Google Scholar] [CrossRef]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013, 41, 1223–1240. [Google Scholar] [CrossRef] [PubMed]
- Vedrenne, V.; Gowher, A.; de Lonlay, P.; Nitschke, P.; Serre, V.; Boddaert, N.; Altuzarra, C.; Mager-Heckel, A.; Chretien, F.; Entelis, N.; et al. Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am. J. Hum. Genet. 2012, 91, 912–918. [Google Scholar] [CrossRef]
- Alodaib, A.; Sobreira, N.; Gold, W.A.; Riley, L.G.; van Bergen, N.J.; Wilson, M.J.; Bennetts, B.; Thorburn, D.R.; Boehm, C.; Christodoulou, J. Whole-exome sequencing identifies novel variants in PNPT1 causing oxidative phosphorylation defects and severe multisystem disease. Eur. J. Hum. Genet. 2016, 25, 79–84. [Google Scholar] [CrossRef]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rötig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Bamborschke, D.; Kreutzer, M.; Koy, A.; Koerber, F.; Lucas, N.; Huenseler, C.; Herkenrath, P.; Lee-Kirsch, M.A.; Cirak, S. PNPT1 mutations may cause Aicardi-Goutières-Syndrome. Brain Dev. 2021, 43, 320–324. [Google Scholar] [CrossRef]
- von Ameln, S.; Wang, G.; Boulouiz, R.; Rutherford, M.A.; Smith, G.M.; Li, Y.; Pogoda, H.M.; Nürnberg, G.; Stiller, B.; Volk, A.E.; et al. A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am. J. Hum. Genet. 2012, 91, 919–927. [Google Scholar] [CrossRef]
- Sato, R.; Arai-Ichinoi, N.; Kikuchi, A.; Matsuhashi, T.; Numata-Uematsu, Y.; Uematsu, M.; Fujii, Y.; Murayama, K.; Ohtake, A.; Abe, T.; et al. Novel biallelic mutations in the PNPT1 gene encoding a mitochondrial-RNA-import protein PNPase cause delayed myelination. Clin. Genet. 2018, 93, 242–247. [Google Scholar] [CrossRef]
- Yu, Y.; Chou, R.; Wu, C.; Wang, Y.; Chang, W.; Tseng, Y.; Chang, W.; Lai, C.; Lee, H.; Huo, L.; et al. Nuclear EGFR suppresses ribonuclease activity of polynucleotide phosphorylase through DNAPK-mediated phosphorylation at serine 776. J. Biol. Chem. 2012, 287, 31015–31026. [Google Scholar] [CrossRef]
