Abstract
ZRSR2 (zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2) is an essential splicing factor involved in 3′ splice-site recognition as a component of both the major and minor spliceosomes that mediate the splicing of U2-type (major) and U12-type (minor) introns, respectively. Studies of ZRSR2-depleted cell lines and ZRSR2-mutated patient samples revealed its essential role in the U12-dependent minor spliceosome. However, the role of ZRSR2 during embryonic development is not clear, as its function is compensated for by Zrsr1 in mice. Here, we utilized the zebrafish model to investigate the role of zrsr2 during embryonic development. Using CRISPR/Cas9 technology, we generated a zrsr2-knockout zebrafish line, termed zrsr2hg129/hg129 (p.Trp167Argfs*9) and examined embryo development in the homozygous mutant embryos. zrsr2hg129/hg129 embryos displayed multiple developmental defects starting at 4 days post fertilization (dpf) and died after 8 dpf, suggesting that proper Zrsr2 function is required during embryonic development. The global transcriptome analysis of 3 dpf zrsr2hg129/hg129 embryos revealed that the loss of Zrsr2 results in the downregulation of essential metabolic pathways and the aberrant retention of minor introns in about one-third of all minor intron-containing genes in zebrafish. Overall, our study has demonstrated that the role of Zrsr2 as a component of the minor spliceosome is conserved and critical for proper embryonic development in zebrafish.
1. Introduction
ZRSR2 (zinc finger CCCH-type, RNA binding motif and serine/arginine rich 2) is an essential splicing factor involved in the recognition of 3′ splice sites during pre-mRNA splicing [1,2]. Pre-mRNA splicing is carried out by the major and minor spliceosomes, which target U2- and U12-type introns, respectively [3,4]. These two types of introns differ in their distinct splice site and branch point sequences [5,6]. Most introns (>99%) in a eukaryotic genome are of a U2-type, while U12-type introns represent <0.5% of the total introns [7,8]. Thus, U2-type and U12-type introns are also termed major and minor introns, respectively. Interestingly, all organisms with minor introns in their genome carry an ortholog of ZRSR2, suggesting its conserved role in their splicing [1].
The human ZRSR2 gene is localized on the X chromosome, and somatic loss-of-function mutations in ZRSR2 are reported in ∼5% of patients with myelodysplastic syndrome (MDS) [9,10,11,12,13,14]. In addition, ZRSR2 mutations are occasionally associated with other hematological diseases, i.e., chronic myelomonocytic leukemia, refractory anemia with ringed sideroblasts, secondary acute myeloid leukemia, and blastic plasmacytoid dendritic cell neoplasm [15,16,17,18]. Transcriptome analyses of ZRSR2-depleted cells and bone marrow samples from MDS patients with ZRSR2 mutations revealed an increase in retention of U12-type introns, while U2-type introns remained largely unaffected [19,20,21]. These findings provide strong support for the essential role of ZRSR2 in the minor spliceosome [19,20,21].
Recently, multiple research groups have investigated the in vivo functions of ZRSR2 using either traditional or conditional knockout mouse models [21,22,23,24]. These studies showed that a functional paralog Zrsr1 compensates for the loss of Zrsr2 in mice [23,24]. ZRSR1 in humans is considered a non-functional pseudogene (ZRSR2P1), but it is functional in mice [22,23,24,25]. While homozygous mutant mice for either Zrsr1 or Zrsr2 developed normally and only exhibited specific phenotypes during adulthood, double knockout mice were embryonic lethal and showed an arrest in embryo development at the 2-cell stage [22,23,24,25]. Thus, studies of these mouse models were unable to reveal specific roles of Zrsr2 in vertebrate embryonic development [21,22,23,24]. Zebrafish are an alternate robust vertebrate model and are suitable for studying the role of genes in embryonic development due to their rapid and external embryonic development and optically clear embryos [26,27,28]. Furthermore, based on the currently available genome sequence, there is no evidence for a Zrsr1 ortholog, and there is a single zrsr2 ortholog in the zebrafish genome. Thus, in this study, we utilized the zebrafish model to investigate the specific roles of zrsr2 in embryonic development.
Here, we report our work on the analysis of zrsr2 expression and the effect of its loss of function during embryonic development. Our data showed that Zrsr2 is essential for multiple processes, including hematopoiesis during embryonic development and its role in the splicing of U12-type introns as a component of the minor spliceosome is conserved in zebrafish. More than one-third of all zebrafish genes that contain U12-type introns (221/612) are dysregulated in the absence of Zrsr2 due to intron retention (IR). Interestingly, a vast majority of these dysregulated genes are involved in RNA and protein processing, thus implicating the importance of these pathways in embryonic development.
2. Results
2.1. The Zebrafish Zrsr2 Protein Has All the Functional Domains of Human ZRSR2 and an Extra C-Terminal Region
The zebrafish genome contains a single copy of the zrsr2 gene localized on chromosome 11. It encodes two major isoforms, isoform 201 (635 amino acids in length; GenBank accession number: XM_021479948) and isoform 204 (483 amino acids in length; GenBank accession number: XM_021479949). Isoform 204 has a longer 5′ untranslated region due to alternate splicing between exons 3 to 6, and its translation start site lies in exon 6 (Figure 1A). Thus, the protein encoded by isoform 204 lacks the first 152 amino acids but is otherwise identical to isoform 201 (Figure 1B). The major ZRSR2 isoform in humans is isoform 201 which contains all known functional domains (482 amino acids in length; GenBank accession number: NM_005089). The alignment of both zebrafish isoforms with human ZRSR2-201 showed an overall protein similarity of 58%, with a higher level of similarity between the functional domains (Figure 1B, Supplementary Figure S1). Interestingly, there is an extra sequence of 140 amino acids at the C-terminus of the zebrafish Zrsr2 compared to the human ZRSR2 (Figure 1B, Supplementary Figure S1). Analysis of this extra C-terminus part by BLASTp (https://blast.ncbi.nlm.nih.gov/ accessed on 21 October 2019) showed that it is also present in the ZRSR2 orthologs from other species of fish. However, this region did not show any recognizable functional domains.
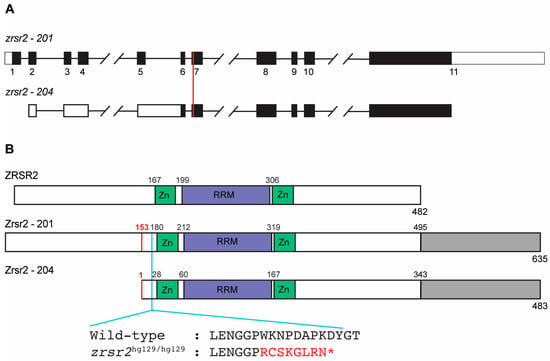
Figure 1.
