Micro-Players of Great Significance—Host microRNA Signature in Viral Infections in Humans and Animals
Abstract
1. Introduction
MicroRNAs and Viral Infections in Humans and Animals
2. MicroRNA Signature in Human Viral Infections
2.1. MicroRNA-155 (miR-155)
2.1.1. Hepatitis Viruses
2.1.2. Hemorrhagic Viruses
2.1.3. Respiratory Viruses
2.2. MicroRNA-223 (miR-223)
2.2.1. Hepatitis Viruses
2.2.2. Hemorrhagic Viruses
2.2.3. Respiratory Viruses
2.2.4. Human Immunodeficiency Viruses
2.3. MicroRNA-146a (miR-146a)
2.3.1. Hepatitis Viruses
2.3.2. Hemorrhagic Viruses
2.3.3. Respiratory Viruses
2.4. MicroRNA-122 (miR-122)
2.4.1. Hepatitis Viruses
2.4.2. Hemorrhagic Viruses
2.4.3. Respiratory Viruses
2.5. MicroRNA-125b (miR-125b)
2.5.1. Hepatitis Viruses
2.5.2. Respiratory Viruses
2.5.3. Human Immunodeficiency Viruses
2.5.4. Neurotropic Viruses
2.6. MicroRNA-132 (miR-132)
2.6.1. Hepatitis Viruses
2.6.2. Respiratory Viruses
2.6.3. Human Herpesviruses
2.7. MicroRNA-34a (miR-34a)
2.7.1. Hepatitis Viruses
2.7.2. Hemorrhagic Viruses
2.7.3. Respiratory Viruses
2.7.4. Human T-Lymphotropic Virus Type 1
2.8. MicroRNA-21 (miR-21)
2.8.1. Hepatitis Viruses
2.8.2. Hemorrhagic Viruses
2.8.3. Respiratory Viruses
2.8.4. Human Immunodeficiency Viruses
2.8.5. Cardiotropic Viruses
2.8.6. Vector-Borne Viruses
2.9. MicroRNA-16 (miR-16)
2.9.1. Hepatitis Viruses
2.9.2. Respiratory Viruses
2.10. MicroRNA-181 Family (miR-181)
2.10.1. Hepatitis Viruses
2.10.2. Respiratory Viruses
2.11. Let-7 Family (Let-7)
2.11.1. Hepatitis Viruses
2.11.2. Hemorrhagic Viruses
2.11.3. Respiratory Viruses
2.11.4. Human Immunodeficiency Viruses
2.11.5. Neurotropic Viruses
2.12. MicroRNA-10a (miR-10a)
2.12.1. Hepatitis Viruses
2.12.2. Respiratory Viruses
2.12.3. Cardiotropic Viruses
3. MicroRNA Signature in Animal Viral Infections
3.1. MicroRNA-155 (miR-155)
3.1.1. Hemorrhagic Viruses
3.1.2. Respiratory Viruses
3.1.3. Lymphotropic Viruses
3.2. MicroRNA-223 (miR-223)
3.2.1. Respiratory Viruses
3.2.2. Lymphotropic Viruses
3.2.3. Vector-Borne Viruses
3.3. MicroRNA-146a (miR-146a)
3.3.1. Respiratory Viruses
3.3.2. Lymphotropic Viruses
3.3.3. Vector-Borne Viruses
3.3.4. Other Viruses
3.4. MicroRNA-145 (miR-145)
3.4.1. Respiratory Viruses
3.4.2. Neurotropic Viruses
3.5. MicroRNA-21 (miR-21)
3.5.1. Respiratory Viruses
3.5.2. Neurotropic Viruses
3.6. MicroRNA-16/miR-15a Cluster (miR-16/miR-15a Cluster)
3.6.1. Hemorrhagic Viruses
3.6.2. Respiratory Viruses
3.7. MicroRNA-181 Family (miR-181 Family)
3.7.1. Respiratory Viruses
3.7.2. Lymphotropic Viruses
3.7.3. Other Viruses
3.8. Let-7 Family (Let-7)
3.8.1. Lymphotropic Viruses
3.8.2. Other Viruses
3.9. MicroRNA-122 (miR-122)
3.9.1. Hemorrhagic Viruses
3.9.2. Respiratory Viruses
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef] [PubMed]
- Leung, A.; Sharp, P.A. MicroRNA Functions in Stress Responses. Mol. Cell 2010, 40, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Rajewsky, N. MicroRNA target predictions in animals. Nat. Genet. 2006, 38, S8–S13. [Google Scholar] [CrossRef]
- Berezikov, E.; Guryev, V.; van de Belt, J.; Wienholds, E.; Plasterk, R.H.; Cuppen, E. Phylogenetic Shadowing and Computational Identification of Human microRNA Genes. Cell 2005, 120, 21–24. [Google Scholar] [CrossRef]
- Ignaz, P. Circulating microRNA in Disease Diagnostics and Their Potential Biological Relevance; Springer: Berlin/Heidelberg, Germany, 2015; Volume 106. [Google Scholar]
- Zhang, Y.; Li, Y.-K. MicroRNAs in the regulation of immune response against infections. J. Zhejiang Univ. Sci. B 2013, 14, 1–7. [Google Scholar] [CrossRef]
- Ardekani, A.M.; Naeini, M.M. The Role of MicroRNAs in Human Diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef]
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143. [Google Scholar] [CrossRef]
- Lee, Y.; Jeon, K.; Lee, J.-T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002, 21, 4663–4670. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of Virus-Encoded MicroRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef]
- Chen, M.; Wang, F.; Xia, H.; Yao, S. MicroRNA-155: Regulation of Immune Cells in Sepsis. Mediat. Inflamm. 2021, 2021, 8874854. [Google Scholar] [CrossRef]
- Tam, W. Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA. Gene 2001, 274, 157–167. [Google Scholar] [CrossRef]
- Mahesh, G.; Biswas, R. MicroRNA-155: A Master Regulator of Inflammation. J. Interf. Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
- Vigorito, E.; Kohlhaas, S.; Lu, D.; Leyland, R. miR-155: An ancient regulator of the immune system. Immunol. Rev. 2013, 253, 146–157. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/reverse-transcribing-dna-and-rna-viruses/w/hepadnaviridae/1317/genus-orthohepadnavirus (accessed on 19 July 2022).
- Rehermann, B.; Nascimbeni, M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat. Rev. Immunol. 2005, 5, 215–229. [Google Scholar] [CrossRef]
- Su, C.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol. J. 2011, 8, 354. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, N.; Panigrahi, R.; Pal, A.; Biswas, A.; Singh, S.P.; Kar, S.K.; Bandopadhyay, M.; Das, D.; Saha, D.; Kanda, T.; et al. Expression of microRNA-155 correlates positively with the expression of Toll-like receptor 7 and modulates hepatitis B virus via C/EBP-β in hepatocytes. J. Viral Hepat. 2015, 22, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-L.; Deng, H.; Li, X.-H.; Huang, Y.-X.; Xie, D.-Y.; Gao, Z.-L. Expression of MicroRNA-155 is Downregulated in Peripheral Blood Mononuclear Cells of Chronic Hepatitis B Patients. Hepat. Mon. 2016, 16, e34483. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Huang, Z.; Liu, H.; Chen, J.; Xie, Z.; Chen, Z.; Peng, J.; Sun, J.; Hou, J.; Zhang, X. Lower Expression of MicroRNA-155 Contributes to Dysfunction of Natural Killer Cells in Patients with Chronic Hepatitis B. Front. Immunol. 2017, 8, 1173. [Google Scholar] [CrossRef]
- Fang, J.; Zhuge, L.; Rao, H.; Huang, S.; Jin, L.; Li, J. Increased Levels of miR-155 are Related to Higher T-Cell Activation in the Peripheral Blood of Patients with Chronic Hepatitis B. Genet. Test. Mol. Biomark. 2019, 23, 118–123. [Google Scholar] [CrossRef]
- Wang, W.; Bian, H.; Li, F.; Li, X.; Zhang, D.; Sun, S.; Song, S.; Zhu, Q.; Ren, W.; Qin, C.; et al. HBeAg induces the expression of macrophage miR-155 to accelerate liver injury via promoting production of inflammatory cytokines. Cell. Mol. Life Sci. 2018, 75, 2627–2641. [Google Scholar] [CrossRef]
- Chen, L.; Ming, X.; Li, W.; Bi, M.; Yan, B.; Wang, X.; Yang, P.; Yang, B. The microRNA-155 mediates hepatitis B virus replication by reinforcing SOCS1 signalling–induced autophagy. Cell Biochem. Funct. 2020, 38, 436–442. [Google Scholar] [CrossRef]
- Xiong, Y.; Chen, S.; Liu, L.; Zhao, Y.; Lin, W.; Ni, J. Increased Serum MicroRNA-155 Level Associated with Nonresponsiveness to Hepatitis B Vaccine. Clin. Vaccine Immunol. 2013, 20, 1089–1091. [Google Scholar] [CrossRef]
- Hefzy, E.M.; Hassuna, N.A.; Shaker, O.G.; Masoud, M.; Abelhameed, T.A.; Ahmed, T.I.; Hemeda, N.F.; Abdelhakeem, M.A.; Mahmoud, R.H. miR-155 T/A (rs767649) and miR-146a A/G (rs57095329) single nucleotide polymorphisms as risk factors for chronic hepatitis B virus infection among Egyptian patients. PLoS ONE 2021, 16, e0256724. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/flaviviridae/362/genus-hepacivirus (accessed on 19 July 2022).