- Fukuhara, N.; Tagawa, H.; Kameoka, Y.; Kasugai, Y.; Karnan, S.; Kameoka, J.; Sasaki, T.; Morishima, Y.; Nakamura, S.; Seto, M. Characterization of target genes at the 2p15-16 amplicon in diffuse large B-cell lymphoma. Cancer Sci. 2006, 97, 499–504. [Google Scholar] [CrossRef]
- Kirschner, L.S.; Taymans, S.E.; Pack, S.; Pak, E.; Pike, B.L.; Chandrasekharappa, S.C.; Zhuang, Z.; Stratakis, C.A. Genomic mapping of chromosomal region 2p15-p21 (D2S378-D2S391): Integration of Genemap’98 within a framework of yeast and bacterial artificial chromosomes. Genomics 1991, 62, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Wang, Y.; Li, S.; Chen, D.; Zhao, X. Novel PNPT1-ALK fusion variant exerted significant benefit to crizotinib in NSCLC. Lung Cancer 2020, 146, 382–384. [Google Scholar] [CrossRef] [PubMed]
- Luczynski, W.; Kowalczuk, O.; Stasiak-Barmuta, A.; Ilendo, E.; Krawczuk-Rybak, M.; Chyczewski, L. Acute lymphoblastic leukemia-derived dendritic cells express tumor associated antigens: PNPT1, PMPCB, RHAMM, BSG and ERCC1. Neoplasma 2009, 56, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Leszczyniecka, M.; Kang, D.; Sarkar, D.; Su, Z.; Holmes, M.; Valerie, K.; Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. USA 2002, 99, 16636–16641. [Google Scholar] [CrossRef]
- Gewartowski, K.; Tomecki, R.; Muchowski, L.; Ska, A.D.; Dzwonek, A.; Malecki, M.; Skurzak, H.; Ostrowski, J.; Stepien, P.P. Up-regulation of human PNPase mRNA by beta-interferon has no effect on protein level in melanoma cell lines. Acta Biochim. Pol. 2006, 53, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.; Wu, L.; Coustry, F.; de Crombrugghe, B.; Maity, S. DNA binding specificity of the CCAAT-binding factor CBF/NF-Y. J. Biol. Chem. 1997, 272, 26562–26572. [Google Scholar] [CrossRef] [PubMed]