Genomic organization and functional domains of zebrafish Zrsr2. (A) A schematic of the genomic structures of zrsr2 isoforms 201 and 204, with exons shown as rectangles and introns as lines connecting the exons. Filled rectangles mark the coding sequence, and white rectangles mark the untranslated regions. The vertical red line marks the genomic target region of the sgRNA used for CRISPR/Cas9 knockout. (B) The alignment of the human ZRSR2 protein with isoforms 201 and 204 of zebrafish Zrsr2, showing the location of conserved functional domains: Zn = Zinc finger domains, RRM = RNA recognition motif. Gray color marks the extra C-terminal region in zebrafish Zrsr2. Black numbers indicate the amino acid number for the start of each domain. Red numbers mark the methionine at position 153 in isoform 201, which is the translation start site of Zrsr2-204. Blue line marks the location of the CRISPR/Cas9-induced mutation and partial amino acid sequences of WT and mutant proteins, with red color marking the frameshift and premature termination codon marked as *.
2.2. Both Isoforms of zrsr2 Are Ubiquitously Expressed during Embryonic Development
To understand the role of zrsr2 in normal embryonic development, we examined the spatial and temporal patterns of the expression of zrsr2 by RT-PCR, quantitative RT-PCR (qRT-PCR), and whole mount in situ hybridization (WISH) in wild-type (WT) embryos from a series of developmental stages. RT-PCR and qRT-PCR were performed using primers unique to the shared regions of isoforms 201 and 204 (Supplementary Table S1). Overall, the traditional and quantitative RT-PCR data were consistent with each other and revealed that zrsr2 is expressed throughout embryonic development, with some variation in the levels of expression of isoforms 201 and 204 at specific stages (Figure 2A, Supplementary Figure S2). Interestingly, the data using isoform-specific primers revealed that isoform 201 is maternally inherited, while the expression of isoform 204 begins after the start of zygotic transcription (Figure 2A, Supplementary Figure S2). The RT-PCR product from the region shared by both isoforms (Supplementary Figure S2) was cloned to generate a zrsr2 riboprobe for WISH (Figure 2B). Our data revealed that, subsequent to the ubiquitous expression at the beginning of zygotic transcription, zrsr2 expression is enriched in the anterior region of the embryo, specifically in the brain from after 24 h post-fertilization (hpf) to 5 dpf (Figure 2B). These data are consistent with the findings from a mouse model in that a large proportion of U12-type intron-containing genes are expressed in the brain [29]. Specific zrsr2 expression was also observed in the ventral side of the tail at 24 hpf and in the intestinal bulb at 5 dpf (Figure 2B). Overall, our WISH data suggested that, in addition to being ubiquitously expressed as an essential splicing factor, zrsr2 is highly expressed in certain developing tissues, which may indicate regions that are actively growing and, thus, more dependent on the splicing machinery.
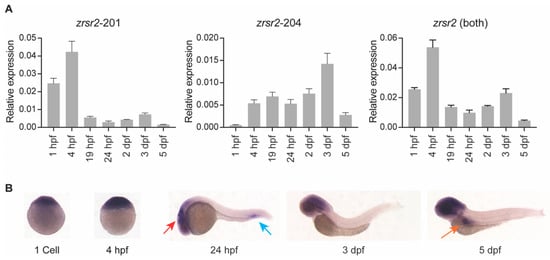
Figure 2.
zrsr2 expression during zebrafish embryonic development. (A) qRT-PCR data showing expression of zrsr2 isoforms using primers unique to each isoform, and total expression using primers common to both isoforms in WT embryos from a series of stages during development as marked across the bottom of each plot. (B) WISH with zrsr2 antisense probe, showing its sites of expression in WT embryos at 1 cell, 4 hpf, 24 hpf, 3 dpf, and 5 dpf as marked. Specific zrsr2 expression is seen in the developing brain (red arrow) and the peripheral blood island (blue arrow) at 24 hpf, and in the intestinal bulb (orange arrow) at 5 dpf.
2.3. zrsr2 Knockout Results in Abnormal Embryonic Development, Hematopoietic Defects, and Lethality by 8 dpf
To understand the in vivo functions of Zrsr2 during vertebrate development, we used CRISPR/Cas9 technology to generate a zebrafish zrsr2-knockout model. To ensure a complete loss-of-function model, a single guide RNA was designed to target exon 7, which is shared by both isoforms (Figure 1A) and is upstream of all known functional domains (Figure 1B). We selected a mutant allele with an 11 bp deletion (c.495delACCCTGGAAAA; p.Trp167Argfs*9), which leads to loss of all functional domains due to a frameshift followed by premature truncation (Figure 1B). The RT-PCR and sequencing of WT and the homozygous mutant embryos confirmed that the mutant allele was expressed, and the deletion did not cause any detectable splicing defects in the zrsr2 transcript (Supplementary Figure S3A). This mutant zebrafish line, referred to as zrsr2hg129/hg129 in subsequent sections, provided a robust knockout model for the investigation of the in vivo functions of Zrsr2 during vertebrate development.
To determine the effect that loss of Zrsr2 function had on embryonic development, we observed the morphological phenotypes of embryos obtained from pairwise crosses of heterozygous fish for several days after fertilization. The zrsr2hg129/hg129 embryos displayed normal morphology for the first three days of development and normal expression of the markers of early development, such as kdrl, myod1, and shh (Supplementary Figure S3B). At 4 dpf, the zrsr2hg129/hg129 embryos could be distinguished from their WT and heterozygous siblings by a smaller head size, lack of a proper jaw, and mild (∼75% of mutant embryos) to moderate (∼25% of mutant embryos) cardiac edema (Figure 3A). Further analysis of the jaw region by Alcian blue staining revealed defects in the mandibular cartilage and pharyngeal arches in zrsr2hg129/hg129 embryos (Figure 3B). Between 5 dpf and 7 dpf, the cardiac edema and jaw defects became more pronounced, and the swim bladder failed to inflate in the mutant embryos (Figure 3A). All zrsr2hg129/hg129 embryos died by 8 dpf (Figure 3C). These findings suggested that Zrsr2 is essential for multiple processes during embryonic development and the normal development of zrsr2hg129/hg129 embryos for the first 3 dpf is most likely due to the compensation by maternally supplied zrsr2 mRNA.
