Molecular Characterization of MHC Class I Alpha 1 and 2 Domains in Asian Seabass (Lates calcarifer)
Abstract
:1. Introduction
2. Results
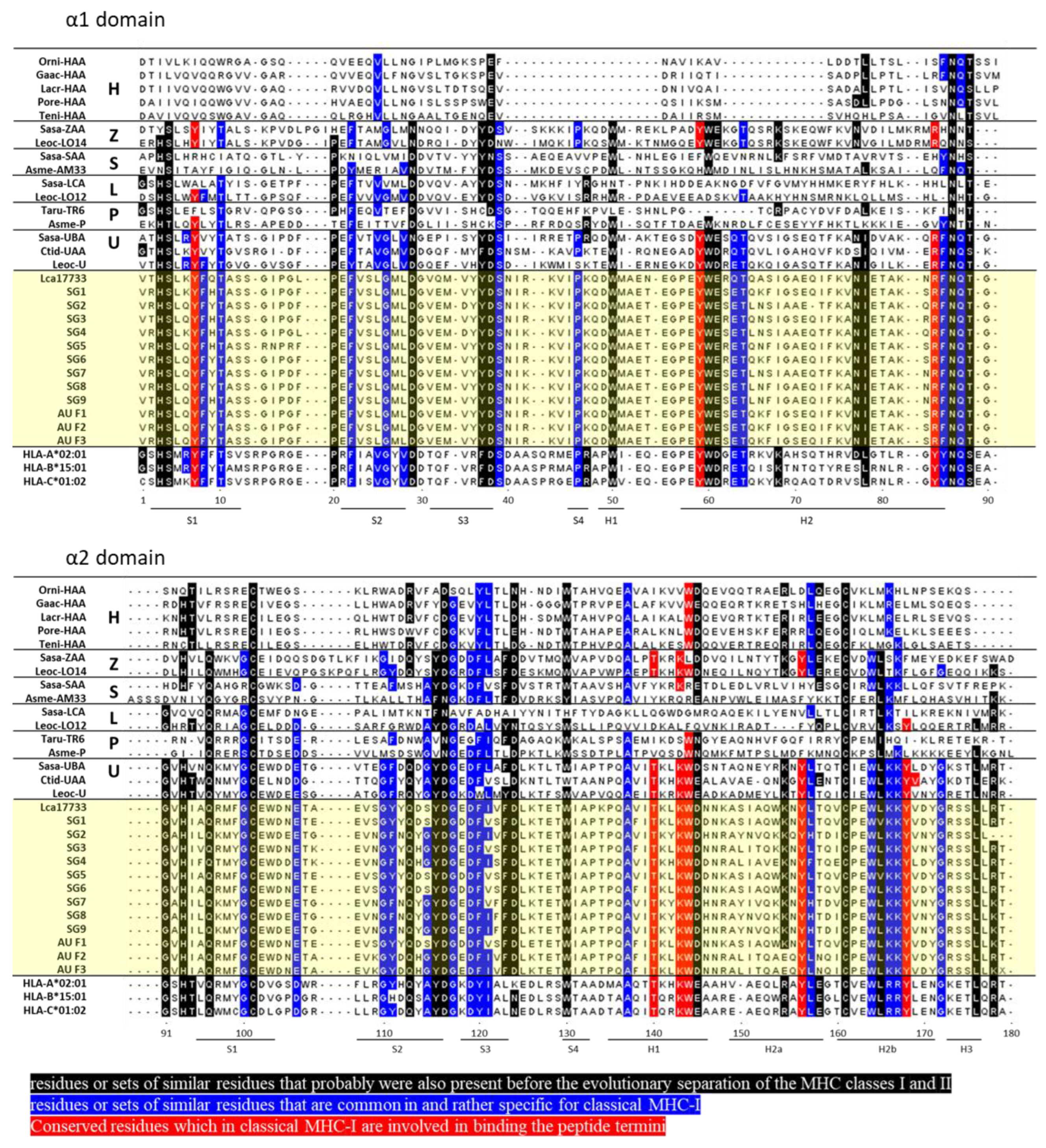
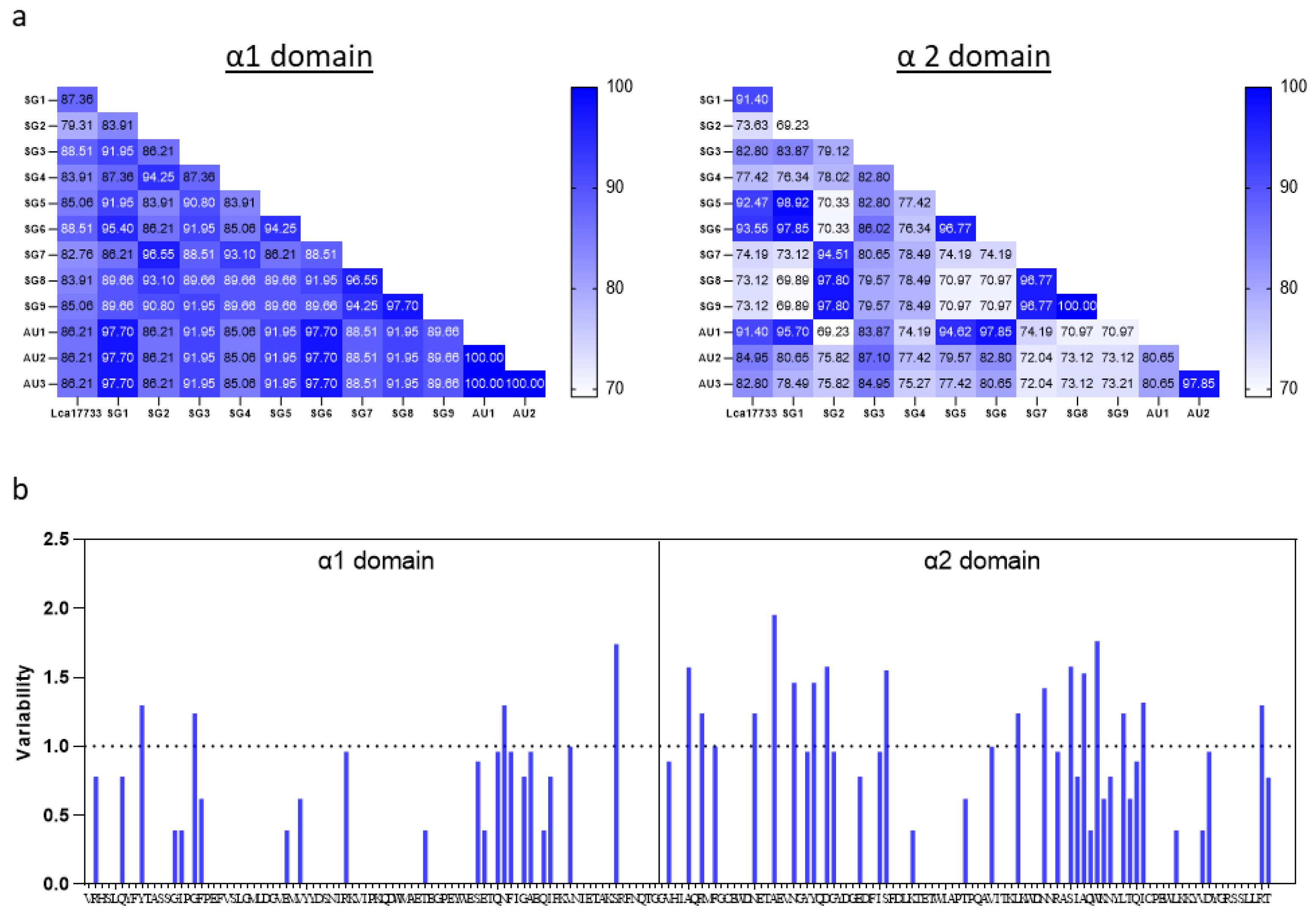
2.1. Amino Acid Sequences of Asian Seabass MHC-I α1 and α2 Domains
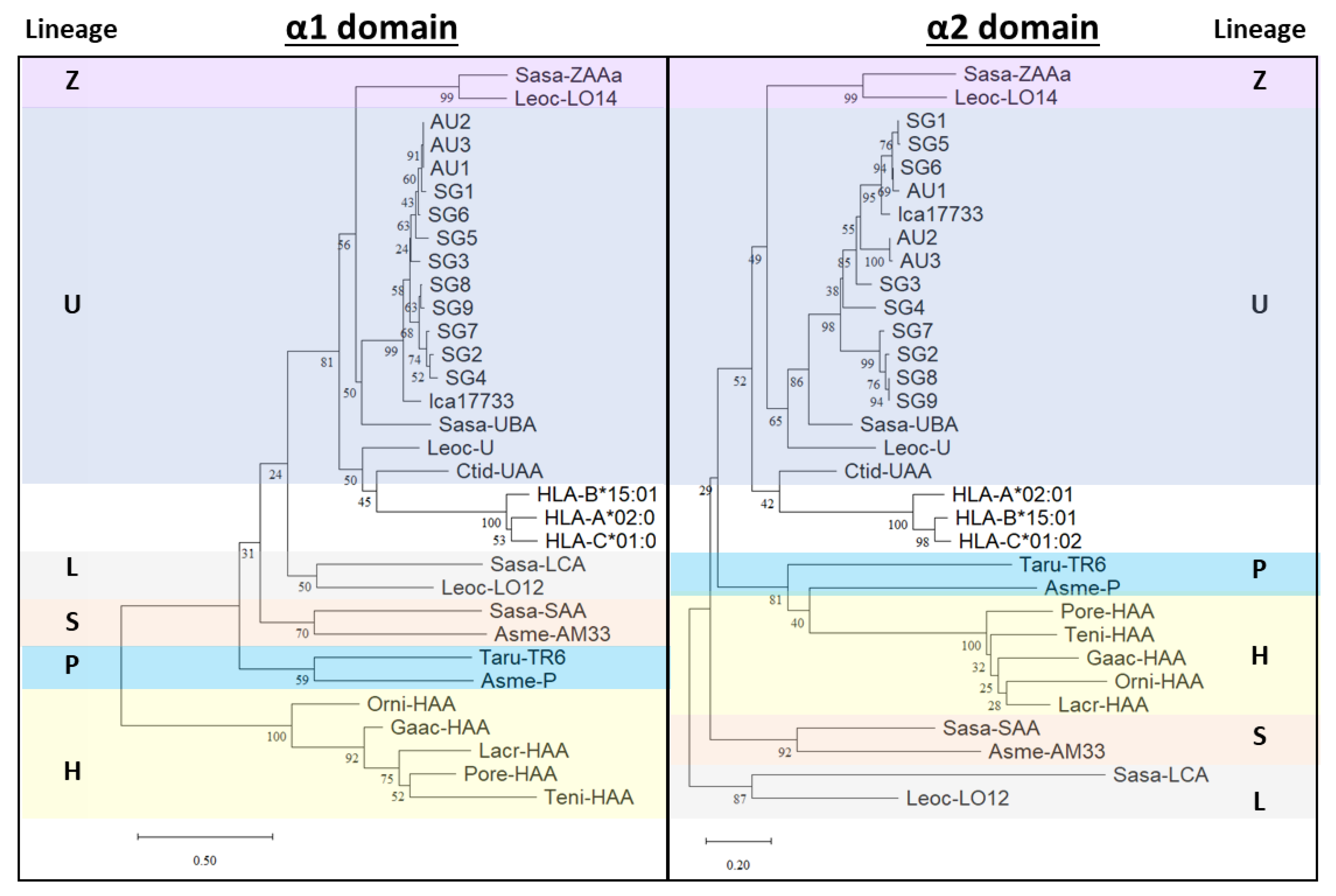
2.2. Phylogenetic Analyses of Asian Seabass MHC-I α1 and α2 Domains
2.3. Protein Modeling
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Data Mining
4.3. Fish Samples
4.4. Experimental Analyses of Asian Seabass Mhc-I α1 and α2 Domains Sequence
4.5. Sequence Alignments, Variability, and Phylogenetic Analysis
4.6. Three-Dimensional Modeling of Human HLA-A2-01 and Grass Carp MHC-I α1 and α2 Domains
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Mathew, G. Taxonomy, Identification and Biology of Seabass (Lates calcarifer). In Course Manual: National Training on Cage Culture of Seabass; CMFRI & NFDB: Kochi, India, 2009. [Google Scholar]
- Gibson-Kueh, S.; Chee, D.; Chen, J.; Wang, Y.H.; Tay, S.; Leong, L.N.; Ng, M.L.; Jones, J.B.; Nicholls, P.K.; Ferguson, H.W. The pathology of ‘scale drop syndrome’ in Asian seabass, Lates calcarifer Bloch, a first description. J. Fish Dis. 2012, 35, 19–27. [Google Scholar] [CrossRef] [PubMed]
- De Groof, A.; Guelen, L.; Deijs, M.; van der Wal, Y.; Miyata, M.; Ng, K.S.; van Grinsven, L.; Simmelink, B.; Biermann, Y.; Grisez, L.; et al. A Novel Virus Causes Scale Drop Disease in Lates calcarifer. PLoS Pathog. 2015, 11, e1005074. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.T.; Jitrakorn, S.; Kayansamruaj, P.; Pirarat, N.; Rodkhum, C.; Rattanarojpong, T.; Senapin, S.; Saksmerprome, V. Infectious spleen and kidney necrosis disease (ISKND) outbreaks in farmed barramundi (Lates calcarifer) in Vietnam. Fish Shellfish Immunol. 2017, 68, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.F.; Ng, K.S.; Grisez, L.; de Groof, A.; Vogels, W.; van der Hoek, L.; Deijs, M. Novel Fish Pathogenic Virus. International Patent Application No. WO2018/029301A1, 15 February 2018. [Google Scholar]
