Rice β-Glucosidase 4 (Os1βGlu4) Regulates the Hull Pigmentation via Accumulation of Salicylic Acid
Abstract
:1. Introduction
2. Results
2.1. bh1 (Brown Hull1) Displayed Brown Hull Pigmentation and Decreased Seed Setting Rate
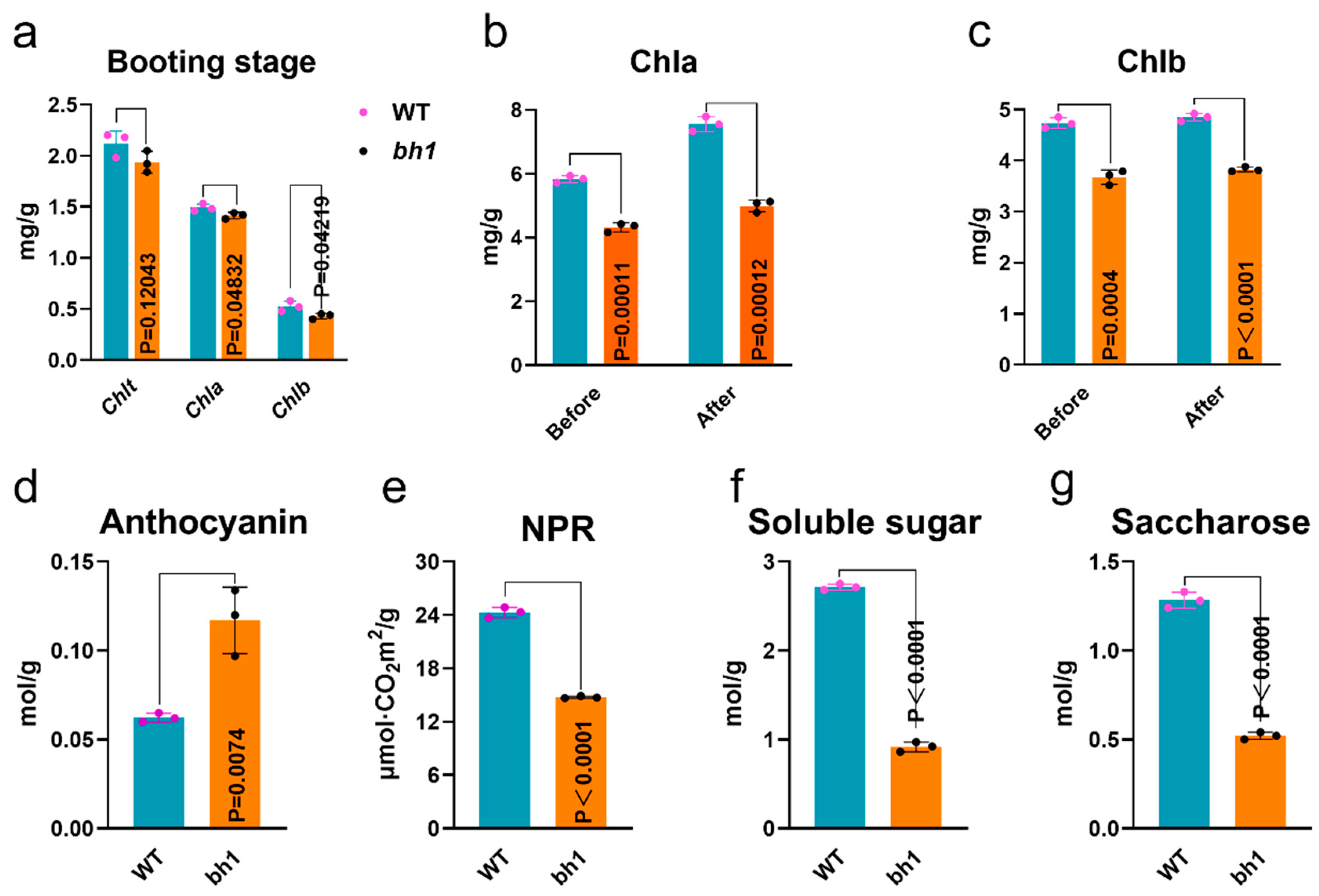
2.2. Hulls of bh1 Have Decreased Activity of Photosynthesis and Sugars
2.3. Accumulation of Reactive Oxygen Species (ROS) in bh1 Panicles
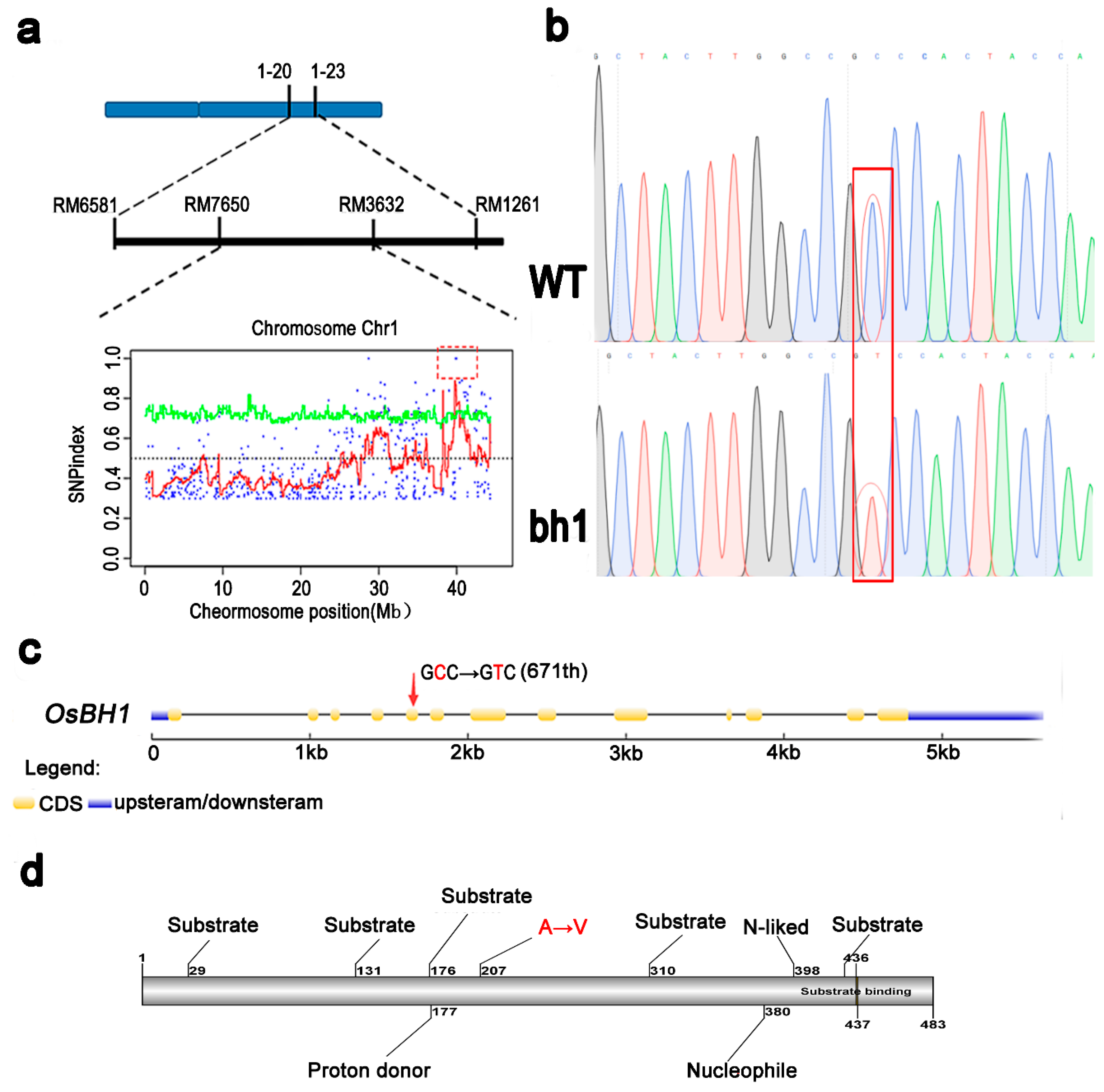
2.4. Genetic Analysis Reveals a Cytoplasmic Os1βGlu4 Encodes bh1 Phenotype
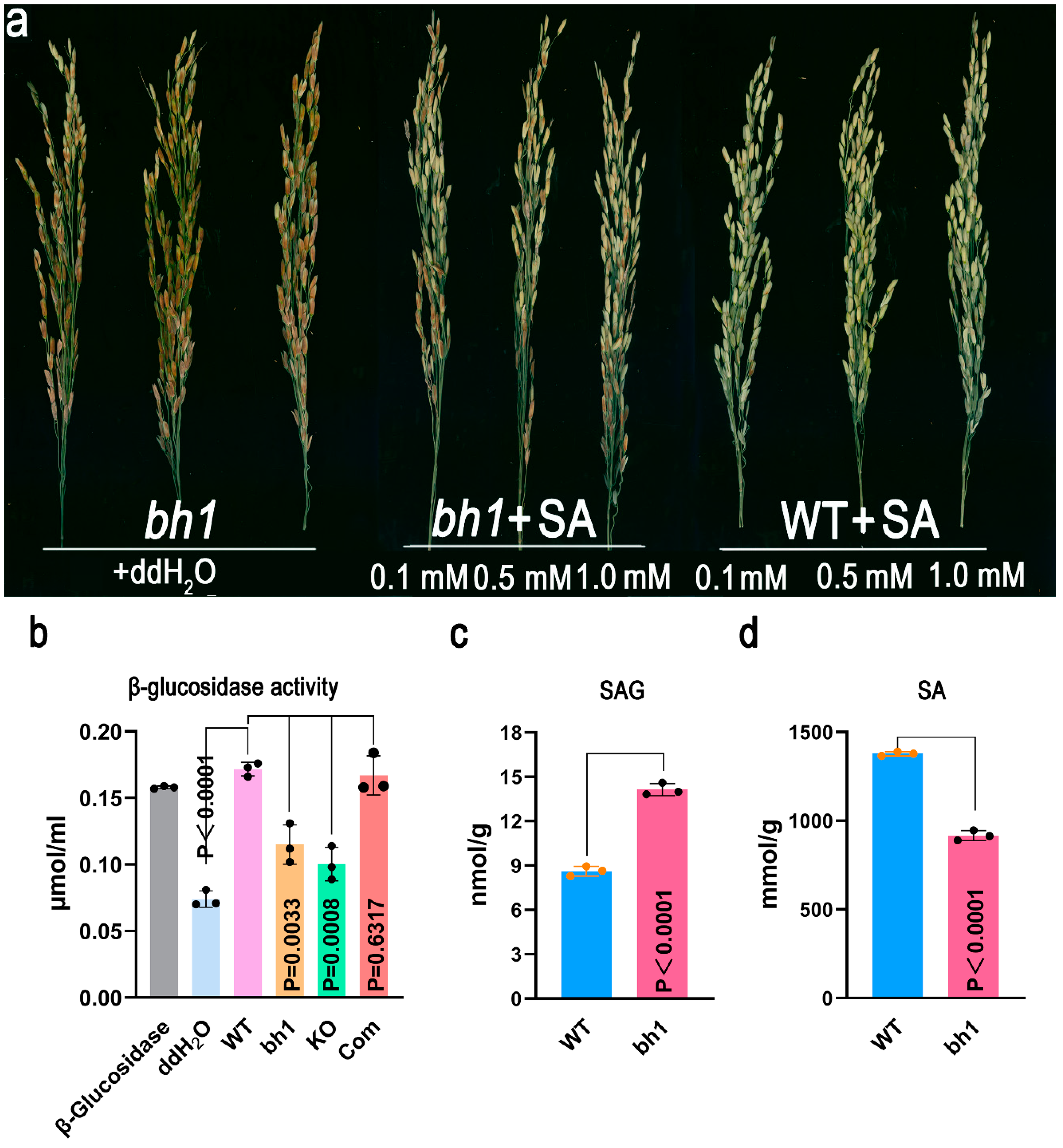
2.5. Os1βGlu4 Can Hydrolyze SAG In-Vitro and InVivo
3. Discussion
3.1. Cytoplasmic Os1βGlu4 Contributes to Cellular SA Pools in Rice
3.2. SA Deficient Plants Have Brown Pigmentation on the Hulls and Showed Decreased Seed Setting Rate in Rice
3.3. Cytoplasmic Os1βGlu4 May Affect SA Pathway and Flavonoid Pathways for Brown Hull Pigmentation
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Measurement of Photosynthetic Pigments
4.3. Determination of Antioxidants and TUNEL Assay
4.4. Genetic Analysis and Map-Based Cloning
4.5. Gene Knock-Out, Complementation and Overexpression Analysis
4.6. Subcellular Localization
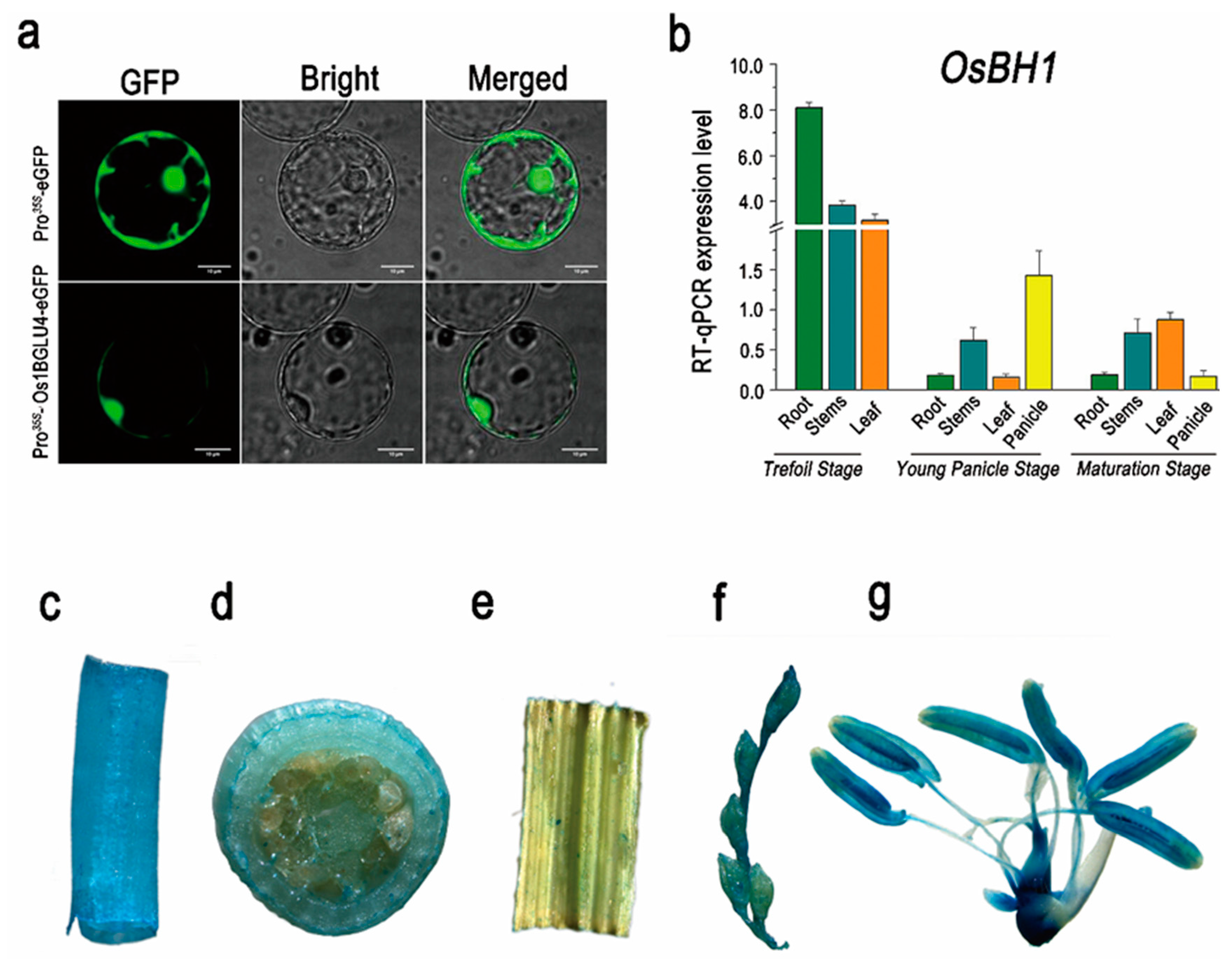
4.7. GUS Activity Staining Assay
4.8. Determination of β-Glucosidase Activity
4.9. In-Vitro/In-Vivo SAG to SA Hydrolysis
4.10. Quantitative Real-Time PCR Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.-Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell 1996, 8, 1809. [Google Scholar] [CrossRef]
- Chow, S.; Gu, K.; Jiang, L.; Nassour, A. Salicylic acid affects swimming, twitching and swarming motility in Pseudomonas aeruginosa, resulting in decreased biofilm formation. J. Exp. Microb. Immun. 2011, 15, 22–29. [Google Scholar]
- Vlot, A.C.; Dempsey, D.M.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Ann. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef]