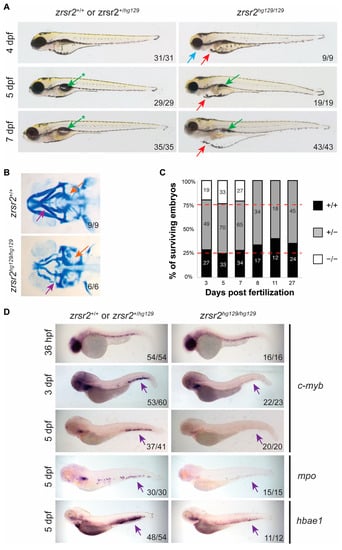
Figure 3.
zrsr2-knockout embryos display morphological and hematopoietic defects. (A) Morphological phenotypes of zrsr2hg129/hg129 embryos at 4, 5, and 7 dpf are shown in the right panel compared to their WT siblings in the left panel. Blue arrow marks defective jaw, red arrows mark cardiac edema, green arrows mark uninflated swim bladder, and green arrows with asterisks mark the normal inflated swim bladder in WT embryos. All images depict lateral views of the embryos. (B) Ventral view of embryos stained with Alcian blue depicting the cartilage of developing jaw of WT embryos (top panel) and zrsr2hg129/hg129 embryos (bottom panel) at 4 dpf with purple arrows marking the mandibular cartilage and orange arrows marking the pharyngeal arches. (C) Survival of embryos collected from adult zrsr2hg129/+ in-cross. The bar graph displays the genotype ratios as a percentage of total sampled embryos, and red lines indicate the expected Mendelian offspring ratios (1:2:1; zrsr2+/+: zrsr2hg129/+: zrsr2hg129/hg129). Numbers in the bars represent the number of embryos of each genotype. Clutches were sampled for genotype makeup at multiple time points as marked on the X-axis. zrsr2hg129/hg129 embryos died by 8 dpf, while zrsr2hg129/+ and zrsr2+/+ embryos survived to adulthood. (D) WISH for definitive hematopoietic markers c-myb (36 hpf, 3 dpf, and 5 dpf), mpo (5 dpf) and hbae1 (5 dpf), showing expression in zrsr2hg129/hg129 embryos (right panel) compared to their siblings (left panel). Lateral views of embryos are shown in all images, with purple arrows marking the CHT region, showing reduced expression in zrsr2hg129/hg129 embryos.
Since recurrent ZRSR2 somatic mutations are found in hematological diseases, we investigated the role of Zrsr2 in hematopoiesis ontogeny by WISH with lineage-specific markers for both primitive and definitive hematopoiesis [30,31]. The onset of primitive hematopoiesis was normal in zrsr2-knockout embryos, as indicated by the normal levels of expression of erythroid and myeloid markers, gata1, scl, l-plastin, and mpo, at 14 to 24 hpf (Supplementary Figures S4A–C). Interestingly, zrsr2hg129/hg129 embryos displayed mild anemia at 2 dpf, as indicated by a significantly reduced o-dianisidine staining compared to their siblings (Supplementary Figure S4D–E). Expression of the hematopoietic stem cell (HSC) markers, runx1 and c-myb, showed that the emergence of HSCs is normal in the mutant embryos (Figure 3D, Supplementary Figure S5). However, no c-myb expression was detected in the caudal hematopoietic tissue (CHT) region in 3 and 5 dpf mutant embryos (Figure 3D). The lack of HSCs in the CHT region is accompanied by a subsequent reduction in the cells of the erythroid (hbae1) and myeloid (mpo) lineages at 5 dpf (Figure 3D). These data suggested that Zrsr2 is important for the maintenance of HSCs during definitive hematopoiesis.
To confirm that the observed phenotypes in our mutant line are specific to the loss of Zrsr2 as opposed to being an off-target effect of CRISPR/Cas9 targeting, we used the crispant strategy, in which mosaic-injected embryos, with phenotypic penetrance based on the frequency of induced indels, are evaluated [32]. An independent sgRNA was designed to target exon 8, and the injected embryos were observed for morphological phenotypes using the original sgRNA as control. As expected, the crispant embryos for both sgRNAs recapitulated the morphological phenotypes observed in the germline mutants (Supplementary Figure S6A). Furthermore, similar to the zrsr2hg129/hg129 embryos, crispants for both sgRNAs displayed a reduced expression of c-myb in the CHT at 3 dpf (Supplementary Figure S6B). Combining these data with our subsequent observations of zrsr2hg129/hg129 embryos for up to five generations, consistently showing the phenotype-to-genotype correlations, we feel confident that our zrsr2hg129/hg129 line is a robust model for the analysis of Zrsr2 deficiency. Overall, our data revealed that Zrsr2 is essential for multiple processes, including hematopoiesis ontogeny during embryonic development in zebrafish.
2.4. Zrsr2 Deficiency Leads to Perturbations in Metabolic Processes
We used RNA sequencing (RNA-Seq) from 3 dpf zrsr2hg129/hg129 embryos before the onset of gross morphological defects to identify the biological processes affected by the loss of Zrsr2 in zebrafish. RNA-Seq was performed in triplicates from WT (zrsr2+/+ embryos) and mutant (zrsr2hg129/hg129) embryos. All raw data were judged to be of good quality by their similar distributions of read counts, genomic coverage, and number of mappable reads in all six samples, indicating there were no outlier samples (Supplementary Figure S7A,B, Supplementary Table S2). RNA-Seq data revealed the reduced expression of both zrsr2 isoforms, indicating a premature stop-codon mediated mRNA decay in the mutant embryos (Supplementary Figure S7C). Analysis of the RNA-Seq data with DESeq2 [33], using a fold change cut-off of 2 and p-adjusted value cutoff of 0.05, identified 1372 differentially expressed genes (DEGs) between zrsr2hg129/hg129 and WT embryos (Figure 4A, Supplementary Table S3). Of these DEGs, 455 genes were upregulated, and 917 genes were downregulated in the zrsr2hg129/hg129 embryos compared to the WT embryos (Supplementary Table S3). Interestingly, gene ontology (GO) analysis of the upregulated genes did not identify any specific biological processes to be significantly affected by the loss of Zrsr2, whereas the downregulated genes affected the metabolic processes, including the transport of vitamins and lipids (Figure 4B). Consistent with our data, widespread changes in metabolic pathways were observed in a Drosophila model with another disrupted minor splicing component, U6atac snRNA [34]. Overall, our data showed that the global transcriptomic changes caused by the Zrsr2 deficiency affected the general metabolic pathways essential for the proper development and homeostasis of embryonic tissues.
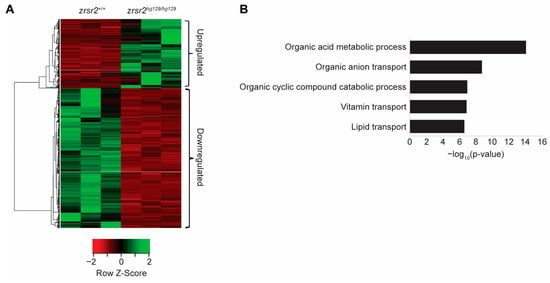
Figure 4.
