Hydroquinones Inhibit Biofilm Formation and Virulence Factor Production in Staphylococcus aureus
Abstract
:1. Introduction
2. Results
2.1. Antibiofilm and Antimicrobial Activities of Hydroquinone (HQ) and the 17 HQ Derivatives against S. aureus
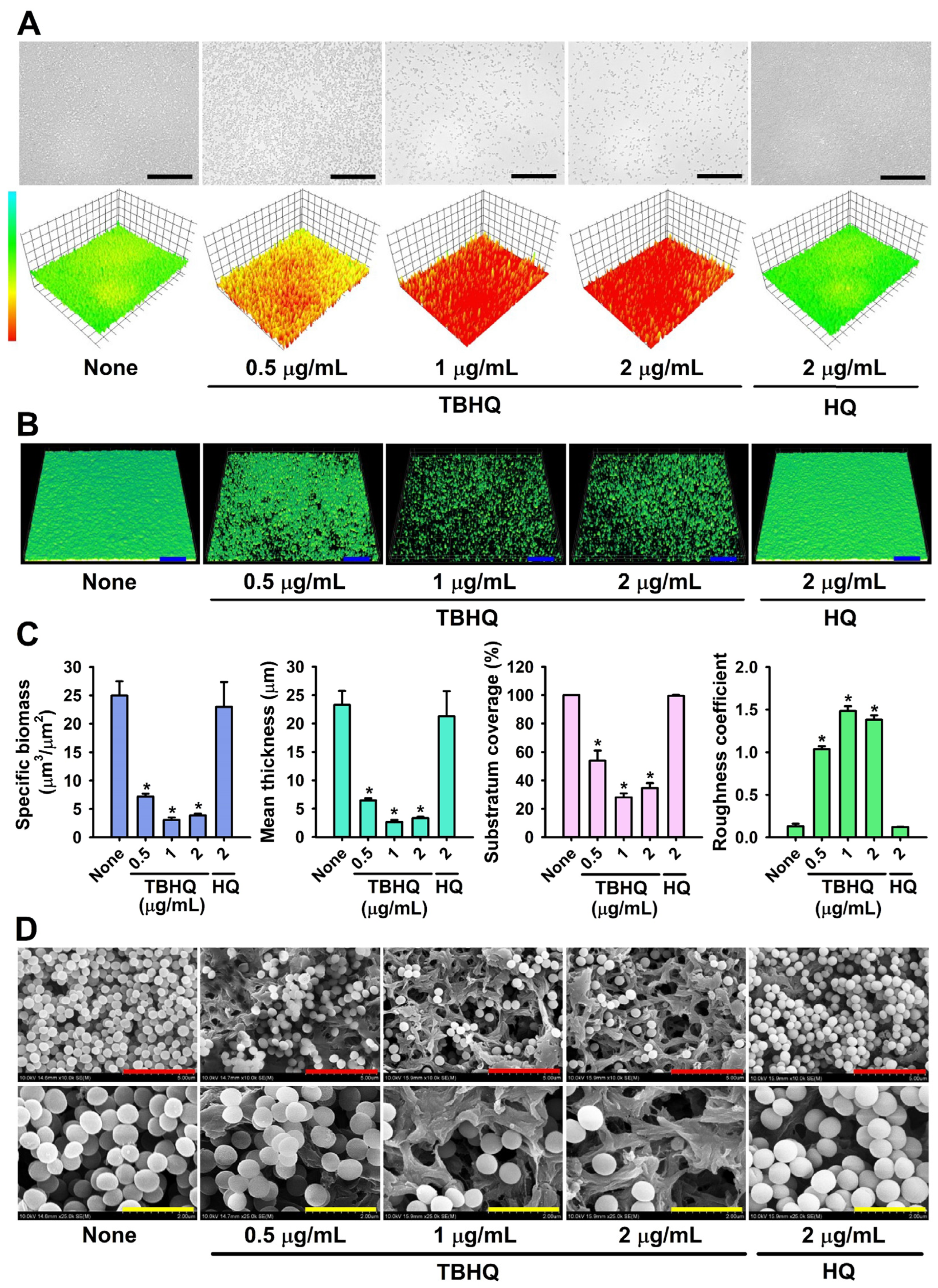
2.2. Microscopic Observations of S. aureus Biofilm Inhibition by TBHQ
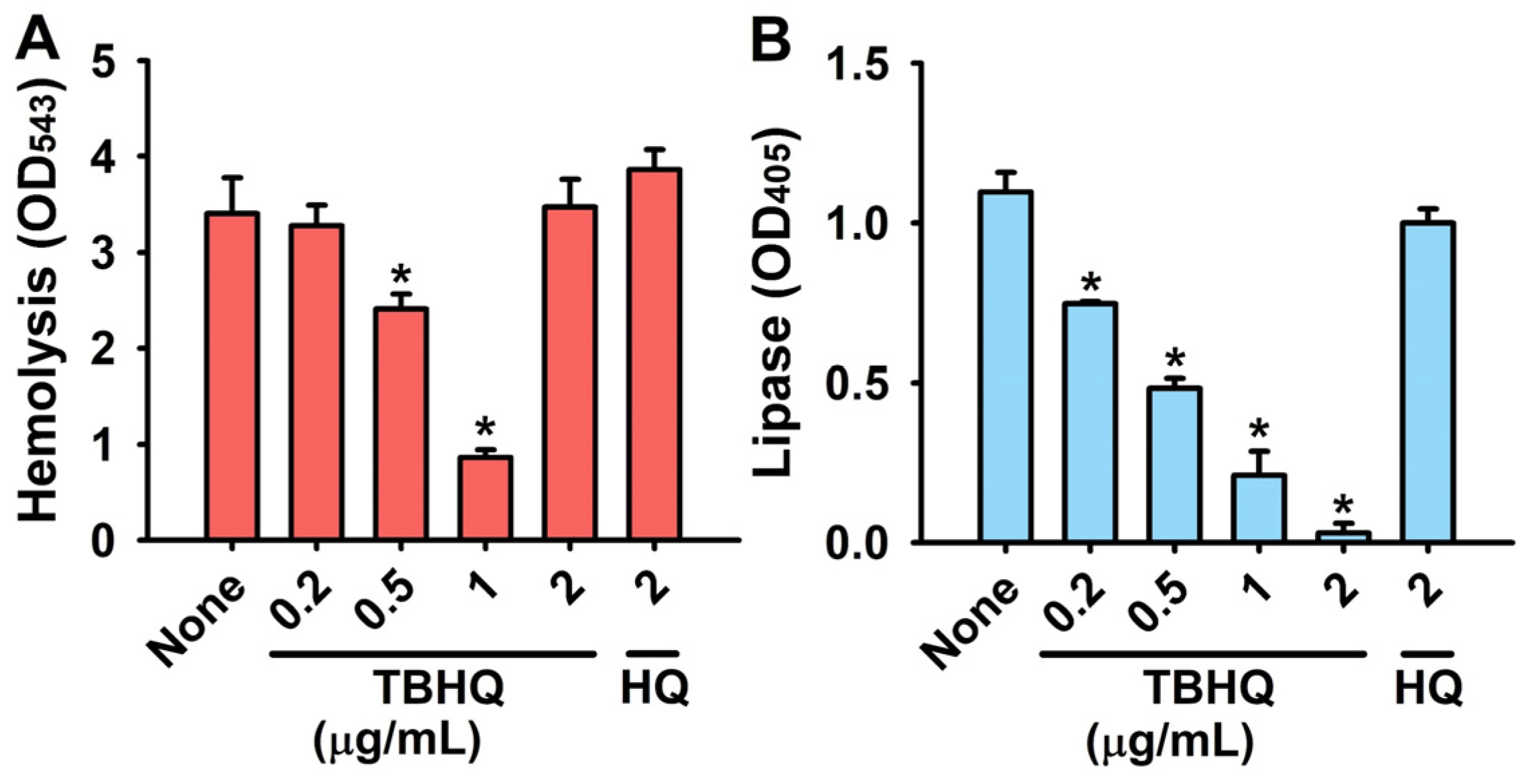
2.3. Inhibitory Effects of TBHQ on Virulence Factors in S. aureus
2.4. TBHQ Repressed the Expressions of Quorum Sensing Gene RNAIII
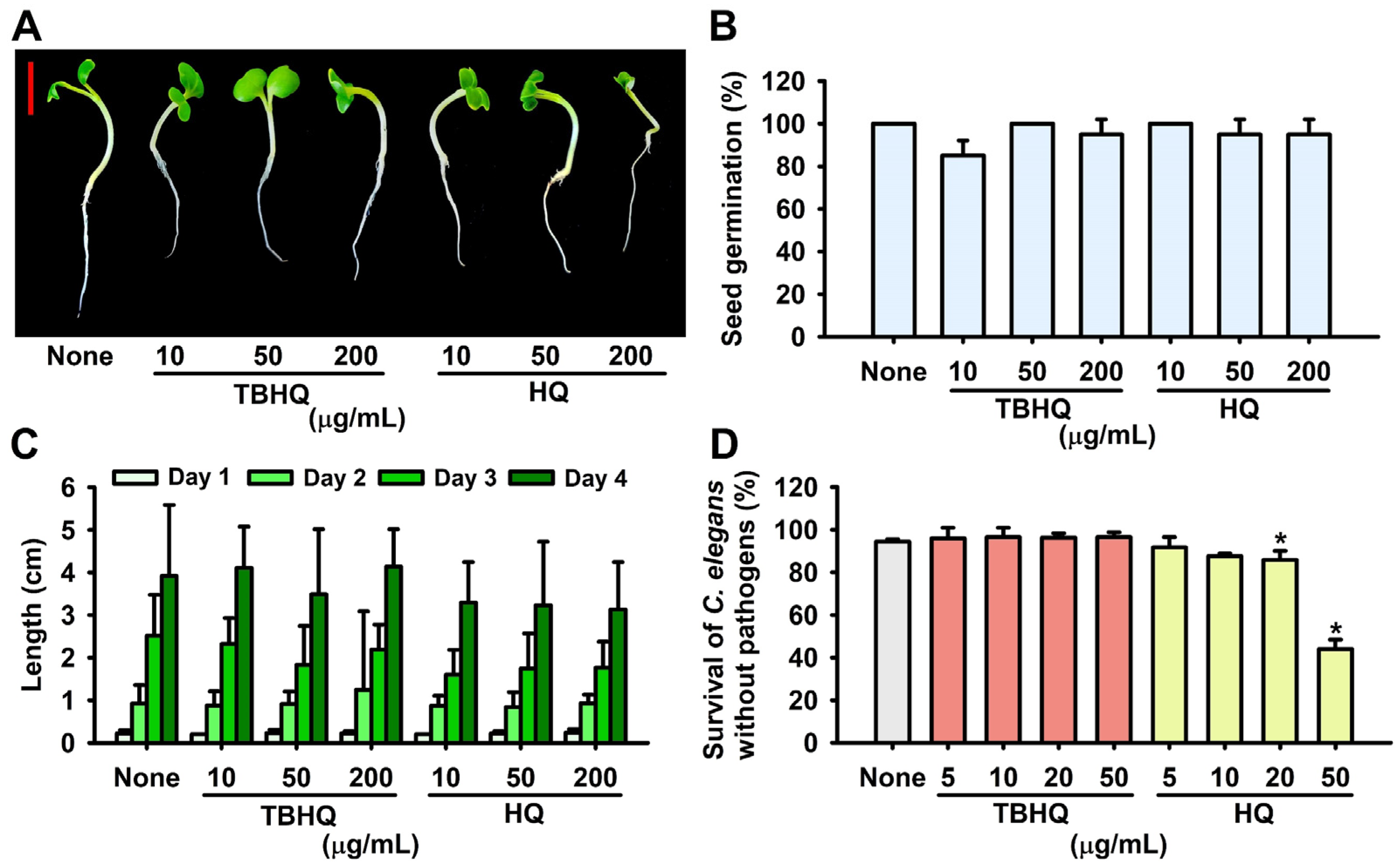
2.5. Toxicity of Hydroquinones in the Plant Germination and Nematode Models
2.6. ADME Profiling of TBHQ
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Growth Measurements, and Materials
4.2. Crystal-Violet Biofilm Assay
4.3. Observations of Biofilm Inhibition by Live Imaging Microscopy, CLSM, and SEM
4.4. Hemolysis Assay
4.5. Lipase Production Assay
4.6. RNA Isolation
4.7. qRT-PCR
4.8. Seed Germination Assay
4.9. Cytotoxicity Assays
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dinges, M.M.; Orwin, P.M.; Schlievert, P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000, 13, 16–34. [Google Scholar] [CrossRef] [PubMed]
- Donlan, R.M.; Costerton, J.W. Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002, 15, 167–193. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Jung, J.; Kim, C.Y.; Kang, H.; Lee, I.H. Antimicrobial peptides encounter resistance of aureolysin during their action on Staphylococcus aureus biofilm. Biotechnol. Bioprocess Eng. 2021, 26, 216–222. [Google Scholar] [CrossRef]
- Xiang, H.; Cao, F.; Ming, D.; Zheng, Y.; Dong, X.; Zhong, X.; Mu, D.; Li, B.; Zhong, L.; Cao, J.; et al. Aloe-emodin inhibits Staphylococcus aureus biofilms and extracellular protein production at the initial adhesion stage of biofilm development. Appl. Microbiol. Biotechnol. 2017, 101, 6671–6681. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Ryu, S.Y.; Lee, J. Calcium-chelating alizarin and other anthraquinones inhibit biofilm formation and the hemolytic activity of Staphylococcus aureus. Sci. Rep. 2016, 6, 19267. [Google Scholar] [CrossRef]