- Bala, S.; Tilahun, Y.; Taha, O.; Alao, H.; Kodys, K.; Catalano, D.; Szabo, G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J. Transl. Med. 2012, 10, 151. [Google Scholar] [CrossRef]
- Jiang, M.; Broering, R.; Trippler, M.; Wu, J.; Zhang, E.; Zhang, X.; Gerken, G.; Lu, M.; Schlaak, J.F. MicroRNA-155 controls Toll-like receptor 3- and hepatitis C virus-induced immune responses in the liver. J. Viral Hepat. 2014, 21, 99–110. [Google Scholar] [CrossRef]
- Sidorkiewicz, M.; Grek, M.; Piekarska, A.; Bartkowiak, J.; Fendler, W.; Kuydowicz, J.; Wroblewski, P.; Paradowski, M. Coordinated increase of miRNA-155 and miRNA-196b expression correlates with the detection of the antigenomic strand of hepatitis C virus in peripheral blood mononuclear cells. Int. J. Mol. Med. 2011, 28, 875–880. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Q.; Ren, J.P.; Zhao, J.; Wang, J.M.; Zhou, Y.; Li, G.Y.; Moorman, J.P.; Yao, Z.Q. MicroRNA-155 regulates interferon-γ production in natural killer cells via Tim-3 signalling in chronic hepatitis C virus infection. Immunology 2015, 145, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Riad, S.E.; El-Ekiaby, N.; Mekky, R.Y.; Ahmed, R.; EL Din, M.A.M.; El-Sayed, M.; Abouelkhair, M.M.; Salah, A.; Zekri, A.R.; Esmat, G.; et al. Expression signature of microRNA-155 in hepatitis C virus genotype 4 infection. Biomed. Rep. 2015, 3, 93–97. [Google Scholar] [CrossRef]
- El-Ekiaby, N.; Hamdi, N.; Negm, M.; Ahmed, R.; Zekri, A.-R.; Esmat, G.; Abdelaziz, A. Repressed induction of interferon-related microRNAs miR-146a and miR-155 in peripheral blood mononuclear cells infected with HCV genotype 4. FEBS Open Bio 2012, 2, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wei, W.; Cheng, N.; Wang, K.; Li, B.; Jiang, X.; Sun, S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology 2012, 56, 1631–1640. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/flaviviridae/360/genus-flavivirus (accessed on 19 July 2022).
- Arboleda, J.F.; Fernandez, G.J.; Urcuqui-Inchima, S. Vitamin D-mediated attenuation of miR-155 in human macrophages infected with dengue virus: Implications for the cytokine response. Infect. Genet. Evol. 2019, 69, 12–21. [Google Scholar] [CrossRef]
- Costa, V.V.; Fagundes, C.T.; Souza, D.G.; Teixeira, M.M. Inflammatory and Innate Immune Responses in Dengue Infection: Protection versus disease induction. Am. J. Pathol. 2013, 182, 1950–1961. [Google Scholar] [CrossRef]
- Su, Y.; Huang, Y.; Wu, Y.; Chen, H.; Hsu, C.; Hsu, Y.; Lee, J. MicroRNA-155 inhibits dengue virus replication by inducing heme oxygenase-1-mediated antiviral interferon responses. FASEB J. 2020, 34, 7283–7294. [Google Scholar] [CrossRef]
- Hayes, E.B.; Komar, N.; Nasci, R.S.; Montgomery, S.P.; O’Leary, D.R.; Campbell, G.L. Epidemiology and Transmission Dynamics of West Nile Virus Disease. Emerg. Infect. Dis. 2005, 11, 1167–1173. [Google Scholar] [CrossRef]
- Kumar, M.; Nerurkar, V.R. Integrated analysis of microRNAs and their disease related targets in the brain of mice infected with West Nile virus. Virology 2014, 452–453, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Park, K.W.; Baik, H.H.; Jin, B.K. IL-13-Induced Oxidative Stress via Microglial NADPH Oxidase Contributes to Death of Hippocampal Neurons In Vivo. J. Immunol. 2009, 183, 4666–4674. [Google Scholar] [CrossRef] [PubMed]
- Natekar, J.P.; Rothan, H.A.; Arora, K.; Strate, P.G.; Kumar, M. Cellular microRNA-155 Regulates Virus-Induced Inflammatory Response and Protects against Lethal West Nile Virus Infection. Viruses 2019, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_9th_report/negative-sense-rna-viruses-2011/w/negrna_viruses/209/orthomyxoviridae (accessed on 19 July 2022).
- Herold, S.; Becker, C.; Ridge, K.M.; Budinger, G.S. Influenza virus-induced lung injury: Pathogenesis and implications for treatment. Eur. Respir. J. 2015, 45, 1463–1478. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Kroeze, E.J.B.V.; Fouchier, R.A.M.; Kuiken, T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 2014, 14, 57–69. [Google Scholar] [CrossRef]
- Shen, S.-M.; Jiang, H.; Zhao, J.-N.; Shi, Y. Down-regulation of miR-155 inhibits inflammatory response in human pulmonary microvascular endothelial cells infected with influenza A virus by targeting sphingosine-1-phosphate receptor. Chin. Med. J. 2020, 133, 2429–2436. [Google Scholar] [CrossRef]
- Izzard, L.; Dlugolenski, D.; Xia, Y.; McMahon, M.; Middleton, D.; Tripp, R.A.; Stambas, J. Enhanced immunogenicity following miR-155 incorporation into the influenza A virus genome. Virus Res. 2017, 235, 115–120. [Google Scholar] [CrossRef]
- Gracias, D.T.; Stelekati, E.; Hope, J.L.; Boesteanu, A.C.; Doering, T.A.; Norton, J.; Mueller, Y.M.; Fraietta, J.A.; Wherry, E.J.; Turner, M.; et al. The microRNA miR-155 controls CD8+ T cell responses by regulating interferon signaling. Nat. Immunol. 2013, 14, 593–602. [Google Scholar] [CrossRef]
- Pociask, D.A.; Robinson, K.M.; Chen, K.; McHugh, K.J.; Clay, M.; Huang, G.T.; Benos, P.V.; Janssen-Heininger, Y.M.; Kolls, J.K.; Anathy, V.; et al. Epigenetic and Transcriptomic Regulation of Lung Repair during Recovery from Influenza Infection. Am. J. Pathol. 2017, 187, 851–863. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/pneumoviridae (accessed on 19 July 2022).