Loss of zrsr2 leads to global gene expression changes in metabolic processes. (A) Heatmap of the differentially expressed genes in zrsr2hg129/hg129 embryos compared to their WT controls at 3 dpf, with the color key shown below the heatmap. An amount of 455 genes were upregulated, and 917 genes were downregulated in the zrsr2hg129/hg129 samples (fold change ≥ 2, p < 0.05). (B) Bar graph of the top 5 biological processes enriched among 917 downregulated genes in zrsr2hg129/hg129 embryos at 3 dpf, with enrichment visualized by −log10 (p-value). Metabolic processes and transport of ions, vitamins, and lipids are among the most significantly enriched pathways among the downregulated genes.
2.5. Loss of Zrsr2 Leads to the Preferential Retention of Minor Introns in Genes Involved in RNA and Protein Processing
As a component of the minor spliceosome, ZRSR2 is known to be essential for the splicing of the U12-type of introns [1]. Its loss of function in other biological models is associated with an increase in the retention of U12-type introns [19,21,23,24,35]. Therefore, we analyzed our RNA-Seq data for IR events with IRFinder [36]. We identified 284 differential IR events between zrsr2hg129/hg129 and WT embryos (Supplementary Table S4). These 284 introns led to the abnormal splicing of 259 unique genes, as more than one intron was retained in some genes (Supplementary Table S4).
Next, we used “The Intron Annotation and Orthology Database” (IAOD) [7] to determine if the retained introns were of the U2- or U12- subtypes. As reported previously in ZRSR2-depleted human and mouse models [19,20,21,24], most of the retained introns (218/284) in the zrsr2hg129/hg129 mutant zebrafish were of the U12-type (Figure 5A). In addition, 19 out of 51 of the preferentially retained U2-type introns were found in those genes that also contained a U12-type intron (Figure 5A). Among the genes with retention of more than one intron, 15 genes showed the retention of both U2- and U12-type introns (Supplementary Table S4). Combining all these data, we found that, out of the 259 genes affected by IR in zrsr2hg129/hg129 mutants, 221 genes contained the U12-type introns (Supplementary Table S4). Thus, our data provide strong support for the notion that the role of Zrsr2 in the splicing of U12-type introns is conserved in zebrafish.
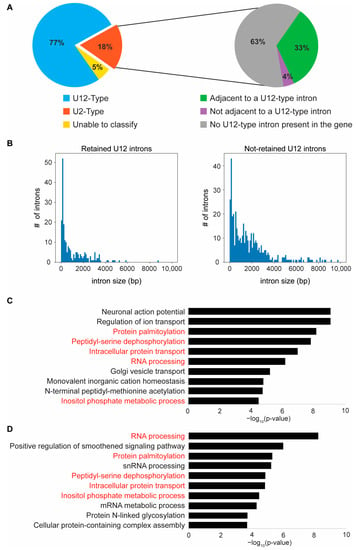
Figure 5.
Loss of zrsr2 leads to the differential retention of U12-type introns in RNA and protein processing genes. (A) Pie charts depicting the classifications for all differentially retained introns between zrsr2hg129/hg129 and WT embryos into U12-type (blue) and U2-type (orange) introns (left) and the further classification of the retained U2-type introns (right) by whether they are adjacent to a known U12-type intron (green) or not adjacent but in a gene with a U12-type intron (purple), or in a gene with no known U12-type introns (grey). Some of the retained introns could not be classified into either category (yellow). (B) Plots of intron size distribution in the retained U12-type introns (left) and the non-retained U12-type introns (right). (C) The top 10 biological processes enriched in all genes (612) containing U12-type introns in the zebrafish genome according to IAOD. (D) The top 10 biological processes enriched in the set of U12-type intron containing genes (221) that have a differential IR between WT and zrsr2hg129/hg129 embryos at 3 dpf, with enrichment visualized by −log10 (p-value). Five of the ten top pathways (written in red) are common in both gene sets (C,D).
A recent study [7] has identified a total of 662 U12-type introns that occur in 612 unique genes in zebrafish. Analysis of our RNA-Seq data showed that the majority of these U12-intron-containing genes are expressed in our dataset. Therefore, to understand why only a subset of all U12-type introns are retained (218 out of 662) in zrsr2 mutants, we compared their sequence features with those not showing significant retention (444 out of 662). Our analysis revealed that while there were no differences in their nucleotide composition, the distribution of the GT-AG versus AT-AC splice sites, or sequence features around both splice sites, showed that the retained introns appeared to be smaller in size (Mean 1077 bp, Median 451 bp) compared to the non-retained introns (Mean 3556 bp, Median 1398 bp) (Figure 5B, Supplementary Figure S8, Supplementary Table S5). These data are consistent with previous findings [19] and suggest that the splicing of smaller U12-type introns is more sensitive to the loss of Zrsr2. To determine if the retention of minor introns affected the expression of corresponding genes, we queried our list of DEGs (Supplementary Table S3) for overlap with those genes showing retention of minor introns (Supplementary Table S4). Interestingly, only 43 genes (26 up, 17 down) showed the retention of minor introns and differential expression in the absence of Zrsr2 function. A similar comparison between genes without minor intron retention and differential expression identified 57 genes (29 up, 28 down). These findings indicate that the global changes in gene expression detected in zrsr2hg129/hg129 mutants are not directly due to changes in the expression of the genes that contain U12-type introns but could represent the downstream effect of their aberrant splicing.
To identify the biological processes that are affected by the retention of the minor introns, we first identified all biological processes that require a properly functioning minor spliceosome by GO analysis of all U12-type intron-containing genes (612) in zebrafish (Figure 5C). Similar to the observations in other organisms [3,5], U12-type introns are present in the genes involved in neuronal action potential, ion transport, protein processing and transport, and RNA processing in zebrafish (Figure 5C). In comparison, the GO analysis of the 221 U12-intron-containing genes with differential IR in zrsr2hg129/hg129 mutants (Figure 5D) showed that 5 of the top 10 biological processes that require the minor spliceosome were affected by the Zrsr2 deficiency, particularly those genes involved in RNA and protein processing pathways (Figure 5C,D). We found that, in the gene set with the retained introns, 48 genes were involved in RNA processing (i.e., RNA binding, RNA modification, RNA splicing, regulation of mRNA processing, RNA metabolic process, etc.), and 46 genes are involved in protein processing (i.e., protein modification by glycosylation, acetylation, phosphorylation, palmitoylation, regulation of protein stability by ubiquitination, deubiquitination, proteolysis and degradation, etc.) (Supplementary Table S4). Thus, our data suggest that Zrsr2 is specifically required for the regulation of genes involved in RNA and protein processing pathways during embryonic development diseases.