- Lee, H.-I.; Leon, J.; Raskin, I. Biosynthesis and metabolism of salicylic acid. Proc. Natl. Acad. Sci. USA 1995, 92, 4076–4079. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.M.A.; Vlot, A.C.; Wildermuth, M.C.; Klessig, D.F. Salicylic acid biosynthesis and metabolism. Arab. Book/Am. Soc. Plant Biol. 2011, 9, e1056. [Google Scholar] [CrossRef]
- Dean, J.V.; Delaney, S.P. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol. Plant 2008, 132, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; D’Auria, J.C.; Tholl, D.; Ross, J.R.; Gershenzon, J.; Noel, J.P.; Pichersky, E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003, 36, 577–588. [Google Scholar] [CrossRef]
- Mol, J.; Grotewold, E.; Koes, R. How genes paint flowers and seeds. Trends Plant Sci. 1998, 3, 212–217. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Giuffrè, A. HPLC-DAD detection of changes in phenol content of red berry skins during grape ripening. Eur. Food Res. Technol. 2013, 237, 555–564. [Google Scholar] [CrossRef]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Aron, P.M.; Kennedy, J.A. Flavan-3-ols: Nature, occurrence and biological activity. Mol. Nutri. Food Res. 2008, 52, 79–104. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Normanly, J.; Tague, B.W.; Peer, W.A.; Taiz, L.; Muday, G.K. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef]
- Thompson, E.P.; Wilkins, C.; Demidchik, V.; Davies, J.M.; Glover, B.J. An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J. Exp. Bot. 2010, 61, 439–451. [Google Scholar] [CrossRef]
- Shim, S.-H.; Mahong, B.; Lee, S.-K.; Kongdin, M.; Lee, C.; Kim, Y.-J.; Qu, G.; Zhang, D.; Ketudat Cairns, J.R.; Jeon, J.-S. Rice β-glucosidase Os12BGlu38 is required for synthesis of intine cell wall and pollen fertility. J. Exp. Bot. 2022, 73, 784–800. [Google Scholar] [CrossRef]
- Chen, X.; Yang, Y.; Yang, X.; Zhu, G.; Lu, X.; Jia, F.; Diao, B.; Yu, S.; Ali, A.; Zhang, H. Investigation of flavonoid components and their associated antioxidant capacity in different pigmented rice varieties. Food Res. Inter. 2022, 161, 111726. [Google Scholar] [CrossRef]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef]