- Chen, J.; Toh, X.; Ong, J.; Wang, Y.; Teo, X.H.; Lee, B.; Wong, P.S.; Khor, D.; Chong, S.M.; Chee, D.; et al. Detection and characterization of a novel marine birnavirus isolated from Asian seabass in Singapore. Virol. J. 2019, 16, 71. [Google Scholar] [CrossRef]
- Girisha, S.; Puneeth, T.; Nithin, M.; Kumar, B.N.; Ajay, S.; Vinay, T.; Suresh, T.; Venugopal, M.; Ramesh, K. Red sea bream iridovirus disease (RSIVD) outbreak in Asian seabass (Lates calcarifer) cultured in open estuarine cages along the west coast of India: First report. Aquaculture 2020, 520, 734712. [Google Scholar] [CrossRef]
- Germain, R.N. MHC-dependent antigen processing and peptide presentation: Providing ligands for T lymphocyte activation. Cell 1994, 76, 287–299. [Google Scholar] [CrossRef]
- Neefjes, J.; Jongsma, M.L.; Paul, P.; Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011, 11, 823–836. [Google Scholar] [CrossRef]
- Madden, D.R. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 1995, 13, 587–622. [Google Scholar] [CrossRef]
- Mungall, A.J.; Palmer, S.A.; Sims, S.K.; Edwards, C.A.; Ashurst, J.L.; Wilming, L.; Jones, M.C.; Horton, R.; Hunt, S.E.; Scott, C.E.; et al. The DNA sequence and analysis of human chromosome 6. Nature 2003, 425, 805–811. [Google Scholar] [CrossRef]
- Norman, P.J.; Norberg, S.J.; Guethlein, L.A.; Nemat-Gorgani, N.; Royce, T.; Wroblewski, E.E.; Dunn, T.; Mann, T.; Alicata, C.; Hollenbach, J.A.; et al. Sequences of 95 human MHC haplotypes reveal extreme coding variation in genes other than highly polymorphic HLA class I and II. Genome Res. 2017, 27, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, J.M.; Katagiri, T.; Hosomichi, K.; Yanagiya, K.; Inoko, H.; Ototake, M.; Aoki, T.; Hashimoto, K.; Shiina, T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics 2007, 59, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Grimholt, U.; Hordvik, I.; Fosse, V.M.; Olsaker, I.; Endresen, C.; Lie, O. Molecular cloning of major histocompatibility complex class I cDNAs from Atlantic salmon (Salmo salar). Immunogenetics 1993, 37, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Nakanishi, T.; Kurosawa, Y. Isolation of carp genes encoding major histocompatibility complex antigens. Proc. Natl. Acad. Sci. USA 1990, 87, 6863–6867. [Google Scholar] [CrossRef] [PubMed]
- Kruiswijk, C.P.; Hermsen, T.T.; Westphal, A.H.; Savelkoul, H.F.; Stet, R.J. A novel functional class I lineage in zebrafish (Danio rerio), carp (Cyprinus carpio), and large barbus (Barbus intermedius) showing an unusual conservation of the peptide binding domains. J. Immunol. 2002, 169, 1936–1947. [Google Scholar] [CrossRef]
- Shum, B.P.; Rajalingam, R.; Magor, K.E.; Azumi, K.; Carr, W.H.; Dixon, B.; Stet, R.J.; Adkison, M.A.; Hedrick, R.P.; Parham, P. A divergent non-classical class I gene conserved in salmonids. Immunogenetics 1999, 49, 479–490. [Google Scholar] [CrossRef]
- Grimholt, U.; Tsukamoto, K.; Hashimoto, K.; Dijkstra, J.M. Discovery of a Novel MHC Class I Lineage in Teleost Fish which Shows Unprecedented Levels of Ectodomain Deterioration while Possessing an Impressive Cytoplasmic Tail Motif. Cells 2019, 8, 1056. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Dijkstra, J.M. Major Histocompatibility Complex (MHC) Genes and Disease Resistance in Fish. Cells 2019, 8, 378. [Google Scholar] [CrossRef]
- Grimholt, U.; Tsukamoto, K.; Azuma, T.; Leong, J.; Koop, B.F.; Dijkstra, J.M. A comprehensive analysis of teleost MHC class I sequences. BMC Evol. Biol. 2015, 15, 32. [Google Scholar] [CrossRef]