- Schoenherr, C.; Paquette, A.; Anderson, D. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. USA 1996, 93, 9881–9886. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Song, L.; Cai, J.; Li, L.; Xu, H.; Li, M.; Wang, J.; Shi, M.; Chen, H.; Jia, H.; et al. The LIM protein Ajuba/SP1 complex forms a feed forward loop to induce SP1 target genes and promote pancreatic cancer cell proliferation. J. Exp. Clin. Cancer Res. 2019, 38, 205. [Google Scholar] [CrossRef]
- Leszczyniecka, M.; Su, Z.; Kang, D.; Sarkar, D.; Fisher, P.B. Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPaseold-35, a Type I interferon inducible early response gene. Gene 2003, 316, 143–156. [Google Scholar] [CrossRef]
- Su, B.; Henley, M. Drugging Fuzzy Complexes in Transcription. Front. Mol. Biosci. 2021, 8, 795743. [Google Scholar] [CrossRef]
- Zaid, A.; Li, R.; Luciakova, K.; Barath, P.; Nery, S.; Nelson, B. On the role of the general transcription factor Sp1 in the activation and repression of diverse mammalian oxidative phosphorylation genes. J. Bioenerg. Biomembr. 1999, 31, 129–135. [Google Scholar] [CrossRef]
- Zanotto, E.; Häkkinen, A.; Teku, G.; Shen, B.; Ribeiro, A.; Jacobs, H. NF-Y influences directionality of transcription from the bidirectional Mrps12/Sarsm promoter in both mouse and human cells. Biochim. Biophys. Acta 2009, 1789, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Suske, G. NF-Y and SP transcription factors—New insights in a long-standing liaison. Biochim. Biophys. Acta. Gene Regul. Mech. 2017, 1860, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Schaufele, F.; Gardner, D. Functional interaction of NF-Y and SP1 is required for type a natriuretic peptide receptor gene transcription. J. Biol. Chem. 2001, 276, 1516–1522. [Google Scholar] [CrossRef]
- Yamada, K.; Tanaka, T.; Miyamoto, K.; Noguchi, T. Sp family members and nuclear factor-Y cooperatively stimulate transcription from the rat pyruvate kinase M gene distal promoter region via their direct interactions. J. Biol. Chem. 2000, 275, 18129–18137. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, C.; Mansilla, S.; Portugal, J. Sp1 transcription factor: A long-standing target in cancer chemotherapy. Pharmacol. Ther. 2015, 152, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Bezzecchi, E.; Ronzio, M.; Semeghini, V.; Andrioletti, V.; Mantovani, R.; Dolfini, D. NF-YA Overexpression in Lung Cancer: LUAD. Genes 2020, 11, 198. [Google Scholar] [CrossRef] [PubMed]
- Bezzecchi, E.; Ronzio, M.; Mantovani, R.; Dolfini, D. NF-Y Overexpression in Liver Hepatocellular Carcinoma (HCC). Int. J. Mol. Sci. 2020, 21, 9157. [Google Scholar] [CrossRef]
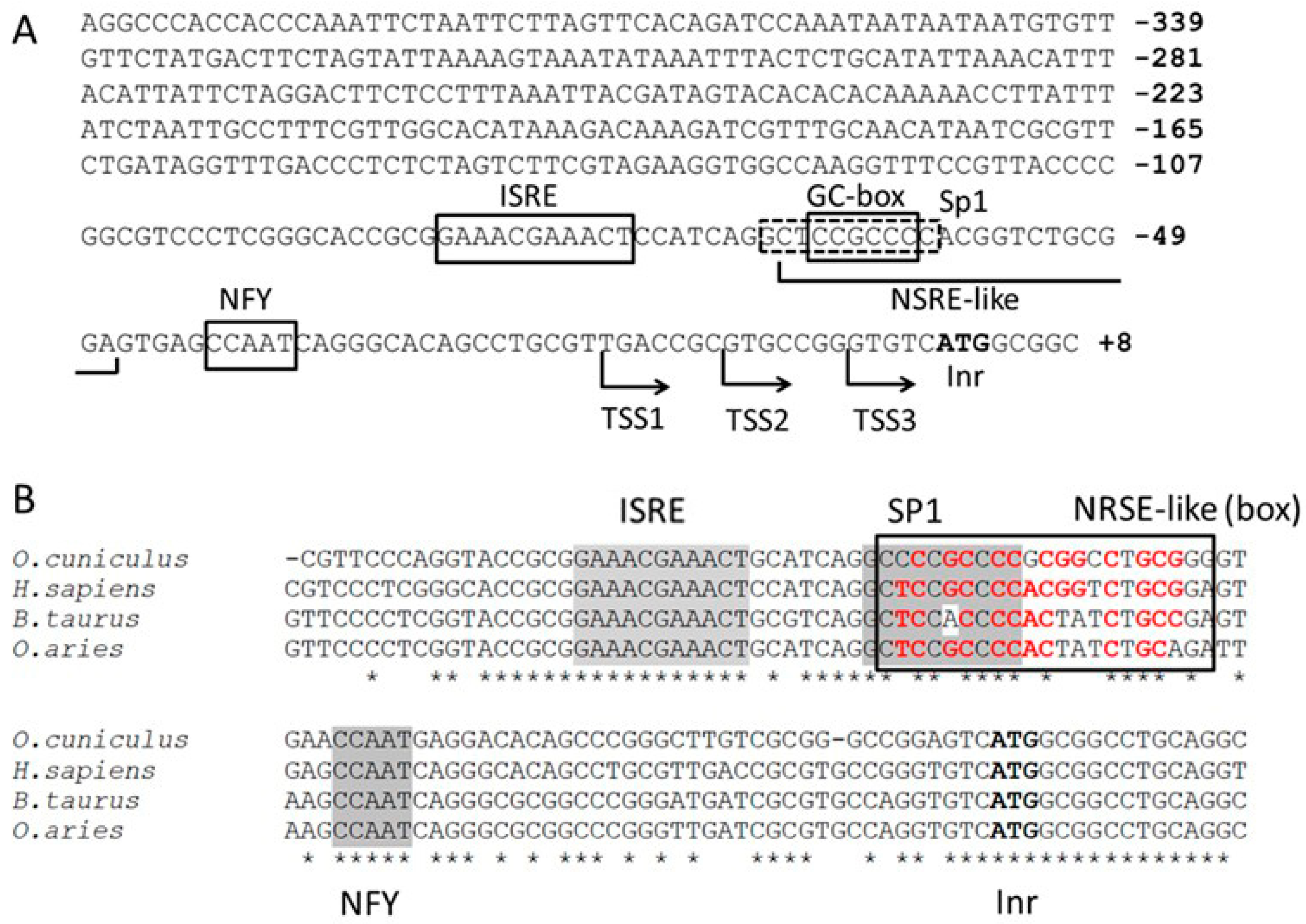
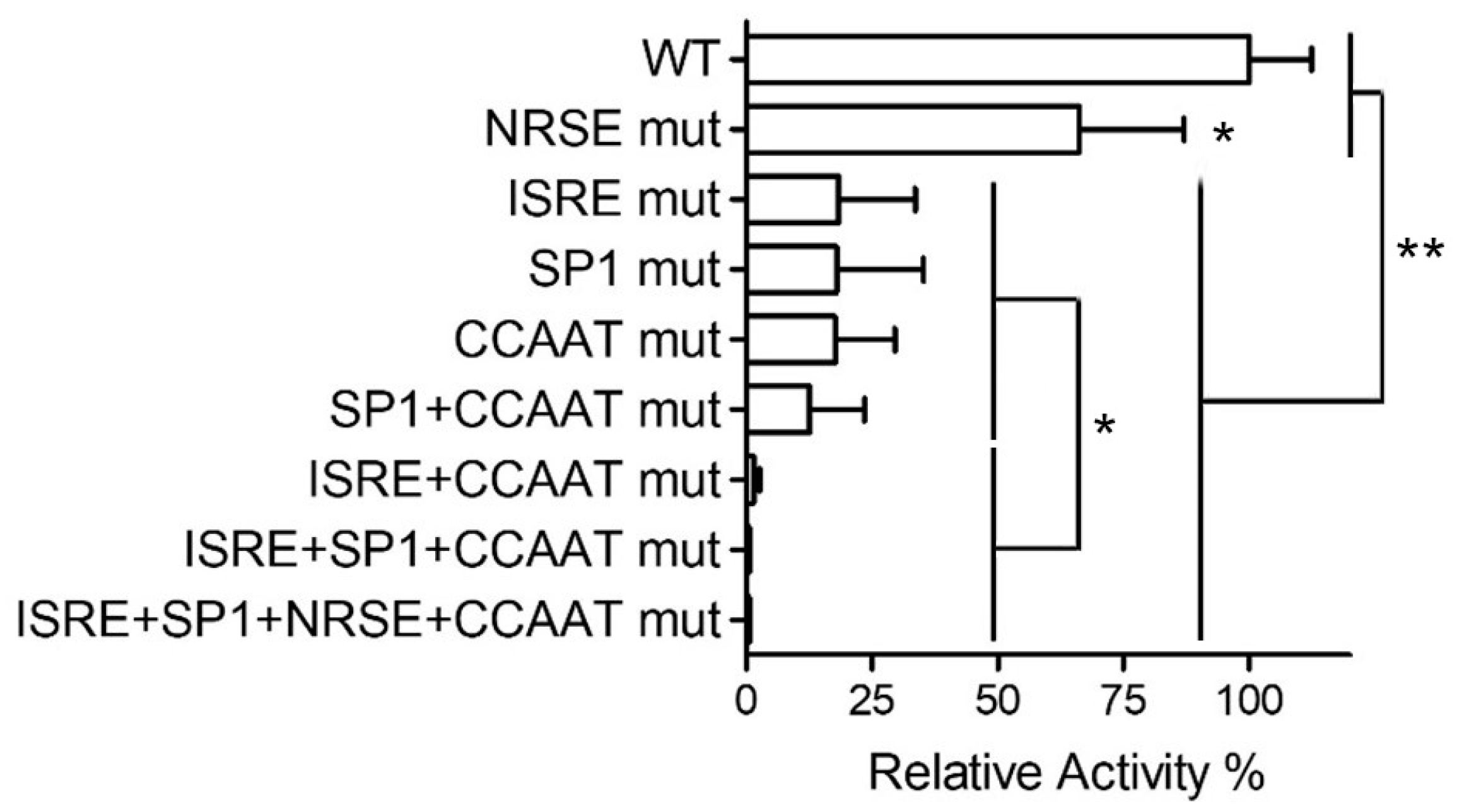
| Construct | Nucleotides −96 to −1 (Mutations in Red) |
|---|---|
| WT | GGGCACCGCGGAAACGAAACTCCATCAGGCTCCGCCCCACGGTCTGCG GAGTGAGCCAATCAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| ISRE mut | GGGCACCGCGCAACTAGAACTCCATCAGGCTCCGCCCCACGGTCTGCG GAGTGAGCCAATCAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| SP1 mut | GGGCACCGCGGAAACGAAACTCCATCAGGCTCCGAACCACGGTCTGCG GAGTGAGCCAATCAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| NRSE mut | GGGCACCGCGGAAACGAAACTCCATCAGGCTCCGCCCCAAAATCTGCG GAGTGAGCCAATCAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| CCAAT mut | GGGCACCGCGGAAACGAAACTCCATCAGGCTCCGCCCCACGGTCTGCG GAGTGAGCCAAACAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| CCAAT+SP1 mut | GGGCACCGCGGAAACGAAACTCCATCAGGCTCCGAACCACGGTCTGCG GAGTGAGCCAAACAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| ISRE+NFYA mut | GGGCACCGCGCAACTAGAACTCCATCAGGCTCCGCCCCACGGTCTGCG GAGTGAGCCAAACAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| ISRE+SP1+NFYA mut | GGGCACCGCGCAACTAGAACTCCATCAGGCTCCGAACCACGGTCTGCG GAGTGAGCCAAACAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGT |
| ISRE+SP1+NRSE+NFYA mut | GGGCACCGCGCAACTAGAACTCCATCAGGCTCCGAACCAAAATCTGCG GAGTGAGCCAAACAGGGCACAGCCTGCGTTGACCGCGTGCCGGGTGTC |
| Site(s) | Name | Sequence (5′-3′) |
|---|---|---|