- Collins, P.L.; Murphy, B.R. Respiratory Syncytial Virus: Reverse Genetics and Vaccine Strategies. Virology 2002, 296, 204–211. [Google Scholar] [CrossRef][Green Version]
- Arroyo, M.; Salka, K.; Chorvinsky, E.; Xuchen, X.; Abutaleb, K.; Perez, G.F.; Weinstock, J.; Gaviria, S.; Gutierrez, M.J.; Nino, G. Airway mir-155 responses are associated with TH1 cytokine polarization in young children with viral respiratory infections. PLoS ONE 2020, 15, e0233352. [Google Scholar] [CrossRef] [PubMed]
- Inchley, C.S.; Sonerud, T.; Fjærli, H.O.; Nakstad, B. Nasal mucosal microRNA expression in children with respiratory syncytial virus infection. BMC Infect. Dis. 2015, 15, 150. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.G.; Chen, Q.; Wang, R.; Xiong, T.T.; Yang, L.X. Relationship between the expression of miR-146a and miR-155 in peripheral blood mononuclear cells and inflammation response in infants with respiratory syncytial virus pneumonia. Chin. J. Child Health Care 2020, 28, 316–319. [Google Scholar] [CrossRef]
- Oshansky, C.M.; Zhang, W.; Moore, E.; Tripp, R.A. The host response and molecular pathogenesis associated with respiratory syncytial virus infection. Futur. Microbiol. 2009, 4, 279–297. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef]
- Lu, C.; Huang, X.; Zhang, X.; Roensch, K.; Cao, Q.; Nakayama, K.I.; Blazar, B.R.; Zeng, Y.; Zhou, X. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood 2011, 117, 4293–4303. [Google Scholar] [CrossRef]
- Wang, S.; Ling, Y.; Yao, Y.; Zheng, G.; Chen, W. Luteolin inhibits respiratory syncytial virus replication by regulating the MiR-155/SOCS1/STAT1 signaling pathway. Virol. J. 2020, 17, 187. [Google Scholar] [CrossRef]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Nemati, M.; Saha, B.; Bansode, Y.D.; Jafarzadeh, S. Protective Potentials of Type III Interferons in COVID-19 Patients: Lessons from Differential Properties of Type I- and III Interferons. Viral Immunol. 2021, 34, 307–320. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Chauhan, P.; Saha, B.; Jafarzadeh, S.; Nemati, M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020, 257, 118102. [Google Scholar] [CrossRef] [PubMed]
- Jafarzadeh, A.; Nemati, M.; Jafarzadeh, S. Contribution of STAT3 to the pathogenesis of COVID-19. Microb. Pathog. 2021, 154, 104836. [Google Scholar] [CrossRef] [PubMed]
- Haroun, R.A.-H.; Osman, W.H.; Amin, R.E.; Hassan, A.K.; Abo-Shanab, W.S.; Eessa, A.M. Circulating plasma miR-155 is a potential biomarker for the detection of SARS-CoV-2 infection. Pathology 2022, 54, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Demirci, Y.M.; Demirci, M.D.S. Circular RNA–MicroRNA–MRNA interaction predictions in SARS-CoV-2 infection. J. Integr. Bioinform. 2021, 18, 45–50. [Google Scholar] [CrossRef]
- Donyavi, T.; Bokharaei-Salim, F.; Baghi, H.B.; Khanaliha, K.; Janat-Makan, M.A.; Karimi, B.; Nahand, J.S.; Mirzaei, H.; Khatami, A.; Garshasbi, S.; et al. Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155–5p, and let-7b-3p in PBMC. Int. Immunopharmacol. 2021, 97, 107641. [Google Scholar] [CrossRef]
- Soni, D.K.; Cabrera-Luque, J.; Kar, S.; Sen, C.; Devaney, J.; Biswas, R. Suppression of miR-155 attenuates lung cytokine storm induced by SARS-CoV-2 infection in human ACE2-transgenic mice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wyler, E.; Mösbauer, K.; Franke, V.; Diag, A.; Gottula, L.T.; Arsiè, R.; Klironomos, F.; Koppstein, D.; Hönzke, K.; Ayoub, S.; et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2021, 24, 102151. [Google Scholar] [CrossRef]
- Chow, J.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef]
- Qi, M.; Liu, B.; Li, S.; Ni, Z.; Li, F. Construction and Investigation of Competing Endogenous RNA Networks and Candidate Genes Involved in SARS-CoV-2 Infection. Int. J. Gen. Med. 2021, 14, 6647–6659. [Google Scholar] [CrossRef]
- Garg, A.; Seeliger, B.; Derda, A.A.; Xiao, K.; Gietz, A.; Scherf, K.; Sonnenschein, K.; Pink, I.; Hoeper, M.M.; Welte, T.; et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021, 23, 468–475. [Google Scholar] [CrossRef]
- Benkő, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kaján, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV Virus Taxonomy Profile: Adenoviridae. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef]
- Kidd, A.H.; Garwicz, D.; Oberg, M. Human and Simian Adenoviruses: Phylogenetic Inferences from Analysis of VA RNA Genes. Virology 1995, 207, 32–45. [Google Scholar] [CrossRef][Green Version]
- Lynch, J.P., 3rd; Kajon, A.E. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention. Semin. Respir. Crit. Care Med. 2016, 37, 586–602. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, M.; Tellgren-Roth, C.; Pettersson, U. Fluctuating expression of microRNAs in adenovirus infected cells. Virology 2015, 478, 99–111. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. Inducible microRNA-155 Feedback Promotes Type I IFN Signaling in Antiviral Innate Immunity by Targeting Suppressor of Cytokine Signaling. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/picornaviridae (accessed on 19 July 2022).
- Bondanese, V.P.; Francisco-Garcia, A.; Bedke, N.; Davies, D.E.; Sanchez-Elsner, T. Identification of host miRNAs that may limit human rhinovirus replication. World J. Biol. Chem. 2014, 5, 437–456. [Google Scholar] [CrossRef]
- Gutierrez, M.J.; Gómez, J.L.; Perez, G.F.; Pancham, K.; Val, S.; Pillai, D.K.; Giri, M.; Ferrante, S.; Freishtat, R.; Rose, M.C.; et al. Airway Secretory microRNAome Changes during Rhinovirus Infection in Early Childhood. PLoS ONE 2016, 11, e0162244. [Google Scholar] [CrossRef]