- Song, Z.M.; Zhang, J.L.; Zhou, K.; Yue, L.M.; Zhang, Y.; Wang, C.Y.; Wang, K.L.; Xu, Y. Anthraquinones as potential antibiofilm agents against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 2021, 12, 709826. [Google Scholar] [CrossRef]
- Hoffman, L.R.; D’Argenio, D.A.; MacCoss, M.J.; Zhang, Z.; Jones, R.A.; Miller, S.I. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 2005, 436, 1171–1175. [Google Scholar] [CrossRef]
- Caiazza, N.C.; O’Toole, G.A. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J. Bacteriol. 2003, 185, 3214–3217. [Google Scholar] [CrossRef]
- Nguyen, M.T.; Luqman, A.; Bitschar, K.; Hertlein, T.; Dick, J.; Ohlsen, K.; Bröker, B.; Schittek, B.; Götz, F. Staphylococcal (phospho)lipases promote biofilm formation and host cell invasion. Int. J. Med. Microbiol. 2018, 308, 653–663. [Google Scholar] [CrossRef]
- Clauditz, A.; Resch, A.; Wieland, K.P.; Peschel, A.; Götz, F. Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect. Immun. 2006, 74, 4950–4953. [Google Scholar] [CrossRef] [Green Version]
- Nordlund, J.J.; Grimes, P.E.; Ortonne, J.P. The safety of hydroquinone. J. Eur. Acad. Dermatol. Venereol. 2006, 20, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Ma, C.; He, N.; Zhao, Y.; Xia, D.; Wei, J.; Kang, W. Antimicrobial mechanism of hydroquinone. Appl. Biochem. Biotechnol. 2019, 189, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Morán, A.; Gutiérrez, S.; Martínez-Blanco, H.; Ferrero, M.A.; Monteagudo-Mera, A.; Rodríguez-Aparicio, L.B. Non-toxic plant metabolites regulate Staphylococcus viability and biofilm formation: A natural therapeutic strategy useful in the treatment and prevention of skin infections. Biofouling 2014, 30, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Van Esch, G.J. Toxicology of tert-butylhydroquinone (TBHQ). Food Chem. Toxicol. 1986, 24, 1063–1065. [Google Scholar] [CrossRef]