- Sarma, A.D.; Sharma, R. Anthocyanin-DNA copigmentation complex: Mutual protection against oxidative damage. Phytochemistry 1999, 52, 1313–1318. [Google Scholar] [CrossRef]
- Dixon, R.A. Engineering of plant natural product pathways. Curr. Opin. Plant Biol. 2005, 8, 329–336. [Google Scholar] [CrossRef]
- Hichri, I.; Barrieu, F.; Bogs, J.; Kappel, C.; Delrot, S.; Lauvergeat, V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011, 62, 2465–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Koes, R.; Verweij, W.; Quattrocchio, F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005, 10, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Hichri, I.; Heppel, S.C.; Pillet, J.; Léon, C.; Czemmel, S.; Delrot, S.; Lauvergeat, V.; Bogs, J. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol. Plant 2010, 3, 509–523. [Google Scholar] [CrossRef]
- Guerriero, G.; Hausman, J.-F.; Ezcurra, I. WD40-repeat proteins in plant cell wall formation: Current evidence and research prospects. Front. Plant Sci. 2015, 6, 1112. [Google Scholar] [CrossRef]
- Zhao, L.; Gao, L.; Wang, H.; Chen, X.; Wang, Y.; Yang, H.; Wei, C.; Wan, X.; Xia, T. The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct. Integr. Genom. 2013, 13, 75–98. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Z.; Chen, C.; Wu, W.; Ren, N.; Jiang, C.; Yu, J.; Zhao, Y.; Zheng, X.; Yang, Q. The C–S–A gene system regulates hull pigmentation and reveals evolution of anthocyanin biosynthesis pathway in rice. J. Exp. Bot. 2018, 69, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Yamaguchi, T.; Omoteno, M.; Takarada, T.; Fujita, K.; Murata, K.; Iyama, Y.; Kojima, Y.; Morikawa, M.; Ozaki, H. Genetic dissection of black grain rice by the development of a near isogenic line. Breed. Sci. 2014, 64, 134–141. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Florindo, R.N.; Souza, V.P.; Mutti, H.S.; Camilo, C.; Manzine, L.R.; Marana, S.R.; Polikarpov, I.; Nascimento, A.S. Structural insights into β-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum. New Biotechnol. 2018, 40, 218–227. [Google Scholar] [CrossRef]
- Himeno, N.; Saburi, W.; Wakuta, S.; Takeda, R.; Matsuura, H.; Nabeta, K.; Sansenya, S.; Cairns, J.R.K.; Mori, H.; Imai, R. Identification of rice β-glucosidase with high hydrolytic activity towards salicylic acid β-d-glucoside. Biosci. Biotechnol. Biochem. 2013, 77, 934–939. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.H.; Piao, H.L.; Kim, H.-Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.-J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.-Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell. 2012, 24, 2184–2199. [Google Scholar] [CrossRef] [PubMed]
- Moellering, E.R.; Muthan, B.; Benning, C. Freezing tolerance in plants requires lipid remodeling at the outer chloroplast membrane. Science 2010, 330, 226–228. [Google Scholar] [CrossRef]
- Hua, Y.; Ekkhara, W.; Sansenya, S.; Srisomsap, C.; Roytrakul, S.; Saburi, W.; Takeda, R.; Matsuura, H.; Mori, H.; Cairns, J.R.K. Identification of rice Os4BGlu13 as a β-glucosidase which hydrolyzes gibberellin A4 1-O-β-d-glucosyl ester, in addition to tuberonic acid glucoside and salicylic acid derivative glucosides. Arch. Biochem. Biophys. 2015, 583, 36–46. [Google Scholar] [CrossRef]
- Opassiri, R.; Maneesan, J.; Akiyama, T.; Pomthong, B.; Jin, S.; Kimura, A.; Cairns, J.R.K. Rice Os4BGlu12 is a wound-induced β-glucosidase that hydrolyzes cell wall-β-glucan-derived oligosaccharides and glycosides. Plant Sci. 2010, 179, 273–280. [Google Scholar] [CrossRef]
- Seshadri, S.; Akiyama, T.; Opassiri, R.; Kuaprasert, B.; Cairns, J.K. Structural and enzymatic characterization of Os3BGlu6, a rice β-glucosidase hydrolyzing hydrophobic glycosides and (1 → 3)-and (1 → 2)-linked disaccharides. Plant Physiol. 2009, 151, 47–58. [Google Scholar] [CrossRef]