- Taïbi, F.; Meuth, V.M.-L.; Massy, Z.A.; Metzinger, L. miR-223: An inflammatory oncomiR enters the cardiovascular field. Biochim. Biophys. Acta 2014, 1842, 1001–1009. [Google Scholar] [CrossRef]
- Jiao, P.; Wang, X.-P.; Luoreng, Z.-M.; Yang, J.; Jia, L.; Ma, Y.; Wei, D.-W. miR-223: An Effective Regulator of Immune Cell Differentiation and Inflammation. Int. J. Biol. Sci. 2021, 17, 2308–2322. [Google Scholar] [CrossRef]
- Yuan, S.; Wu, Q.; Wang, Z.; Che, Y.; Zheng, S.; Chen, Y.; Zhong, X.; Shi, F. miR-223: An Immune Regulator in Infectious Disorders. Front. Immunol. 2021, 12, 781815. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating MicroRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Wong, Q.W.; Lung, R.W.; Law, P.T.; Lai, P.B.-S.; Chan, K.Y.Y.; To, K.; Wong, N. MicroRNA-223 Is Commonly Repressed in Hepatocellular Carcinoma and Potentiates Expression of Stathmin1. Gastroenterology 2008, 135, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Giray, B.G.; Emekdas, G.; Tezcan, S.; Ulger, M.; Serin, M.S.; Sezgin, O.; Altintas, E.; Tiftik, E.N. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol. Biol. Rep. 2014, 41, 4513–4519. [Google Scholar] [CrossRef]
- Yu, G.; Chen, X.; Chen, S.; Ye, W.; Hou, K.; Liang, M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J. Transl. Med. 2016, 14, 122. [Google Scholar] [CrossRef] [PubMed]
- Hyrina, A.; Olmstead, A.D.; Steven, P.; Krajden, M.; Tam, E.; Jean, F. Treatment-Induced Viral Cure of Hepatitis C Virus-Infected Patients Involves a Dynamic Interplay among three Important Molecular Players in Lipid Homeostasis: Circulating microRNA (miR)-24, miR-223, and Proprotein Convertase Subtilisin/Kexin Type 9. eBioMedicine 2017, 23, 68–78. [Google Scholar] [CrossRef]
- El-Guendy, N.M.; Helwa, R.; El-Halawany, M.S.; Ali, S.A.R.; Aly, M.T.; Alieldin, N.H.; Fouad, S.A.H.; Saeid, H.; Abdel-Wahab, A.-H.A. The Liver MicroRNA Expression Profiles Associated With Chronic Hepatitis C Virus (HCV) Genotype-4 Infection: A Preliminary Study. Hepat. Mon. 2016, 16, e33881. [Google Scholar] [CrossRef]
- Wu, N.; Gao, N.; Fan, D.; Wei, J.; Zhang, J.; An, J. miR-223 inhibits dengue virus replication by negatively regulating the microtubule-destabilizing protein STMN1 in EAhy926 cells. Microbes Infect. 2014, 16, 911–922. [Google Scholar] [CrossRef]
- Li, Y.; Chan, E.Y.; Li, J.; Ni, C.; Peng, X.; Rosenzweig, E.; Tumpey, T.M.; Katze, M.G. MicroRNA Expression and Virulence in Pandemic Influenza Virus-Infected Mice. J. Virol. 2010, 84, 3023–3032. [Google Scholar] [CrossRef]
- Vela, E.M.; Kasoji, M.D.; Wendling, M.Q.; Price, J.A.; Knostman, K.A.B.; Bresler, H.S.; Long, J.P. MicroRNA expression in mice infected with seasonal H1N1, swine H1N1 or highly pathogenic H5N. J. Med Microbiol. 2014, 63, 1131–1142. [Google Scholar] [CrossRef]
- Wu, Z.; Hao, R.; Li, P.; Zhang, X.; Liu, N.; Qiu, S.; Wang, L.; Wang, Y.; Xue, W.; Liu, K.; et al. MicroRNA Expression Profile of Mouse Lung Infected with 2009 Pandemic H1N1 Influenza Virus. PLoS ONE 2013, 8, e74190. [Google Scholar] [CrossRef] [PubMed]
- Tambyah, P.A.; Sepramaniam, S.; Ali, J.M.; Chai, S.C.; Swaminathan, P.; Armugam, A.; Jeyaseelan, K. microRNAs in Circulation Are Altered in Response to Influenza A Virus Infection in Humans. PLoS ONE 2013, 8, e76811. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-J.; Kim, H.B.; Baek, Y.H.; Kim, E.-H.; Pascua, P.N.Q.; Park, S.-J.; Kwon, H.-I.; Lim, G.-J.; Kim, S.; Kim, Y.-I.; et al. Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 2014, 14, 252. [Google Scholar] [CrossRef] [PubMed]
- Moschos, S.A.; Williams, A.E.; Perry, M.M.; Birrell, M.A.; Belvisi, M.G.; Lindsay, M.A. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genom. 2007, 8, 240. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-C.; Miao, Y.; Chen, R.-L.; Zhang, Y.-Q.; Wu, H.; Yang, S.-M.; Shang, L.-Q. STIM1 mediates IAV-induced inflammation of lung epithelial cells by regulating NLRP3 and inflammasome activation via targeting miR-223. Life Sci. 2021, 266, 118845. [Google Scholar] [CrossRef] [PubMed]
- Micaroni, M.; Stanley, A.C.; Khromykh, T.; Venturato, J.; Wong, C.X.F.; Lim, J.P.; Marsh, B.J.; Storrie, B.; Gleeson, P.A.; Stow, J.L. Rab6a/a’ Are Important Golgi Regulators of Pro-Inflammatory TNF Secretion in Macrophages. PLoS ONE 2013, 8, e57034. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xu, Y.; Bao, L.; Zhang, L.; Yu, P.; Qu, Y.; Zhu, H.; Zhao, W.; Han, Y.; Qin, C. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses 2019, 11, 59. [Google Scholar] [CrossRef]
- Mallick, B.; Ghosh, Z.; Chakrabarti, J. MicroRNome Analysis Unravels the Molecular Basis of SARS Infection in Bronchoalveolar Stem Cells. PLoS ONE 2009, 4, e7837. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, X.; Jiang, X.-M.; Guo, J.; Fu, Z.; Zhou, Z.; Yang, P.; Guo, H.; Guo, X.; Liang, G.; et al. Decreased inhibition of exosomal miRNAs on SARS-CoV-2 replication underlies poor outcomes in elderly people and diabetic patients. Signal Transduct. Target. Ther. 2021, 6, 300. [Google Scholar] [CrossRef]
- Demiray, A.; Sarı, T.; Çalışkan, A.; Nar, R.; Aksoy, L.; Akbubak, I.H. Serum microRNA signature is capable of predictive and prognostic factor for SARS-COV-2 virulence. Turk. J. Biochem. 2021, 46, 245–253. [Google Scholar] [CrossRef]
- Houshmandfar, S.; Saeedi-Boroujeni, A.; Rashno, M.; Khodadadi, A.; Mahmoudian-Sani, M.-R. miRNA-223 as a regulator of inflammation and NLRP3 inflammasome, the main fragments in the puzzle of immunopathogenesis of different inflammatory diseases and COVID-19. Naunyn-Schmiedebergs Arch. Pharmakol. 2021, 394, 2187–2195. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/reverse-transcribing-dna-and-rna-viruses/w/retroviridae (accessed on 19 July 2022).