3. Discussion
ZRSR2 is an essential component of both the major and minor spliceosomes and is frequently mutated in hematopoietic disorders, particularly MDS. Recent studies have uncovered its requirement in the proper splicing of U12-type introns as a component of the minor spliceosome [19,20,21]. However, its role during vertebrate embryonic development is not known. In this study, we took advantage of the zebrafish model to study the impact of Zrsr2 deficiency on vertebrate embryonic development. We also showed that role of ZRSR2 in the splicing of the U12-type introns is conserved in zebrafish.
It has been suggested that minor introns are evolutionarily conserved as a mechanism for the regulation of cellular differentiation and multicellular growth [37]. The tissue-specific expression of genes with U12-type introns is regulated by their alternative splicing in both mice and humans [29]. Consistent with these findings, we observed that zrsr2 is expressed throughout early zebrafish development when the cells are rapidly dividing and differentiating. The major isoform (zrsr2-201) is both maternally inherited and zygotically expressed throughout embryogenesis, while an alternate isoform (zrsr2-204) is expressed after the start of zygotic transcription. Our data on their differential expression during development suggests that the proteins encoded by each isoform perform specific functions which can be explored further using isoform-specific antibodies. zrsr2 is highly expressed in the anterior region of the embryos and intestinal bulb, as has also been reported for other components of the minor spliceosome, e.g., dhx15 and rnpc3 [38,39]. The Zrsr2-deficient embryos developed normally for the first 3 days after fertilization, likely due to the presence of maternally-supplied WT zrsr2 mRNA. At 4 dpf, they started to display pleiotropic phenotypes, indicating the requirement of proper Zrsr2 function in several developing tissues. It is possible that these tissues are more dependent on the minor spliceosome to produce appropriate mRNA transcripts for their growth and differentiation. Similar to the embryonic lethality observed upon disruption of the other components of the minor spliceosome in Drosophila and zebrafish [34,38,39], zrsr2-knockout fish displayed lethality by 8 dpf. Thus, our study adds to the growing evidence that the proper functioning of the minor spliceosome is critical for early animal development.
Our zrsr2-knockout zebrafish model allowed us to investigate the role of ZRSR2 during vertebrate embryonic hematopoiesis, as the previous studies in mice were inconclusive due to the compensation by its functional paralog, Zrsr1 [23,24]. zrsr2-knockout zebrafish embryos displayed a lack of HSCs in the CHT region at 3 dpf and reduced numbers of cells from the erythroid and myeloid lineages at 5 dpf, suggesting that a functional Zrsr2 is required for normal hematopoietic development. However, we did not observe significant dysregulation of the genes involved in the hematopoietic pathway via RNA-Seq. This is probably due to our approach of using the whole embryo (minus the heads) for RNA-Seq.
Our study also demonstrated that the critical role of Zrsr2 in the regulation of the expression of genes with U12-type introns as a component of the minor spliceosome is conserved in zebrafish. Similar to human MDS bone marrow samples from patients with ZRSR2 mutations, zrsr2-deficient zebrafish showed preferential retention of more than one-third of all U12-type introns (218/662). Specifically, the e2f and mapk gene families seem to be most affected by retained introns when the minor spliceosome function is disrupted [19,20,24,35,39]. Among the genes with retained introns in zrsr2hg129/hg129 mutants, we also identified multiple genes from both the e2f and mapk gene families (e2f4, mapk3, mapk8a, mapk12a, mapk14a). A recent study has provided strong evidence for a link between the retention of a specific U12-type intron in LZTR1 and the pathogenesis of MDS [21]. Therefore, we checked our list of genes with differential IR events and confirmed that this U12-type intron in lztr1 was conserved in zebrafish and was differentially retained in zrsr2hg129/hg129 embryos (p < 0.0001) (Supplementary Table S4).
Furthermore, we found that the genes affected by IR in the absence of a functional Zrsr2 in zebrafish during embryonic development are important for the processing of RNA and proteins and protein transport. These findings suggest that Zrsr2 is important for the regulation of these biological processes during embryonic development in zebrafish. Thus, it is possible that the morphological and hematopoietic defects observed in zrsr2-knockout embryos are due to the indirect effect of the dysregulation of these important cellular functions in tissues that are actively growing and differentiating. In conclusion, our study of zrsr2-deficient zebrafish provides strong support for the critical role of Zrsr2 in the splicing of minor introns in RNA and protein processing genes that play a crucial role in multiple developmental processes during embryogenesis. Further studies using hematopoietic cells from our zrsr2-knockout model for RNA-Seq will lead to a better understanding of the role of Zrsr2 in hematological diseases.
4. Materials and Methods
4.1. Zebrafish Husbandry and Ethics Statement
Zebrafish (Danio rerio) of WT strain TAB5 maintained in our laboratory were used in this study. All experiments were performed in compliance with the National Institutes of Health guidelines for animal handling and research using an Animal Care and Use Committee-approved protocol (G-05-5 assigned to RS). Zebrafish husbandry and embryo staging were performed as previously described [40]. All embryos were raised in an incubator at 28.5 °C until 5–6 dpf. Larvae and adult zebrafish were housed in a recirculating aquatic system with a light/dark cycle (14/10 h, respectively) and a water temperature of 28 °C.
4.2. Generation of zrsr2 Mutants, Crispants and Genotyping
Two single-guide RNAs targeting exons 7 (5′–GGAGCATCTGGGTTTTTCCA–3′) and 8 (5′–TGCCAGTCTGGAGTACAGTG–3′) were designed and synthesized as described previously [41]. WT zebrafish embryos were injected with 50 pg of each single guide RNA mixed with 150 pg of Cas9 mRNA. Embryos injected with sgRNA to exon 7 were grown to adulthood for generation of zrsr2 knockout zebrafish model. Founder screening, identification of heterozygous F1 progeny, and all subsequent genotyping during line maintenance and phenotype analysis were performed by fluorescent PCR, as described previously [41], using primers listed in Supplementary Table S1. For crispant analysis, injected embryos were either harvested and fixed at 3 dpf for WISH or observed for morphological phenotypes up to 5 dpf, and harvested for DNA extraction. Fluorescent PCR and CRISPR-STAT [42] were performed on all crispant embryos to correlate the observed phenotypes with the reduction of WT peak and the presence of indel alleles.
4.3. RNA Extraction, RT-PCR, qRT-PCR, and Sequencing
RT-PCR and qRT-PCR were used to determine the expression of both isoforms of zrsr2 throughout embryonic development using primers described in Supplementary Table S1. RNA was extracted from WT embryos collected at 1 hpf, 4 hpf, 19 hpf, 24 hpf, 2 dpf, 3 dpf, and 5 dpf, using Direct-zol RNA Microprep kit (Zymo Research, Irvine, CA, USA). cDNA for RT-PCR was synthesized using SuperScript IV One-Step RT-PCR System (Thermo Fisher Scientific, Waltham, MA, USA). RT-PCR products were analyzed by gel electrophoresis using 1.5% agarose gels. cDNA for qRT-PCR was generated with iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). qPCR was then performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s instruction, using actb2 primers (Supplementary Table S1) as internal controls. RT-PCR with primers shared by isoforms 201 and 204 was used to confirm the presence of CRISPR/Cas9-induced deletion in RNA from 5 dpf, WT, and mutant embryos. These RT-PCR products were sequenced using Sanger sequencing as described previously [43].