- Koenig, R.L.; Ray, J.L.; Maleki, S.J.; Smeltzer, M.S.; Hurlburt, B.K. Staphylococcus aureus AgrA binding to the RNAIII-agr regulatory region. J. Bacteriol. 2004, 186, 7549–7555. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-independent target gene control by the agr quorum-sensing system: Insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Horswill, A.R.; Gordon, C.P. Structure-activity relationship studies of small molecule modulators of the staphylococcal accessory gene regulator. J. Med. Chem. 2020, 63, 2705–2730. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef]
- Giacometti, A.; Cirioni, O.; Gov, Y.; Ghiselli, R.; Del Prete, M.S.; Mocchegiani, F.; Saba, V.; Orlando, F.; Scalise, G.; Balaban, N.; et al. RNAIII inhibiting peptide inhibits in vivo biofilm formation by drug-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 1979–1983. [Google Scholar] [CrossRef] [Green Version]
- Ciulla, M.; Di Stefano, A.; Marinelli, L.; Cacciatore, I.; Di Biase, G. RNAIII inhibiting peptide (RIP) and derivatives as potential tools for the treatment of S. aureus biofilm infections. Curr. Top. Med. Chem. 2018, 18, 2068–2079. [Google Scholar] [CrossRef] [PubMed]
- Hammoud Mahdi, D.; Hubert, J.; Renault, J.H.; Martinez, A.; Schubert, A.; Engel, K.M.; Koudogbo, B.; Vissiennon, Z.; Ahyi, V.; Nieber, K.; et al. Chemical profile and antimicrobial activity of the fungus-growing termite strain Macrotermes Bellicosus used in traditional medicine in the republic of benin. Molecules 2020, 25, 5015. [Google Scholar] [CrossRef] [PubMed]
- Mikolasch, A.; Hildebrandt, O.; Schlüter, R.; Hammer, E.; Witt, S.; Lindequist, U. Targeted synthesis of novel β-lactam antibiotics by laccase-catalyzed reaction of aromatic substrates selected by pre-testing for their antimicrobial and cytotoxic activity. Appl. Microbiol. Biotechnol. 2016, 100, 4885–4899. [Google Scholar] [CrossRef] [PubMed]