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Purohit, V.; Rapaka, R.S.; Rutter, J.; Shurtleff, D. Do Opioids Activate Latent HIV-1 by Down-Regulating Anti-HIV microRNAs? J. Neuroimmune Pharmacol. 2012, 7, 519–523. [Google Scholar] [CrossRef]
- Sun, G.; Li, H.; Wu, X.; Covarrubias, M.; Scherer, L.; Meinking, K.; Luk, B.; Chomchan, P.; Alluin, J.; Gombart, A.F.; et al. Interplay between HIV-1 infection and host microRNAs. Nucleic Acids Res. 2012, 40, 2181–2196. [Google Scholar] [CrossRef]
- Monticelli, S.; Ansel, K.M.; Xiao, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.-H.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, R71. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Curtale, G.; Citarella, F.; Carissimi, C.; Goldoni, M.; Carucci, N.; Fulci, V.; Franceschini, D.; Meloni, F.; Barnaba, V.; Macino, G. An emerging player in the adaptive immune response: MicroRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 2010, 115, 265–273. [Google Scholar] [CrossRef]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a Feedback Inhibits RIG-I-Dependent Type I IFN Production in Macrophages by Targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Ju, Y.; Zhao, B.; Yan, X.; Hu, J.; Shi, L.; Yang, L.; Ma, Z.; Chen, L.; et al. MicroRNA-146a Feedback Suppresses T Cell Immune Function by Targeting Stat1 in Patients with Chronic Hepatitis B. J. Immunol. 2013, 191, 293–301. [Google Scholar] [CrossRef]
- Fu, L.; Fu, X.; Mo, J.; Li, X.; Li, R.; Peng, S. miR-146a-5p enhances hepatitis B virus replication through autophagy to promote aggravation of chronic hepatitis B. IUBMB Life 2019, 71, 1336–1346. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y. miR-146 promotes HBV replication and expression by targeting ZEB2. Biomed. Pharmacother. 2018, 99, 576–582. [Google Scholar] [CrossRef]
- Ji, M.; Mei, X.; Jing, X.; Xu, X.; Chen, X.; Pan, W. The cooperative complex of Argonaute-2 and microRNA-146a regulates hepatitis B virus replication through flap endonuclease. Life Sci. 2020, 257, 118089. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Dai, X.-P.; Zhang, W.; Sun, S.-H.; Zeng, Y.; Zhao, G.-Y.; Kou, Z.-H.; Guo, Y.; Yu, H.; Du, L.-Y.; et al. Upregulation of MicroRNA-146a by Hepatitis B Virus X Protein Contributes to Hepatitis Development by Downregulating Complement Factor H. mBio 2015, 6, e02459–e002473. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; He, X.; Li, J.; Xie, Q.; Lin, J.; Chang, Y. Association of a single-nucleotide polymorphism within the miR-146a gene with susceptibility for acute-on-chronic hepatitis B liver failure. Immunogenetics 2013, 65, 257–263. [Google Scholar] [CrossRef]
- He, Q.; Li, W.; Ren, J.; Huang, Y.; Huang, Y.; Hu, Q.; Chen, J.; Chen, W. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget 2016, 7, 16003–16011. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.Y.; Ge, X.; Shan, X.; Lei, X.; Ke, C.; Huang, A.; Hu, J. Effect of structure-specific nucleic acid enzyme FEN1 on HBV replication. Third. Mil. Med. Univ. 2012, 34, 1935–1938. [Google Scholar]
- Ren, J.P.; Ying, R.S.; Cheng, Y.Q.; Wang, L.; Elgazzar, M.A.; Li, G.Y.; Ning, S.B.; Moorman, J.P.; Yao, Z.Q. HCV-induced miR146a controls SOCS1/STAT3 and cytokine expression in monocytes to promote regulatory T-cell development. J. Viral Hepat. 2016, 23, 755–766. [Google Scholar] [CrossRef]
- Zhang, W.-J.; Wang, H.; Tong, Q.-X.; Jie, S.-H.; Yang, D.-L.; Peng, C. Involvement of TLR2-MyD88 in abnormal expression of miR-146a in peripheral blood monocytes of patients with chronic hepatitis C. J. Huazhong Univ. Sci. Technol. 2015, 35, 219–224. [Google Scholar] [CrossRef]
- Bandiera, S.; Pernot, S.; El Saghire, H.; Durand, S.C.; Thumann, C.; Crouchet, E.; Ye, T.; Fofana, I.; Oudot, M.A.; Barths, J.; et al. Hepatitis C Virus-Induced Upregulation of MicroRNA miR-146a-5p in Hepatocytes Promotes Viral Infection and Deregulates Metabolic Pathways Associated with Liver Disease Pathogenesis. J. Virol. 2016, 90, 6387–6400. [Google Scholar] [CrossRef]
- Srikiatkhachorn, A.; Mathew, A.; Rothman, A.L. Immune-mediated cytokine storm and its role in severe dengue. Semin. Immunopathol. 2017, 39, 563–574. [Google Scholar] [CrossRef]
- Wu, S.; He, L.; Li, Y.; Wang, T.; Feng, L.; Jiang, L.; Zhang, P.; Huang, X. miR-146a facilitates replication of dengue virus by dampening interferon induction by targeting TRAF. J. Infect. 2013, 67, 329–341. [Google Scholar] [CrossRef]
- Pu, J.; Wu, S.; Xie, H.; Li, Y.; Yang, Z.; Wu, X.; Huang, X. miR-146a Inhibits dengue-virus-induced autophagy by targeting TRAF6. Arch. Virol. 2017, 162, 3645–3659. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.F.; de Andrade, A.A.S.; Pagliari, C.; de Carvalho, L.V.; Silveira, T.S.; Cardoso, J.F.; e Silva, A.L.T.; de Vasconcelos, J.M.; Moreira-Nunes, C.A.; Burbano, R.M.R.; et al. Differential expression analysis and profiling of hepatic miRNA and isomiRNA in dengue hemorrhagic fever. Sci. Rep. 2021, 11, 5554. [Google Scholar] [CrossRef]
- Ouyang, X.; Jiang, X.; Gu, D.; Zhang, Y.; Kong, S.K.; Jiang, C.; Xie, W. Dysregulated Serum MiRNA Profile and Promising Biomarkers in Dengue-infected Patients. Int. J. Med. Sci. 2016, 13, 195–205. [Google Scholar] [CrossRef]
- Zhang, F.; Sun, X.; Zhu, Y.; Qin, W. Downregulation of miR-146a inhibits influenza A virus replication by enhancing the type I interferon response in vitro and in vivo. Biomed. Pharmacother. 2019, 111, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Buggele, W.A.; Johnson, K.E.; Horvath, C.M. Influenza A Virus Infection of Human Respiratory Cells Induces Primary MicroRNA Expression. J. Biol. Chem. 2012, 287, 31027–31040. [Google Scholar] [CrossRef]
- Terrier, O.; Textoris, J.; Carron, C.; Marcel, V.; Bourdon, J.-C.; Rosa-Calatrava, M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J. Gen. Virol. 2013, 94, 985–995. [Google Scholar] [CrossRef]
- Deng, Y.; Yan, Y.; Tan, K.S.; Liu, J.; Chow, V.T.; Tao, Z.-Z.; Wang, D.-Y. MicroRNA-146a induction during influenza H3N2 virus infection targets and regulates TRAF6 levels in human nasal epithelial cells (hNECs). Exp. Cell Res. 2017, 352, 184–192. [Google Scholar] [CrossRef]
- Eilam-Frenkel, B.; Naaman, H.; Brkic, G.; Veksler-Lublinsky, I.; Rall, G.; Shemer-Avni, Y.; Gopas, J. MicroRNA 146-5p, miR-let-7c-5p, miR-221 and miR-345-5p are differentially expressed in Respiratory Syncytial Virus (RSV) persistently infected HEp-2 cells. Virus Res. 2018, 251, 34–39. [Google Scholar] [CrossRef]
- Lubkowska, A.; Pluta, W.; Strońska, A.; Lalko, A. Role of Heat Shock Proteins (HSP70 and HSP90) in Viral Infection. Int. J. Mol. Sci. 2021, 22, 9366. [Google Scholar] [CrossRef]
- Tang, H.; Gao, Y.; Li, Z.; Miao, Y.; Huang, Z.; Liu, X.; Xie, L.; Li, H.; Wen, W.; Zheng, Y.; et al. The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19. Clin. Transl. Med. 2020, 10, e200. [Google Scholar] [CrossRef] [PubMed]
- Sabbatinelli, J.; Giuliani, A.; Matacchione, G.; Latini, S.; Laprovitera, N.; Pomponio, G.; Ferrarini, A.; Baroni, S.S.; Pavani, M.; Moretti, M.; et al. Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021, 193, 111413. [Google Scholar] [CrossRef] [PubMed]