4.4. Generation of zrsr2 Riboprobe, WISH, and o-Dianisidine Staining
To generate a template for zrsr2 riboprobe synthesis for WISH, we performed RT-PCR using RNA from 5 dpf WT embryos and primers from the region shared by both isoforms (Supplementary Table S1). The RT-PCR product was cloned into a pCR4-TOPO vector (Thermo Fisher Scientific, Waltham, MA, USA) and sequence-verified by Sanger sequencing. The plasmid DNA was extracted using QIAprep Spin Miniprep Kit (Qiagen, Germantown, MD, USA), digested with SpeI, and labeled with a DIG labeling kit (Sigma-Aldrich, St. Louis, MO, USA) using T7 polymerase. Antisense riboprobes for all other WISH markers used in this study were also generated using a DIG labeling kit (Sigma-Aldrich, St. Louis, MO, USA). Zrsr2hg129/+ fish were in-crossed, and embryos were collected at various developmental stages. Collected embryos were euthanized and fixed overnight in 4% PFA at 4 °C. WISH was performed as described previously [44]. For o-dianisidine staining, euthanized embryos were incubated in o-dianisidine (MP Biomedicals, Costa Mesa, CA, USA) solution for 15 min in the dark [45], followed by fixation in 4% PFA prior to imaging. Embryos used for WISH and o-dianisidine staining were genotyped via fluorescent PCR, as described previously [41], using REDExtract-N-Amp PCR ReadyMix (Sigma-Aldrich, St. Louis, MO, USA) for product amplification.
4.5. Alcian Blue Staining
Embryos from zrsr2hg129/+ in-crosses were euthanized and collected at 4 dpf, fixed in 4% PFA for 2 h at room temperature, and then stained in 0.15% Alcian Blue (Sigma-Aldrich, St. Louis, MO, USA), 50% EtOH, 0.1M HCl (pH 1) overnight, as described previously [46]. Embryos were then rinsed in 100% EtOH and rehydrated in a series of three 5 min EtOH/1x PBS washes. Embryos were then digested in 0.05% trypsin dissolved in 35% saturated NaBO4 for 2.5 h, followed by bleaching in a 3% H2O2 1% KOH solution. Embryos were stored in 70% glycerol at 4 °C, imaged, and genotyped via fluorescent PCR, as described above.
4.6. Imaging and Image Analysis
Images of embryos for morphological phenotypes, Alcian blue staining, o-dianisidine staining, and WISH were acquired with a Leica M205 microscope, Leica DFC7000G camera, and the Leica Application Suite X Imaging Software Suite version 3.4.1 (Leica Microsystems, Deerfield, IL, USA). FIJI imaging software version 2.1.0 was used to quantify the staining intensity for o-dianisidine signals, as described in [47]. Differences between zrsr2hg129/hg129 and control embryos were determined using an unpaired t-test with Welch’s correction.
4.7. RNA Extraction and Sequencing (RNA-Seq)
zrsr2hg129/+ and WT sibling adult fish were in-crossed to collect mutant and WT embryos respectively. At 3 dpf, embryos from each clutch were euthanized with MS-222 (0.4 g/L) and decapitated to collect the heads for genotyping to identify homozygous mutant embryos. The embryo bodies were preserved in DNA/RNA Shield (Zymo Research, Irvine, CA, USA) in a corresponding 96-well plate for RNA extraction. After genotyping, 3 pools of 10 preserved embryos were collected from mutant and WT crosses. The pooled embryos were homogenized with a glass homogenizer, and the RNA was extracted using miRNeasy kit (Qiagen, Germantown, MD, USA) according to manufacturer’s instructions. RNA quantity and integrity were checked by the Bioanalyzer (Agilent, Santa Clara, CA, USA). The RNA Integrity number (RIN) values for samples used for RNA-Seq were as follows: WT triplicates (9.7, 9.7, 9.2), Mutant triplicates (9, 8.9, 9). An amount of 500 nanograms of total RNA was used in conjunction with the TruSeq® Stranded Total RNA Library Prep kit (Illumina, San Diego, CA, USA). The libraries were checked for quality via the Bioanalyzer and quantitated by the Qubit (Thermo Fisher Scientific, Waltham, MA, USA). Equimolar amounts of each library were pooled and run on an Illumina Next-Seq 550/500 High Output Kit v2.5 (300 cycles). All RNA-Seq data are available at GEO (https://www.ncbi.nlm.nih.gov/geo/ accessed on 10 January 2022) under the accession number: GSE193365.
4.8. Bioinformatics Analysis of RNA-Seq Data
Low quality reads, identified by having N-base ratios above 10% or low-quality bases ratio above 50%, and reads containing adapter sequences were removed. The clean reads were aligned to GRCz11 with hisat2 (2.2.1.0) [48] and sorted with sambamba (0.7.1) [49]. Reads were counted with Stringtie (2.0.3) [50] based on GRCz11.99 annotation. Differentially expressed genes were identified using DESeq2 [33], using 0.05 and 1 as the cutoff for p-adjusted and absolute Log2FoldChange values, respectively. A heatmap was generated with Heatmapper [51], using Average Linkage clustering and Pearson distance measurement. IRFinder was used to identify the introns that were differentially spliced between WT and mutant transcripts [36], and intron types were classified using the Intron Annotation and Orthology Database [7]. Enriched pathways associated with differentially expressed genes, and differentially spliced genes between WT and zrsr2hg129/hg129 samples were identified in Metascape [52], which uses hypergeometric testing and a Benjamini-Hochberg p-value correction algorithm to identify statistically significant pathways. Comparison of U12-type (retained versus non-retained) introns was performed by extracting the sequences for each group and an analysis of their size distribution, base content, and terminal dinucleotides. For the analysis of the splice sites, the first and last 20 bases of all introns were extracted and illustrated with WebLogo [53].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810668/s1.
Author Contributions
Conceptualization, R.W., E.B. (Elizabeth Broadbridge), S.M.B., P.L., E.B. (Erica Bresciani) and R.S.; Data curation, K.Y. and A.E.; Formal analysis, R.W., K.B., E.B. (Elizabeth Broadbridge), K.Y., A.E. and R.S.; Funding acquisition, R.S.; Investigation, R.W., K.B., E.B. (Elizabeth Broadbridge), B.C. and T.Z.; Methodology, W.P.; Software, R.W. and K.Y.; Supervision, S.M.B., P.L., E.B. (Erica Bresciani) and R.S.; Validation, K.B.; Visualization, R.W. and B.C.; Writing—original draft, R.W. and R.S.; Writing—review and editing, K.B., B.C., S.M.B., P.L., E.B. (Erica Bresciani) and R.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health (ZIC HG200351 to R.S.).