- Wang, C.; Chen, S.; Dong, Y.; Ren, R.; Chen, D.; Chen, X. Chloroplastic Os3BGlu6 contributes significantly to cellular ABA pools and impacts drought tolerance and photosynthesis in rice. New Phytol. 2020, 226, 1042–1054. [Google Scholar] [CrossRef]
- Rouyi, C.; Baiya, S.; Lee, S.-K.; Mahong, B.; Jeon, J.-S.; Ketudat-Cairns, J.R.; Ketudat-Cairns, M. Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PLoS ONE 2014, 9, e96712. [Google Scholar] [CrossRef]
- Jiaxiang, S.; Linghua, T.; Qingsen, Z. Relationship between chlorophyll content of rice glume and grain filling. Jiangsu Nongye Xuebao 2001, 1, 24–27. [Google Scholar]
- Abe, A.; Kosugi, S.; Yoshida, K.; Natsume, S.; Takagi, H.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Mitsuoka, C.; Tamiru, M. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012, 30, 174. [Google Scholar] [CrossRef] [PubMed]
- Jerie, P. Milestones of cardivascular pharmacotherapy: Salicylates and aspirin. Cas. Lek. Ceskych 2006, 145, 901–904. [Google Scholar]
- Rekhter, D.; Lüdke, D.; Ding, Y.; Feussner, K.; Zienkiewicz, K.; Lipka, V.; Wiermer, M.; Zhang, Y.; Feussner, I. Isochorismate-derived biosynthesis of the plant stress hormone salicylic acid. Science 2019, 365, 498–502. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, H.; Hua, H.; Wang, L.; Song, C.-P. A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chin. Sci. Bullet. 2011, 56, 3538–3546. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, K.; Jang, K.-M.; Kim, S.-G.; Park, J.-H.; Kim, S. Single-dose oral toxicity study of β-glucosidase 1 (Atbg1) protein introduced into genetically modified rapeseed (Brassica napus L.). J. Life Sci. 2017, 27, 194–201. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, M.; Li, Q.; Wang, L.; Wang, J.; Jeon, J.-S.; Qu, N.; Zhang, Y.; He, Z. OsRAR1 and OsSGT1 physically interact and function in rice basal disease resistance. Mol. Plant-Microbe. Interact. 2008, 21, 294–303. [Google Scholar] [CrossRef]