- Vij, S.; Kuhl, H.; Kuznetsova, I.S.; Komissarov, A.; Yurchenko, A.A.; Van Heusden, P.; Singh, S.; Thevasagayam, N.M.; Prakki, S.R.; Purushothaman, K.; et al. Chromosomal-Level Assembly of the Asian Seabass Genome Using Long Sequence Reads and Multi-layered Scaffolding. PLoS Genet. 2016, 12, e1005954. [Google Scholar] [CrossRef]
- Schellens, I.M.; Hoof, I.; Meiring, H.D.; Spijkers, S.N.; Poelen, M.C.; van Gaans-van den Brink, J.A.; van der Poel, K.; Costa, A.I.; van Els, C.A.; van Baarle, D.; et al. Comprehensive Analysis of the Naturally Processed Peptide Repertoire: Differences between HLA-A and B in the Immunopeptidome. PLoS ONE 2015, 10, e0136417. [Google Scholar] [CrossRef]
- Rammensee, H.G.; Friede, T.; Stevanoviíc, S. MHC ligands and peptide motifs: First listing. Immunogenetics 1995, 41, 178–228. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Okamura, K.; Yamaguchi, H.; Ototake, M.; Nakanishi, T.; Kurosawa, Y. Conservation and diversification of MHC class I and its related molecules in vertebrates. Immunol. Rev. 1999, 167, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, J.; Salomonsen, J.; Flajnik, M. Evolutionary conservation of MHC class I and class II molecules—Different yet the same. Semin. Immunol. 1994, 6, 411–424. [Google Scholar] [CrossRef]
- Saper, M.A.; Bjorkman, P.J.; Wiley, D.C. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J. Mol. Biol. 1991, 219, 277–319. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, N.; Wei, X.; Lu, S.; Li, S.; Hashimoto, K.; Dijkstra, J.M.; Xia, C. The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between β2-Microglobulin and H Chain Conserved in Evolution. J. Immunol. 2021, 207, 308–321. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Yamaguchi, T.; Grimholt, U. Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species. Immunogenetics 2018, 70, 459–476. [Google Scholar] [CrossRef]