- Roganović, J. Downregulation of microRNA-146a in diabetes, obesity and hypertension may contribute to severe COVID-19. Med Hypotheses 2021, 146, 110448. [Google Scholar] [CrossRef]
- Roganović, J.R. microRNA-146a and -155, upregulated by periodontitis and type 2 diabetes in oral fluids, are predicted to regulate SARS-CoV-2 oral receptor genes. J. Periodontol. 2021, 92, e35–e43. [Google Scholar] [CrossRef] [PubMed]
- Lagos-Quintana, M.; Rauhut, R.; Yalcin, A.; Meyer, J.; Lendeckel, W.; Tuschl, T. Identification of Tissue-Specific MicroRNAs from Mouse. Curr. Biol. 2002, 12, 735–739. [Google Scholar] [CrossRef]
- Girard, M.; Jacquemin, E.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008, 48, 648–656. [Google Scholar] [CrossRef]
- Xu, H.; He, J.-H.; Xiao, Z.-D.; Zhang, Q.-Q.; Chen, Y.-Q.; Zhou, H.; Qu, L.-H. Liver-enriched transcription factors regulate MicroRNA-122 that targets CUTL1 during liver development. Hepatology 2010, 52, 1431–1442. [Google Scholar] [CrossRef]
- Laudadio, I.; Manfroid, I.; Achouri, Y.; Schmidt, D.; Wilson, M.D.; Cordi, S.; Thorrez, L.; Knoops, L.; Jacquemin, P.; Schuit, F.; et al. A Feedback Loop Between the Liver-Enriched Transcription Factor Network and Mir-122 Controls Hepatocyte Differentiation. Gastroenterology 2012, 142, 119–129. [Google Scholar] [CrossRef]
- Bandiera, S.; Pfeffer, S.; Baumert, T.F.; Zeisel, M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015, 62, 448–457. [Google Scholar] [CrossRef]
- Arataki, K.; Hayes, C.N.; Akamatsu, S.; Akiyama, R.; Abe, H.; Tsuge, M.; Miki, D.; Ochi, H.; Hiraga, N.; Imamura, M.; et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med Virol. 2013, 85, 789–798. [Google Scholar] [CrossRef]
- Wang, S.; Qiu, L.; Yan, X.; Jin, W.; Wang, Y.; Chen, L.; Wu, E.; Ye, X.; Gao, G.F.; Wang, F.; et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G1-modulated P53 activity. Hepatology 2012, 55, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Gao, C.; Cai, S.; Xia, M.; Liao, G.; Zhang, X.; Peng, J. Circulating miR-122 Is a Predictor for Virological Response in CHB Patients with High Viral Load Treated with Nucleos(t)ide Analogs. Front. Genet. 2019, 10, 243. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimifard, M.; Zandi, M.; Moradi, A. Evaluation of miR-122 levels in chronic HBV and liver cirrhosis patients. Arch. Med. Lab. Sci. 2013, 2, 3. [Google Scholar]
- Van der Ree, M.H.; Jansen, L.; Kruize, Z.; van Nuenen, A.C.; van Dort, K.A.; Takkenberg, R.B.; Reesink, H.W.; Kootstra, N.A. Plasma MicroRNA Levels Are Associated with Hepatitis B e Antigen Status and Treatment Response in Chronic Hepatitis B Patients. J. Infect. Dis. 2017, 215, 1421–1429. [Google Scholar] [CrossRef]
- Chen, S.; Ni, M.; Yu, B.; Lv, T.; Lu, M.; Gong, F. Construction and identification of a human liver specific microRNA eukaryotic expression vector. Cell. Mol. Immunol. 2007, 4, 473–477. [Google Scholar]
- Chen, Y.; Shen, A.; Rider, P.J.; Yu, Y.; Wu, K.; Mu, Y.; Hao, Q.; Liu, Y.; Gong, H.; Zhu, Y.; et al. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 2011, 25, 4511–4521. [Google Scholar] [CrossRef]
- Li, R.-F.; Fan, C.-G.; Wang, C.-M.; Tian, C.; Wang, Y.; Li, L.; Sun, W.-S.; Liu, Y.-G. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol. Rep. 2011, 26, 1281–1286. [Google Scholar] [CrossRef]
- Wu, D.-B.; Liu, F.-W.; Li, J.; Liu, C.; Liu, L.; Chen, E.-Q.; Zhao, L.-S.; Tang, H.; Zhou, T.-Y. Intrahepatic IFN-alpha expression in liver specimens from HBV-infected patients with different outcomes. Eur. Rev. Med Pharmacol. Sci. 2013, 17, 2474–2480. [Google Scholar]
- Koeberlein, B.; Hausen, A.Z.; Bektas, N.; Zentgraf, H.; Chin, R.; Toan, N.L.; Kandolf, R.; Torresi, J.; Bock, C.T. Hepatitis B virus overexpresses suppressor of cytokine signaling-3 (SOCS3) thereby contributing to severity of inflammation in the liver. Virus Res. 2010, 148, 51–59. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Xi, Y.; Zhu, W.-N.; Zeng, C.; Zhang, Z.-Q.; Guo, Z.-C.; Hao, D.-L.; Liu, G.; Feng, L.; Chen, H.-Z.; et al. Positive regulation of hepatic miR-122 expression by HNF4α. J. Hepatol. 2011, 55, 602–611. [Google Scholar] [CrossRef]
- Song, K.; Han, C.; Dash, S.; Balart, L.A.; Wu, T. MiR-122 in hepatitis B virus and hepatitis C virus dual infection. World J. Hepatol. 2015, 7, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Wu, J.; Zhai, A.; Qian, J.; Wang, X.; Qaria, M.A.; Zhang, Q.; Li, Y.; Fang, Y.; Kao, W.; et al. HBV triggers APOBEC2 expression through miR-122 regulation and affects the proliferation of liver cancer cells. Int. J. Oncol. 2019, 55, 1137–1148. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Han, C.; Zhang, J.; Lu, N.; Dash, S.; Feitelson, M.; Lim, K.; Wu, T. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology 2013, 58, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Xiao, X.; Jiang, Y.; Luo, K.; Tian, Y.; Peng, M.; Zhang, M.; Xu, Y.; Gong, G. HBx Down-Regulated Gld2 Plays a Critical Role in HBV-Related Dysregulation of miR-122. PLoS ONE 2014, 9, e92998. [Google Scholar] [CrossRef]
- Ding, J.; Ding, X.; Ning, J.; Yi, F.; Chen, J.; Zhao, D.; Zheng, J.; Liang, Z.; Hu, Z.; Du, Q. Circulating microRNA-122 as a potential biomarker for liver injury. Mol. Med. Rep. 2012, 5, 1428–1432. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Fang, S.; Wang, M.; Xiong, A.; Zheng, C.; Wang, J.; Yin, C. Diagnostic value of circulating miRNA-122 for hepatitis B virus and/or hepatitis C virus-associated chronic viral hepatitis. Biosci. Rep. 2019, 39, BSR20190900. [Google Scholar] [CrossRef]
- Shimakami, T.; Yamane, D.; Jangra, R.K.; Kempf, B.J.; Spaniel, C.; Barton, D.J.; Lemon, S.M. Stabilization of hepatitis C virus RNA by an Ago2–miR-122 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 941–946. [Google Scholar] [CrossRef]
- Jopling, C.L.; Yi, M.; Lancaster, A.M.; Lemon, S.M.; Sarnow, P. Modulation of Hepatitis C Virus RNA Abundance by a Liver-Specific MicroRNA. Science 2005, 309, 1577–1581. [Google Scholar] [CrossRef]
- Chang, J.; Guo, J.-T.; Jiang, D.; Guo, H.; Taylor, J.M.; Block, T.M. Liver-Specific MicroRNA miR-122 Enhances the Replication of Hepatitis C Virus in Nonhepatic Cells. J. Virol. 2008, 82, 8215–8223. [Google Scholar] [CrossRef]
- Narbus, C.M.; Israelow, B.; Sourisseau, M.; Michta, M.L.; Hopcraft, S.E.; Zeiner, G.M.; Evans, M.J. HepG2 Cells Expressing MicroRNA miR-122 Support the Entire Hepatitis C Virus Life Cycle. J. Virol. 2011, 85, 12087–12092. [Google Scholar] [CrossRef]
- Henke, J.I.; Goergen, D.; Zheng, J.; Song, Y.; Schüttler, C.G.; Fehr, C.; Jünemann, C.; Niepmann, M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008, 27, 3300–3310. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Zheng, J.; Lambrecht, R.W.; Bonkovsky, H.L. Reciprocal Effects of Micro-RNA-122 on Expression of Heme Oxygenase-1 and Hepatitis C Virus Genes in Human Hepatocytes. Gastroenterology 2007, 133, 1166–1174. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.M.; Cheng, G.; Wieland, S.; Volinia, S.; Croce, C.M.; Chisari, F.V.; David, M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 2007, 449, 919–922. [Google Scholar] [CrossRef]