| ISRE | PNPT1-ISRE-F PNPT1-ISRE-R | CACCGCGGAAACGAAACTCCATCAGG CCTGATGGAGTTTCGTTTCCGCGGTG |
| PNPT1-ISRE-mut-F PNPT1-ISRE-mut-R | CACCGCGCAACTAGAACTCCATCAGG CCTGATGGAGTTCTAGTTGCGCGGTG | |
| SP1/NRSE | PNPT1-SP1/NRSE-F PNPT1-SP1/NRSE-R | CATCAGGCTCCGCCCCACGGTCTGCGGA TCCGCAGACCGTGGGGCGGAGCCTGATG |
| PNPT1-SP1-mut-F PNPT1-SP1-mut-R | CATCAGGCTCCGAACCACGGTCTGCGGA TCCGCAGACCGTGGTTCGGAGCCTGATG | |
| PNPT1-NRSE-mut-F PNPT1-NRSE-mut-R | CATCAGGCTCCGCCCCAAAATCTGCGGA TCCGCAGATTTTGGGGCGGAGCCTGATG | |
| PNPT1-NRSE+SP1-mut-F PNPT1-NRSE+SP1-mut-R | CATCAGGCTCCGAACCAAAATCTGCGGA TCCGCAGATTTTGGTTCGGAGCCTGATG | |
| CCAAT box | PNPT1-CAATbox-F PNPT1-CAATbox-R | GGAGTGAGCCAATCAGGGCACAGCCTGC GCAGGCTGTGCCCTGATTGGCTCACTCC |
| PNPT1-CAATbox-mut-F PNPT1-CAATbox-mut-R | GGAGTGAGCCAAACAGGGCACAGCCTGC GCAGGCTGTGCCCTGTTTGGCTCACTCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ventura, I.; Revert, F.; Revert-Ros, F.; Gómez-Tatay, L.; Prieto-Ruiz, J.A.; Hernández-Andreu, J.M. SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer. Int. J. Mol. Sci. 2022, 23, 11399. https://doi.org/10.3390/ijms231911399
Ventura I, Revert F, Revert-Ros F, Gómez-Tatay L, Prieto-Ruiz JA, Hernández-Andreu JM. SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer. International Journal of Molecular Sciences. 2022; 23(19):11399. https://doi.org/10.3390/ijms231911399
Chicago/Turabian StyleVentura, Ignacio, Fernando Revert, Francisco Revert-Ros, Lucía Gómez-Tatay, Jesús A. Prieto-Ruiz, and José Miguel Hernández-Andreu. 2022. "SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer" International Journal of Molecular Sciences 23, no. 19: 11399. https://doi.org/10.3390/ijms231911399
APA StyleVentura, I., Revert, F., Revert-Ros, F., Gómez-Tatay, L., Prieto-Ruiz, J. A., & Hernández-Andreu, J. M. (2022). SP1 and NFY Regulate the Expression of PNPT1, a Gene Encoding a Mitochondrial Protein Involved in Cancer. International Journal of Molecular Sciences, 23(19), 11399. https://doi.org/10.3390/ijms231911399