- Kaufman, J. Vertebrates and the evolution of the major histocompatibility complex (MHC) class I and class II molecules. Verh. Dtsch. Zool. Ges. 1988, 81, 131–144. [Google Scholar]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving Genes and Proteins; Bryson, V., Vogel, H.J., Eds.; Academic Press: New York, NY, USA, 1965. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Reiser, J.B.; Legoux, F.; Gras, S.; Trudel, E.; Chouquet, A.; Léger, A.; Le Gorrec, M.; Machillot, P.; Bonneville, M.; Saulquin, X.; et al. Analysis of relationships between peptide/MHC structural features and naive T cell frequency in humans. J. Immunol. 2014, 193, 5816–5826. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, N.; Ma, L.; Zhang, L.; Meng, G.; Xia, C. The Mechanism of β2m Molecule-Induced Changes in the Peptide Presentation Profile in a Bony Fish. iScience 2020, 23, 101119. [Google Scholar] [CrossRef]
- Ellis, J.M.; Henson, V.; Slack, R.; Ng, J.; Hartzman, R.J.; Katovich Hurley, C. Frequencies of HLA-A2 alleles in five U.S. population groups. Predominance Of A*02011 and identification of HLA-A*0231. Hum. Immunol. 2000, 61, 334–340. [Google Scholar] [CrossRef]
- Van Tienderen, P.H.; de Haan, A.A.; van der Linden, C.G.; Vosman, B. Biodiversity assessment using markers for ecologically important traits. Trends Ecol. Evol. 2002, 17, 577–582. [Google Scholar] [CrossRef]
- Sommer, S. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2005, 2, 16. [Google Scholar] [CrossRef]
- Miller, K.M.; Withler, R.E. Mhc diversity in Pacific salmon: Population structure and trans-species allelism. Hereditas 1997, 127, 83–95. [Google Scholar] [CrossRef]
- Landry, C.; Bernatchez, L. Comparative analysis of population structure across environments and geographical scales at major histocompatibility complex and microsatellite loci in Atlantic salmon (Salmo salar). Mol. Ecol. 2001, 10, 2525–2539. [Google Scholar] [CrossRef]