- Wang, X.; Peng, J.; Wang, J.; Li, M.; Wu, D.; Wu, S.; Liao, J.; Dou, J. Hepatitis C virus core impacts expression of miR122 and miR204 involved in carcinogenic progression via regulation of TGFBRAP1 and HOTTIP expression. OncoTargets Ther. 2018, 11, 1173–1182. [Google Scholar] [CrossRef] [PubMed]
- Bihrer, V.; Friedrich-Rust, M.; Kronenberger, B.; Forestier, N.; Haupenthal, J.; Shi, Y.; Peveling-Oberhag, J.; Radeke, H.; Sarrazin, C.; Herrmann, E.; et al. Serum miR-122 as a Biomarker of Necroinflammation in Patients With Chronic Hepatitis C Virus Infection. Am. J. Gastroenterol. 2011, 106, 1663–1669. [Google Scholar] [CrossRef]
- Tambyah, P.A.; Ching, C.S.; Sepramaniam, S.; Ali, J.M.; Armugam, A.; Jeyaseelan, K. microRNA expression in blood of dengue patients. Ann. Clin. Biochem. 2016, 53, 466–476. [Google Scholar] [CrossRef]
- Lee, T.-C.; Lin, Y.-L.; Liao, J.-T.; Su, C.-M.; Lin, C.-C.; Lin, W.-P.; Liao, C.-L. Utilizing liver-specific microRNA-122 to modulate replication of dengue virus replicon. Biochem. Biophys. Res. Commun. 2010, 396, 596–601. [Google Scholar] [CrossRef]
- Diallo, I.; Ho, J.; Laffont, B.; Laugier, J.; Benmoussa, A.; Lambert, M.; Husseini, Z.; Soule, G.; Kozak, R.; Kobinger, G.; et al. Altered microRNA Transcriptome in Cultured Human Liver Cells upon Infection with Ebola Virus. Int. J. Mol. Sci. 2021, 22, 3792. [Google Scholar] [CrossRef]
- Madara, J.J.; Han, Z.; Ruthel, G.; Freedman, B.D.; Harty, R.N. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Futur. Virol. 2015, 10, 537–546. [Google Scholar] [CrossRef]
- Wang, S.; Liu, P.; Yang, P.; Zheng, J.; Zhao, D. Peripheral blood microRNAs expression is associated with infant respiratory syncytial virus infection. Oncotarget 2017, 8, 96627–96635. [Google Scholar] [CrossRef]
- Ioannidis, I.; McNally, B.; Willette, M.; Peeples, M.E.; Chaussabel, D.; Durbin, J.E.; Ramilo, O.; Mejias, A.; Flaño, E. Plasticity and Virus Specificity of the Airway Epithelial Cell Immune Response during Respiratory Virus Infection. J. Virol. 2012, 86, 5422–5436. [Google Scholar] [CrossRef] [PubMed]
- Kurt-Jones, E.A.; Popova, L.; Kwinn, L.A.; Haynes, L.M.; Jones, L.P.; Tripp, R.; Walsh, E.E.; Freeman, M.W.; Golenbock, D.T.; Anderson, L.J.; et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000, 1, 398–401. [Google Scholar] [CrossRef]
- Haynes, L.M.; Moore, D.D.; Kurt-Jones, E.A.; Finberg, R.W.; Anderson, L.J.; Tripp, R.A. Involvement of Toll-Like Receptor 4 in Innate Immunity to Respiratory Syncytial Virus. J. Virol. 2001, 75, 10730–10737. [Google Scholar] [CrossRef]
- Thornburg, N.J.; Shepherd, B.; Crowe, J.E., Jr. Transforming Growth Factor Beta Is a Major Regulator of Human Neonatal Immune Responses following Respiratory Syncytial Virus Infection. J. Virol. 2010, 84, 12895–12902. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Salice, P.; Bosis, S.; Ghiglia, S.; Tremolati, E.; Tagliabue, C.; Gualtieri, L.; Barbier, P.; Galeone, C.; Marchisio, P.; et al. Altered cardiac rhythm in infants with bronchiolitis and respiratory syncytial virus infection. BMC Infect. Dis. 2010, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.A.E.-G.; Zayed, K.M. Impact of Acute Bronchiolitis on Cardiac Functions and Serum microRNA-122 and 499. Am. J. Infect. Dis. 2016, 12, 11–19. [Google Scholar] [CrossRef]
- Collison, A.M.; Sokulsky, L.A.; Kepreotes, E.; de Siqueira, A.P.; Morten, M.; Edwards, M.R.; Walton, R.P.; Bartlett, N.W.; Yang, M.; Nguyen, T.H.; et al. miR-122 promotes virus-induced lung disease by targeting SOCS1. JCI Insight 2021, 6, e127933. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.; et al. Modulation of miR-155 and miR-125b Levels following Lipopolysaccharide/TNF-α Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef]
- Murphy, A.J.; Guyre, P.M.; Pioli, P.A. Estradiol Suppresses NF-κB Activation through Coordinated Regulation of let-7a and miR-125b in Primary Human Macrophages. J. Immunol. 2010, 184, 5029–5037. [Google Scholar] [CrossRef]
- Busch, S.; Auth, E.; Scholl, F.; Huenecke, S.; Koehl, U.; Suess, B.; Steinhilber, D. 5-Lipoxygenase Is a Direct Target of miR-19a-3p and miR-125b-5p. J. Immunol. 2015, 194, 1646–1653. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; So, A.Y.-L.; Sinha, N.; Gibson, W.S.J.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b Potentiates Macrophage Activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Kong, N.; Ye, L.; Han, W.; Zhou, J.; Zhang, Q.; He, C.; Pan, H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014, 344, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Zhang, J.-P.; Li, B.; Zeng, C.; You, K.; Chen, M.-X.; Yuan, Y.; Zhuang, S.-M. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013, 32, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.E.; Schlamp, C.L.; Nickells, R.W. BAX to basics: How the BCL2 gene family controls the death of retinal ganglion cells. Prog. Retin. Eye Res. 2017, 57, 1–25. [Google Scholar] [CrossRef]
- Deng, W.; Zhang, X.; Ma, Z.; Lin, Y.; Lu, M. MicroRNA-125b-5p mediates post-transcriptional regulation of hepatitis B virus replication via the LIN28B/let-7 axis. RNA Biol. 2017, 14, 1389–1398. [Google Scholar] [CrossRef]
- Li, F.; Zhou, P.; Deng, W.; Wang, J.; Mao, R.; Zhang, Y.; Li, J.; Yu, J.; Yang, F.; Huang, Y.; et al. Serum microRNA-125b correlates with hepatitis B viral replication and liver necroinflammation. Clin. Microbiol. Infect. 2016, 22, 384.e1–384.e10. [Google Scholar] [CrossRef]
- Chen, S.; Chen, H.; Gao, S.; Qiu, S.; Zhou, H.; Yu, M.; Tu, J. Differential expression of plasma microRNA-125b in hepatitis B virus-related liver diseases and diagnostic potential for hepatitis B virus-induced hepatocellular carcinoma. Hepatol. Res. 2017, 47, 312–320. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; He, Y.; Zhan, X.; Zhao, R.; Huang, Y.; Xu, H.; Zhu, Z.; Liu, Q. miR-125b inhibits hepatitis B virus expression in vitro through targeting of the SCNN1A gene. Arch. Virol. 2014, 159, 3335–3343. [Google Scholar] [CrossRef]
- Peng, C.; Wang, H.; Zhang, W.-J.; Jie, S.-H.; Tong, Q.-X.; Lu, M.-J.; Yang, D.-L. Inhibitory effect of miR-125b on hepatitis C virus core protein-induced TLR2/MyD88 signaling in THP-1 cells. World J. Gastroenterol. 2016, 22, 4354–4361. [Google Scholar] [CrossRef]
- Dai, C.-Y.; Tsai, Y.-S.; Chou, W.-W.; Liu, T.; Huang, C.-F.; Wang, S.-C.; Tsai, P.-C.; Yeh, M.-L.; Hsieh, M.-Y.; Huang, C.-I.; et al. The IL-6/STAT3 pathway upregulates microRNA-125b expression in hepatitis C virus infection. Oncotarget 2018, 9, 11291–11302. [Google Scholar] [CrossRef] [PubMed]
- Shwetha, S.; Sharma, G.; Raheja, H.; Goel, A.; Aggarwal, R.; Das, S. Interaction of miR-125b-5p with Human antigen R mRNA: Mechanism of controlling HCV replication. Virus Res. 2018, 258, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sultana, C.; Rosca, A.; Ruta, S. Correlation Between miR-125b Expression and Liver Fibrosis in Patients with Chronic Hepatitis C. Hepat. Mon. 2019, 19, e84615. [Google Scholar] [CrossRef]
- Liu, W.; Hu, J.; Zhou, K.; Chen, F.; Wang, Z.; Liao, B.; Dai, Z.; Cao, Y.; Fan, J.; Zhou, J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. OncoTargets Ther. 2017, 10, 3843–3851. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/hepeviridae (accessed on 19 July 2022).