- Ooi, N.; Chopra, I.; Eady, A.; Cove, J.; Bojar, R.; O’Neill, A.J. Antibacterial activity and mode of action of tert-butylhydroquinone (TBHQ) and its oxidation product, tert-butylbenzoquinone (TBBQ). J. Antimicrob. Chemother. 2013, 68, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Ganjun, Y.; Xiaoyi, Z.; Peibo, L.; Qinghua, Z.; Jincheng, C. New activity for old drug: In vitro activities of vitamin K3 and menadione sodium bisulfite against methicillin-resistant Staphylococcus aureus. Afr. J. Pharmacy Pharmacol. 2014, 8, 451–454. [Google Scholar] [CrossRef]
- Wei, G.; He, Y. Antibacterial and antibiofilm activities of novel cyclic peptides against methicillin-resistant Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 8029. [Google Scholar] [CrossRef]
- Gobin, M.; Proust, R.; Lack, S.; Duciel, L.; Des Courtils, C.; Pauthe, E.; Gand, A.; Seyer, D. A combination of the natural molecules gallic acid and carvacrol eradicates P. aeruginosa and S. aureus mature biofilms. Int. J. Mol. Sci. 2022, 23, 7118. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, S.; Cho, K.H.; Lee, J.-H.; Lee, J. Antibiofilm activities of cinnamaldehyde analogs against Uropathogenic Escherichia coli and Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 7225. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Khadke, S.K.; Lee, J. Antibiofilm and antifungal activities of medium-chain fatty acids against Candida albicans via mimicking of the quorum-sensing molecule farnesol. Microb. Biotechnol. 2021, 14, 1353–1366. [Google Scholar] [CrossRef]
- Weston, S.A.; Parish, C.R. New fluorescent dyes for lymphocyte migration studies: Analysis by flow cytometry and fluorescence microscopy. J. Immunol. Methods 1990, 133, 87–97. [Google Scholar] [CrossRef]