- Miller, K.M.; Kaukinen, K.H.; Beacham, T.D.; Withler, R.E. Geographic heterogeneity in natural selection on an MHC locus in sockeye salmon. Genetica 2001, 111, 237–257. [Google Scholar] [CrossRef]
- Miller, K.M.; Winton, J.R.; Schulze, A.D.; Purcell, M.K.; Ming, T.J. Major histocompatibility complex loci are associated with susceptibility of Atlantic salmon to infectious hematopoietic necrosis virus. In Genetics of Subpolar Fish and Invertebrates; Springer: Berlin/Heidelberg, Germany, 2004; pp. 307–316. [Google Scholar]
- Robinson, J.; Malik, A.; Parham, P.; Bodmer, J.G.; Marsh, S.G. IMGT/HLA database—A sequence database for the human major histocompatibility complex. Tissue Antigens 2000, 55, 280–287. [Google Scholar] [CrossRef]
- Reche, P.A.; Reinherz, E.L. Sequence variability analysis of human class I and class II MHC molecules: Functional and structural correlates of amino acid polymorphisms. J. Mol. Biol. 2003, 331, 623–641. [Google Scholar] [CrossRef] [Green Version]
- Klein, J.; Sato, A.; Nagl, S.; O’hUigín, C. Molecular trans-species polymorphism. Annu. Rev. Ecol. Syst. 1998, 29, 1–21+C1. [Google Scholar] [CrossRef]
- Ratcliffe, F.C.; Garcia de Leaniz, C.; Consuegra, S. MHC class I-α population differentiation in a commercial fish, the European sea bass (Dicentrarchus labrax). Anim. Genet. 2022, 53, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Lukacs, M.F.; Harstad, H.; Bakke, H.G.; Beetz-Sargent, M.; McKinnel, L.; Lubieniecki, K.P.; Koop, B.F.; Grimholt, U. Comprehensive analysis of MHC class I genes from the U-, S-, and Z-lineages in Atlantic salmon. BMC Genom. 2010, 11, 154. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The PhyloFish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef]