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee (ACUC) of the National Human Genome Research Institute (protocol G-05-5 assigned to R.S., approved on 6 September 2017, and renewed on 6 August 2020).
Informed Consent Statement
Not applicable.
Data Availability Statement
All RNA-Seq data are available at GEO (https://www.ncbi.nlm.nih.gov/geo/ accessed on 10 January 2022) under the accession number: GSE193365.
Acknowledgments
We are thankful for the excellent animal care provided by the Charles River staff of our zebrafish facility. We would like to thank WeiWei Wu for her expert advice and help in the RNA extraction for RNA-seq, Alberto Rissone for his expert advice on WISH, and Jamie Diemer for carefully reading the manuscript. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov accessed multiple times starting 20 February 2020).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shen, H.; Zheng, X.; Luecke, S.; Green, M.R. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010, 24, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Tronchere, H.; Wang, J.; Fu, X.D. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 1997, 388, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Turunen, J.J.; Niemela, E.H.; Verma, B.; Frilander, M.J. The significant other: Splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 2013, 4, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Will, C.L.; Luhrmann, R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol. Chem. 2005, 386, 713–724. [Google Scholar] [CrossRef]
- Burge, C.B.; Padgett, R.A.; Sharp, P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell 1998, 2, 773–785. [Google Scholar] [CrossRef]
- Dietrich, R.C.; Incorvaia, R.; Padgett, R.A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12-dependent introns. Mol. Cell 1997, 1, 151–160. [Google Scholar] [CrossRef]
- Moyer, D.C.; Larue, G.E.; Hershberger, C.E.; Roy, S.W.; Padgett, R.A. Comprehensive database and evolutionary dynamics of U12-type introns. Nucleic Acids Res. 2020, 48, 7066–7078. [Google Scholar] [CrossRef]
- Alioto, T.S. U12DB: A database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007, 35, D110–D115. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, S.A.; Goswami, M.; Tang, G.; Routbort, M.J.; Patel, K.P.; Luthra, R.; Medeiros, L.J.; Ok, C.Y. Comparison of therapy-related myelodysplastic syndrome with ring sideroblasts and de novo myelodysplastic syndrome with ring sideroblasts. Leuk. Res. 2019, 86, 106227. [Google Scholar] [CrossRef]
- Damm, F.; Kosmider, O.; Gelsi-Boyer, V.; Renneville, A.; Carbuccia, N.; Hidalgo-Curtis, C.; Della Valle, V.; Couronne, L.; Scourzic, L.; Chesnais, V.; et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood 2012, 119, 3211–3218. [Google Scholar] [CrossRef]
- Haferlach, T.; Nagata, Y.; Grossmann, V.; Okuno, Y.; Bacher, U.; Nagae, G.; Schnittger, S.; Sanada, M.; Kon, A.; Alpermann, T.; et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia 2014, 28, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.Y.; Seo, J.Y.; Kim, S.H.; Jung, H.A.; Park, S.; Kim, K.; Jung, C.W.; Kim, J.S.; Park, J.S.; Kim, H.J.; et al. Mutations in the Spliceosomal Machinery Genes SRSF2, U2AF1, and ZRSR2 and Response to Decitabine in Myelodysplastic Syndrome. Anticancer Res. 2015, 35, 3081–3089. [Google Scholar] [PubMed]
- Thol, F.; Kade, S.; Schlarmann, C.; Loffeld, P.; Morgan, M.; Krauter, J.; Wlodarski, M.W.; Kolking, B.; Wichmann, M.; Gorlich, K.; et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood 2012, 119, 3578–3584. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Lindsley, R.C.; Mar, B.G.; Mazzola, E.; Grauman, P.V.; Shareef, S.; Allen, S.L.; Pigneux, A.; Wetzler, M.; Stuart, R.K.; Erba, H.P.; et al. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 2015, 125, 1367–1376. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Tefferi, A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016, 6, e393. [Google Scholar] [CrossRef]
- Srinath, M.; Coberly, E.; Ebersol, K.; Binz, K.; Laziuk, K.; Gunning, W.T.; Gruner, B.; Hammer, R.; Sathi, B.K. ZRSR2 mutation in a child with refractory macrocytic anemia and Down Syndrome. Pediatr. Hematol. Oncol. 2019, 36, 236–243. [Google Scholar] [CrossRef]
- Togami, K.; Chung, S.S.; Madan, V.; Booth, C.A.G.; Kenyon, C.M.; Cabal-Hierro, L.; Taylor, J.; Kim, S.S.; Griffin, G.K.; Ghandi, M.; et al. Sex-Biased ZRSR2 Mutations in Myeloid Malignancies Impair Plasmacytoid Dendritic Cell Activation and Apoptosis. Cancer Discov. 2022, 12, 522–541. [Google Scholar] [CrossRef]
- Madan, V.; Kanojia, D.; Li, J.; Okamoto, R.; Sato-Otsubo, A.; Kohlmann, A.; Sanada, M.; Grossmann, V.; Sundaresan, J.; Shiraishi, Y.; et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat. Commun. 2015, 6, 6042. [Google Scholar] [CrossRef]
- Madan, V.; Li, J.; Zhou, S.; Teoh, W.W.; Han, L.; Meggendorfer, M.; Malcovati, L.; Cazzola, M.; Ogawa, S.; Haferlach, T.; et al. Distinct and convergent consequences of splice factor mutations in myelodysplastic syndromes. Am. J. Hematol. 2020, 95, 133–143. [Google Scholar] [CrossRef]
- Inoue, D.; Polaski, J.T.; Taylor, J.; Castel, P.; Chen, S.; Kobayashi, S.; Hogg, S.J.; Hayashi, Y.; Pineda, J.M.B.; El Marabti, E.; et al. Minor intron retention drives clonal hematopoietic disorders and diverse cancer predisposition. Nat. Genet. 2021, 53, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Redondo, I.; Pericuesta, E.; Navarrete-Lopez, P.; Ramos-Ibeas, P.; Planells, B.; Fonseca-Balvis, N.; Vaquero-Rey, A.; Fernandez-Gonzalez, R.; Laguna-Barraza, R.; Horiuchi, K.; et al. Zrsr2 and functional U12-dependent spliceosome are necessary for follicular development. iScience 2022, 25, 103860. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Redondo, I.; Ramos-Ibeas, P.; Pericuesta, E.; Fernandez-Gonzalez, R.; Laguna-Barraza, R.; Gutierrez-Adan, A. Minor Splicing Factors Zrsr1 and Zrsr2 Are Essential for Early Embryo Development and 2-Cell-Like Conversion. Int. J. Mol. Sci. 2020, 21, 4115. [Google Scholar] [CrossRef]