- Ali, A.; Wu, T.; Xu, Z.; Riaz, A.; Alqudah, A.M.; Iqbal, M.Z.; Zhang, H.; Liao, Y.; Chen, X.; Liu, Y. Phytohormones and Transcriptome Analyses Revealed the Dynamics Involved in Spikelet Abortion and Inflorescence Development in Rice. Int. J. Mol. Sci. 2022, 23, 7887. [Google Scholar] [CrossRef]
- Ali, A.; Wu, T.; Zhang, H.; Xu, P.; Zafar, S.A.; Liao, Y.; Chen, X.; Zhou, H.; Liu, Y.; Wang, W.; et al. A putative SUBTILISIN-LIKE SERINE PROTEASE 1 (SUBSrP1) regulates anther cuticle biosynthesis and panicle development in rice. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Ali, A.; Xu, P.; Riaz, A.; Wu, X. Current Advances in Molecular Mechanisms and Physiological Basis of Panicle Degeneration in Rice. Int. J. Mol. Sci. 2019, 20, 1613. [Google Scholar] [CrossRef]
- Couture, J.-F.; Collazo, E.; Trievel, R.C. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006, 13, 698–703. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.-P.; Zhang, L.; Yan, P.; Ahammed, G.J.; Han, W.-Y. Methyl salicylate enhances flavonoid biosynthesis in tea leaves by stimulating the phenylpropanoid pathway. Molecules 2019, 24, 362. [Google Scholar] [CrossRef] [PubMed]
- Gondor, O.K.; Janda, T.; Soós, V.; Pál, M.; Majláth, I.; Adak, M.K.; Balázs, E.; Szalai, G. Salicylic acid induction of flavonoid biosynthesis pathways in wheat varies by treatment. Front. Plant Sci. 2016, 7, 1447. [Google Scholar] [CrossRef] [PubMed]
- Shoeva, O.Y.; Mock, H.-P.; Kukoeva, T.V.; Börner, A.; Khlestkina, E.K. Regulation of the flavonoid biosynthesis pathway genes in purple and black grains of Hordeum vulgare. PLoS ONE 2016, 11, e0163782. [Google Scholar]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Carey, C.C.; Strahle, J.T.; Selinger, D.A.; Chandler, V.L. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 2004, 16, 450–464. [Google Scholar] [CrossRef]