- Davidson, W.S.; Koop, B.F.; Jones, S.J.; Iturra, P.; Vidal, R.; Maass, A.; Jonassen, I.; Lien, S.; Omholt, S.W. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol. 2010, 11, 403. [Google Scholar] [CrossRef]
- Kai, W.; Kikuchi, K.; Tohari, S.; Chew, A.K.; Tay, A.; Fujiwara, A.; Hosoya, S.; Suetake, H.; Naruse, K.; Brenner, S.; et al. Integration of the genetic map and genome assembly of fugu facilitates insights into distinct features of genome evolution in teleosts and mammals. Genome Biol. Evol. 2011, 3, 424–442. [Google Scholar] [CrossRef]
- Balas, A.; García-Sánchez, F.; Gómez-Reino, F.; Vicario, J.L. HLA class I allele (HLA-A2) expression defect associated with a mutation in its enhancer B inverted CAT box in two families. Hum. Immunol. 1994, 41, 69–73. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, Z.; Huan, X.; Awate, S.; Schrittwieser, M.; Renia, L.; Ren, E.C. Molecular Characterization of MHC Class I Alpha 1 and 2 Domains in Asian Seabass (Lates calcarifer). Int. J. Mol. Sci. 2022, 23, 10688. https://doi.org/10.3390/ijms231810688
Loh Z, Huan X, Awate S, Schrittwieser M, Renia L, Ren EC. Molecular Characterization of MHC Class I Alpha 1 and 2 Domains in Asian Seabass (Lates calcarifer). International Journal of Molecular Sciences. 2022; 23(18):10688. https://doi.org/10.3390/ijms231810688
Chicago/Turabian StyleLoh, Zhixuan, Xuelu Huan, Sunita Awate, Markus Schrittwieser, Laurent Renia, and Ee Chee Ren. 2022. "Molecular Characterization of MHC Class I Alpha 1 and 2 Domains in Asian Seabass (Lates calcarifer)" International Journal of Molecular Sciences 23, no. 18: 10688. https://doi.org/10.3390/ijms231810688
APA StyleLoh, Z., Huan, X., Awate, S., Schrittwieser, M., Renia, L., & Ren, E. C. (2022). Molecular Characterization of MHC Class I Alpha 1 and 2 Domains in Asian Seabass (Lates calcarifer). International Journal of Molecular Sciences, 23(18), 10688. https://doi.org/10.3390/ijms231810688