- Kim, Y.-G.; Lee, J.-H.; Park, S.; Lee, J. The anticancer agent 3,3’-diindolylmethane inhibits multispecies biofilm formation by acne-causing bacteria and Candida albicans. Microbiol. Spectr. 2022, 10, e0205621. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, J.-H.; Kim, Y.-G.; Hu, L.; Lee, J. Fatty acids as aminoglycoside antibiotic adjuvants against Staphylococcus aureus. Front. Microbiol. 2022, 13, 876932. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-G.; Lee, J.-H.; Raorane, C.J.; Oh, S.T.; Park, J.G.; Lee, J. Herring oil and omega fatty acids inhibit Staphylococcus aureus biofilm formation and virulence. Front. Microbiol. 2018, 9, 1241. [Google Scholar] [CrossRef] [PubMed]
- Tharmalingam, N.; Rajmuthiah, R.; Kim, W.; Fuchs, B.B.; Jeyamani, E.; Kelso, M.J.; Mylonakis, E. Antibacterial properties of four novel hit compounds from a methicillin-resistant Staphylococcus aureus-Caenorhabditis elegans high-throughput screen. Microb. Drug. Resist. 2018, 24, 666–674. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, Y.-G.; Lee, J. Inhibition of Staphylococcus aureus biofilm formation and virulence factor production by petroselinic acid and other unsaturated C18 fatty acids. Microbiol. Spectr. 2022, 10, e0133022. [Google Scholar] [CrossRef]
- Lin, L.L.; Wu, C.C.; Huang, H.C.; Chen, H.J.; Hsieh, H.L.; Juan, H.F. Identification of microRNA 395a in 24-epibrassinolide-regulated root growth of Arabidopsis thaliana using microRNA arrays. Int. J. Mol. Sci. 2013, 14, 14270–14286. [Google Scholar] [CrossRef] [Green Version]


| Hydroquinones | Structures | MIC | Growth (%) | |
|---|---|---|---|---|
| 10 | 50 | |||
| µg/mL | ||||
| 2,5-Bis(1,1,3,3-tetramethylbutyl)hydroquinone (TBHQ) |  | 5 | 15 | 14 |
| Chlorohydroquinone | 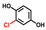 | >400 | 82 | 53 |
| Deoxyarbutin | 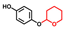 | >400 | 96 | 95 |
| 2,5-Dibromohydroquinone |  | >400 | 86 | 70 |
| 2,3-Dicyanohydroquinone | 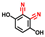 | >400 | 95 | 95 |
| 2,3-Dimethylhydroquinone | 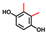 | 50 | 64 | 14 |
| 2,6-Dimethylhydroquinone | 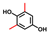 | 50 | 84 | 17 |