- Madan, V.; Cao, Z.; Teoh, W.W.; Dakle, P.; Han, L.; Shyamsunder, P.; Jeitany, M.; Zhou, S.; Li, J.; Nordin, H.B.M.; et al. ZRSR1 co-operates with ZRSR2 in regulating splicing of U12-type introns in murine hematopoietic cells. Haematologica 2022, 107, 680–689. [Google Scholar] [CrossRef]
- Horiuchi, K.; Perez-Cerezales, S.; Papasaikas, P.; Ramos-Ibeas, P.; Lopez-Cardona, A.P.; Laguna-Barraza, R.; Fonseca Balvis, N.; Pericuesta, E.; Fernandez-Gonzalez, R.; Planells, B.; et al. Impaired Spermatogenesis, Muscle, and Erythrocyte Function in U12 Intron Splicing-Defective Zrsr1 Mutant Mice. Cell Rep. 2018, 23, 143–155. [Google Scholar] [CrossRef]
- Fuentes, R.; Letelier, J.; Tajer, B.; Valdivia, L.E.; Mullins, M.C. Fishing forward and reverse: Advances in zebrafish phenomics. Mech. Dev. 2018, 154, 296–308. [Google Scholar] [CrossRef]
- Pickart, M.A.; Klee, E.W. Zebrafish approaches enhance the translational research tackle box. Transl. Res. 2014, 163, 65–78. [Google Scholar] [CrossRef]
- Varshney, G.K.; Burgess, S.M. Mutagenesis and phenotyping resources in zebrafish for studying development and human disease. Brief. Funct. Genom. 2014, 13, 82–94. [Google Scholar] [CrossRef]
- Olthof, A.M.; Hyatt, K.C.; Kanadia, R.N. Minor intron splicing revisited: Identification of new minor intron-containing genes and tissue-dependent retention and alternative splicing of minor introns. BMC Genom. 2019, 20, 686. [Google Scholar] [CrossRef]
- Gore, A.V.; Pillay, L.M.; Venero Galanternik, M.; Weinstein, B.M. The zebrafish: A fintastic model for hematopoietic development and disease. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e312. [Google Scholar] [CrossRef]
- Davidson, A.J.; Zon, L.I. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene 2004, 23, 7233–7246. [Google Scholar] [CrossRef] [PubMed]
- Burger, A.; Lindsay, H.; Felker, A.; Hess, C.; Anders, C.; Chiavacci, E.; Zaugg, J.; Weber, L.M.; Catena, R.; Jinek, M.; et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development 2016, 143, 2025–2037. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pessa, H.K.; Greco, D.; Kvist, J.; Wahlstrom, G.; Heino, T.I.; Auvinen, P.; Frilander, M.J. Gene expression profiling of U12-type spliceosome mutant Drosophila reveals widespread changes in metabolic pathways. PLoS ONE 2010, 5, e13215. [Google Scholar] [CrossRef]
- Gault, C.M.; Martin, F.; Mei, W.; Bai, F.; Black, J.B.; Barbazuk, W.B.; Settles, A.M. Aberrant splicing in maize rough endosperm3 reveals a conserved role for U12 splicing in eukaryotic multicellular development. Proc. Natl. Acad. Sci. USA 2017, 114, E2195–E2204. [Google Scholar] [CrossRef]
- Middleton, R.; Gao, D.; Thomas, A.; Singh, B.; Au, A.; Wong, J.J.; Bomane, A.; Cosson, B.; Eyras, E.; Rasko, J.E.; et al. IRFinder: Assessing the impact of intron retention on mammalian gene expression. Genome Biol. 2017, 18, 51. [Google Scholar] [CrossRef]
- Baumgartner, M.; Drake, K.; Kanadia, R.N. An Integrated Model of Minor Intron Emergence and Conservation. Front. Genet. 2019, 10, 1113. [Google Scholar] [CrossRef]
- McElderry, J.; Carrington, B.; Bishop, K.; Kim, E.; Pei, W.; Chen, Z.; Ramanagoudr-Bhojappa, R.; Prakash, A.; Burgess, S.M.; Liu, P.P.; et al. Splicing factor DHX15 affects tp53 and mdm2 expression via alternate splicing and promoter usage. Hum. Mol. Genet. 2019, 28, 4173–4185. [Google Scholar] [CrossRef]
- Markmiller, S.; Cloonan, N.; Lardelli, R.M.; Doggett, K.; Keightley, M.C.; Boglev, Y.; Trotter, A.J.; Ng, A.Y.; Wilkins, S.J.; Verkade, H.; et al. Minor class splicing shapes the zebrafish transcriptome during development. Proc. Natl. Acad. Sci. USA 2014, 111, 3062–3067. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 5th ed.; University of Oregon Press: Eugene, OR, USA, 2007. [Google Scholar]
- Varshney, G.K.; Carrington, B.; Pei, W.; Bishop, K.; Chen, Z.; Fan, C.; Xu, L.; Jones, M.; LaFave, M.C.; Ledin, J.; et al. A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 2016, 11, 2357–2375. [Google Scholar] [CrossRef]
- Carrington, B.; Varshney, G.K.; Burgess, S.M.; Sood, R. CRISPR-STAT: An easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015, 43, e157. [Google Scholar] [CrossRef] [PubMed]
- Sood, R.; Carrington, B.; Bishop, K.; Jones, M.; Rissone, A.; Candotti, F.; Chandrasekharappa, S.C.; Liu, P. Efficient methods for targeted mutagenesis in zebrafish using zinc-finger nucleases: Data from targeting of nine genes using CompoZr or CoDA ZFNs. PLoS ONE 2013, 8, e57239. [Google Scholar] [CrossRef]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Detrich, H.W., 3rd; Kieran, M.W.; Chan, F.Y.; Barone, L.M.; Yee, K.; Rundstadler, J.A.; Pratt, S.; Ransom, D.; Zon, L.I. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 1995, 92, 10713–10717. [Google Scholar] [CrossRef] [PubMed]
- Schoenebeck, J.J.; Hutchinson, S.A.; Byers, A.; Beale, H.C.; Carrington, B.; Faden, D.L.; Rimbault, M.; Decker, B.; Kidd, J.M.; Sood, R.; et al. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 2012, 8, e1002849. [Google Scholar] [CrossRef]
- Dobrzycki, T.; Krecsmarik, M.; Monteiro, R. Genotyping and Quantification of In Situ Hybridization Staining in Zebrafish. J. Vis. Exp. 2020, 155, e59956. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).