- Walker, A.R.; Davison, P.A.; Bolognesi-Winfield, A.C.; James, C.M.; Srinivasan, N.; Blundell, T.L.; Esch, J.J.; Marks, M.D.; Gray, J.C. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 1999, 11, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Grain, D.; Bobet, S.; Le Gourrierec, J.; Thévenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB–b HLH–WDR complexes and their targets in A rabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Li, C.; Liu, C.-Q.; Zhang, H.-S.; Chen, C.-P.; Yang, X.-R.; Chen, L.-F.; Liu, Q.-S.; Guo, J.; Sun, C.-H.; Wang, P.-R.; et al. LPS1, encoding iron-sulfur subunit sdh2-1 of succinate dehydrogenase, affects leaf senescence and grain yield in rice. Int. J. Mol. Sci. 2021, 22, 157. [Google Scholar] [CrossRef]
- Gan, N.; Jia, L.; Zheng, L. A novel sandwich electrochemical immunosensor based on the DNA-derived magnetic nanochain probes for alpha-fetoprotein. J. Anal. Chem. 2011, 2011, 957805. [Google Scholar] [CrossRef]
- Xia, X.; Xing, Y.; Li, G.; Wu, J.; Kan, J. Antioxidant activity of whole grain Qingke (Tibetan Hordeum vulgare L.) toward oxidative stress in d-galactose induced mouse model. J. Funct. Foods 2018, 45, 355–362. [Google Scholar] [CrossRef]
- Wang, Y.; Liang, C.; Meng, Z.; Li, Y.; Abid, M.A.; Askari, M.; Wang, P.; Wang, Y.; Sun, G.; Cai, Y. Leveraging atriplex hortensis choline monooxygenase to improve chilling tolerance in cotton. Environ. Exp. Bot. 2019, 162, 364–373. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Sun, C.; Ma, Q.; Bu, H.; Chong, K.; Xu, Y. A C2H2 zinc-finger protein OsZFP213 interacts with OsMAPK3 to enhance salt tolerance in rice. J. Plant Physiol. 2018, 229, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Ali, A.; Xue, Z.; Zhou, X.; Ye, W.; Guo, D.; Liao, Y.; Jiang, P.; Wu, T.; Zhang, H. Disruption of LLM9428/OsCATC Represses Starch Metabolism and Confers Enhanced Blast Resistance in Rice. Int. J. Mol. Sci. 2022, 23, 3827. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y. CRISPR/Cas9 Platforms for Genome Editing in Plants: Developments and Applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Su, J.; Duan, S.; Ao, Y.; Dai, J.; Liu, J.; Wang, P.; Li, Y.; Liu, B.; Feng, D. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Meth. 2011, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-L.; Chen, Y.-C.; Lu, M.-Y.J.; Chang, J.-J.; Wang, H.-T.C.; Ke, H.-M.; Wang, T.-Y.; Ruan, S.-K.; Wang, T.-Y.; Hung, K.-Y. A highly efficient β-glucosidase from the buffalo rumen fungus Neocallimastix patriciarum W5. Biotechnol. Biofuels 2012, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, P.; Wu, T.; Ali, A.; Wang, J.; Fang, Y.; Qiang, R.; Liu, Y.; Tian, Y.; Liu, S.; Zhang, H.; et al. Rice β-Glucosidase 4 (Os1βGlu4) Regulates the Hull Pigmentation via Accumulation of Salicylic Acid. Int. J. Mol. Sci. 2022, 23, 10646. https://doi.org/10.3390/ijms231810646
Xu P, Wu T, Ali A, Wang J, Fang Y, Qiang R, Liu Y, Tian Y, Liu S, Zhang H, et al. Rice β-Glucosidase 4 (Os1βGlu4) Regulates the Hull Pigmentation via Accumulation of Salicylic Acid. International Journal of Molecular Sciences. 2022; 23(18):10646. https://doi.org/10.3390/ijms231810646
Chicago/Turabian StyleXu, Peizhou, Tingkai Wu, Asif Ali, Jinhao Wang, Yongqiong Fang, Runrun Qiang, Yutong Liu, Yunfeng Tian, Su Liu, Hongyu Zhang, and et al. 2022. "Rice β-Glucosidase 4 (Os1βGlu4) Regulates the Hull Pigmentation via Accumulation of Salicylic Acid" International Journal of Molecular Sciences 23, no. 18: 10646. https://doi.org/10.3390/ijms231810646
APA StyleXu, P., Wu, T., Ali, A., Wang, J., Fang, Y., Qiang, R., Liu, Y., Tian, Y., Liu, S., Zhang, H., Liao, Y., Chen, X., Shoaib, F., Sun, C., Xu, Z., Xia, D., Zhou, H., & Wu, X. (2022). Rice β-Glucosidase 4 (Os1βGlu4) Regulates the Hull Pigmentation via Accumulation of Salicylic Acid. International Journal of Molecular Sciences, 23(18), 10646. https://doi.org/10.3390/ijms231810646