| 2,5-Di-tert-butylhydroquinone | 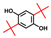 | >400 | 78 | 65 |
| Hydroquinone (HQ) | 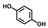 | >400 | 93 | 77 |
| Hydroquinone monobenzyl ether | 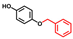 | >400 | 101 | 90 |
| Hydroquinone-O,O′-diacetic acid | 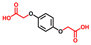 | >400 | 96 | 96 |
| Methylhydroquinone | 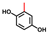 | >400 | 90 | 84 |
| 2-Methoxyhydroquinone | 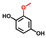 | >400 | 86 | 60 |
| 4-Methoxyphenol | 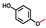 | >400 | 92 | 78 |
| tert-Butylhydroquinone | 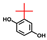 | 200 | 81 | 69 |
| Tetrachlorohydroquinone |  | 200 | 93 | 76 |
| Tetrafluorohydroquinone | 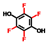 | 400 | 96 | 88 |
| Trimethylhydroquinone | 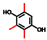 | 100 | 86 | 70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Lee, J.-H.; Kim, Y.-G.; Tan, Y.; Lee, J. Hydroquinones Inhibit Biofilm Formation and Virulence Factor Production in Staphylococcus aureus. Int. J. Mol. Sci. 2022, 23, 10683. https://doi.org/10.3390/ijms231810683
Kim S, Lee J-H, Kim Y-G, Tan Y, Lee J. Hydroquinones Inhibit Biofilm Formation and Virulence Factor Production in Staphylococcus aureus. International Journal of Molecular Sciences. 2022; 23(18):10683. https://doi.org/10.3390/ijms231810683
Chicago/Turabian StyleKim, Sanghun, Jin-Hyung Lee, Yong-Guy Kim, Yulong Tan, and Jintae Lee. 2022. "Hydroquinones Inhibit Biofilm Formation and Virulence Factor Production in Staphylococcus aureus" International Journal of Molecular Sciences 23, no. 18: 10683. https://doi.org/10.3390/ijms231810683
APA StyleKim, S., Lee, J.-H., Kim, Y.-G., Tan, Y., & Lee, J. (2022). Hydroquinones Inhibit Biofilm Formation and Virulence Factor Production in Staphylococcus aureus. International Journal of Molecular Sciences, 23(18), 10683. https://doi.org/10.3390/ijms231810683