- Harms, D.; Choi, M.; Allers, K.; Wang, B.; Pietsch, H.; Papp, C.-P.; Hanisch, L.; Kurreck, J.; Hofmann, J.; Bock, C.-T. Specific circulating microRNAs during hepatitis E infection can serve as indicator for chronic hepatitis E. Sci. Rep. 2020, 10, 5337. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, F.; Yang, D.; Peng, T.; Lu, G. Identification of miRNA-mRNA Crosstalk in Respiratory Syncytial Virus- (RSV-) Associated Pediatric Pneumonia through Integrated miRNAome and Transcriptome Analysis. Mediat. Inflamm. 2020, 2020, 8919534. [Google Scholar] [CrossRef]
- Makkoch, J.; Poomipak, W.; Saengchoowong, S.; Khongnomnan, K.; Praianantathavorn, K.; Jinato, T.; Poovorawan, Y.; Payungporn, S. Human microRNAs profiling in response to influenza A viruses (subtypes pH1N1, H3N2, and H5N1). Exp. Biol. Med. 2016, 241, 409–420. [Google Scholar] [CrossRef]
- Chen, L.; Liu, Y.; Wu, J.; Deng, C.; Tan, J.; Liu, H.; Zhong, L. Lung adenocarcinoma patients have higher risk of SARS-CoV-2 infection. Aging 2021, 13, 1620–1632. [Google Scholar] [CrossRef]
- Chaudhuri, E.; Dash, S.; Balasubramaniam, M.; Padron, A.; Holland, J.; Sowd, G.A.; Villalta, F.; Engelman, A.N.; Pandhare, J.; Dash, C. The HIV-1 capsid-binding host factor CPSF6 is post-transcriptionally regulated by the cellular microRNA miR-125b. J. Biol. Chem. 2020, 295, 5081–5094. [Google Scholar] [CrossRef]
- Mantri, C.K.; Dash, J.P.; Mantri, J.V.; Dash, C.C.V. Cocaine Enhances HIV-1 Replication in CD4+ T Cells by Down-Regulating MiR-125b. PLoS ONE 2012, 7, e51387. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/flaviviridae (accessed on 19 July 2022).
- Ghosh, D.; Basu, A. Japanese Encephalitis—A Pathological and Clinical Perspective. PLOS Negl. Trop. Dis. 2009, 3, e437. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-W.; Tsai, K.-N.; Chen, Y.-S.; Chang, R.-Y. Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus. Int. J. Mol. Sci. 2021, 22, 4218. [Google Scholar] [CrossRef] [PubMed]
- Mziaut, H.; Henniger, G.; Ganss, K.; Hempel, S.; Wolk, S.; McChord, J.; Chowdhury, K.; Ravassard, P.; Knoch, K.-P.; Krautz, C.; et al. MiR-132 controls pancreatic beta cell proliferation and survival through Pten/Akt/Foxo3 signaling. Mol. Metab. 2020, 31, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, A.; Liu, X.; Meisgen, F.; Grünler, J.; Botusan, I.R.; Narayanan, S.; Erikci, E.; Blomqvist, L.; Du, L.; et al. MicroRNA-132 enhances transition from inflammation to proliferation during wound healing. J. Clin. Investig. 2015, 125, 3008–3026. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tong, Z.; Chen, K.; Hu, X.; Jin, H.; Hou, M. The Role of miRNA-132 against Apoptosis and Oxidative Stress in Heart Failure. BioMed Res. Int. 2018, 2018, 3452748. [Google Scholar] [CrossRef] [PubMed]
- Strum, J.C.; Johnson, J.H.; Ward, J.; Xie, H.; Feild, J.; Hester, A.; Alford, A.; Waters, K.M. MicroRNA 132 Regulates Nutritional Stress-Induced Chemokine Production through Repression of SirT1. Mol. Endocrinol. 2009, 23, 1876–1884. [Google Scholar] [CrossRef]
- Wei, X.; Tan, C.; Tang, C.; Ren, G.; Xiang, T.; Qiu, Z.; Liu, R.; Wu, Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell. Signal. 2013, 25, 1037–1043. [Google Scholar] [CrossRef]
- Liu, B.; Yang, X.-F.; Liang, X.-P.; Wang, L.; Shao, M.-M.; Han, W.-X.; Wu, Y.-H. Expressions of MiR-132 in patients with chronic hepatitis B, posthepatitic cirrhosis and hepatitis B virus-related hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 8431–8437. [Google Scholar]
- Zhang, F.; Lin, X.; Yang, X.; Lu, G.; Zhang, Q.; Zhang, C. MicroRNA-132-3p suppresses type I IFN response through targeting IRF1 to facilitate H1N1 influenza A virus infection. Biosci. Rep. 2019, 39, BSR20192769. [Google Scholar] [CrossRef]
- Hsu, A.C.-Y.; Parsons, K.; Moheimani, F.; Knight, D.A.; Hansbro, P.M.; Fujita, T.; Wark, P.A. Impaired Antiviral Stress Granule and IFN-β Enhanceosome Formation Enhances Susceptibility to Influenza Infection in Chronic Obstructive Pulmonary Disease Epithelium. Am. J. Respir. Cell Mol. Biol. 2016, 55, 117–127. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/herpesviridae (accessed on 19 July 2022).
- Tognarelli, E.I.; Palomino, T.F.; Corrales, N.; Bueno, S.M.; Kalergis, A.; González, P.A. Herpes Simplex Virus Evasion of Early Host Antiviral Responses. Front. Cell. Infect. Microbiol. 2019, 9, 127. [Google Scholar] [CrossRef] [PubMed]
- Pepose, J.S. Herpes simplex keratitis: Role of viral infection versus immune response. Surv. Ophthalmol. 1991, 35, 345–352. [Google Scholar] [CrossRef]
- Mulik, S.; Xu, J.; Reddy, P.B.; Rajasagi, N.K.; Gimenez, F.; Sharma, S.; Lu, P.Y.; Rouse, B.T. Role of miR-132 in Angiogenesis after Ocular Infection with Herpes Simplex Virus. Am. J. Pathol. 2012, 181, 525–534. [Google Scholar] [CrossRef]
- Taheri, F.; Ebrahimi, S.O.; Shareef, S.; Reiisi, S. Regulatory and immunomodulatory role of miR-34a in T cell immunity. Life Sci. 2020, 262, 118209. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, H.; Li, J.; Xue, R.; Liu, H.; Zhu, Z.; Pan, C.; Lin, Y.; Hu, A.; Gou, P.; et al. Elevated HMGB1 expression induced by hepatitis B virus X protein promotes epithelial-mesenchymal transition and angiogenesis through STAT3/miR-34a/NF-kappaB in primary liver cancer. Am. J. Cancer Res. 2021, 11, 479–494. [Google Scholar]
- FeiLi, X.; Wu, S.; Ye, W.; Tu, J.; Lou, L. MicroRNA-34a-5p inhibits liver fibrosis by regulating TGF-β1/Smad3 pathway in hepatic stellate cells. Cell Biol. Int. 2018, 42, 1370–1376. [Google Scholar] [CrossRef]
- Rana, M.A.; Ijaz, B.; Daud, M.; Tariq, S.; Nadeem, T.; Husnain, T. Interplay of Wnt β-catenin pathway and miRNAs in HBV pathogenesis leading to HCC. Clin. Res. Hepatol. Gastroenterol. 2019, 43, 373–386. [Google Scholar] [CrossRef]
- Yang, P.; Li, Q.-J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y.; et al. TGF-β-miR-34a-CCL22 Signaling-Induced Treg Cell Recruitment Promotes Venous Metastases of HBV-Positive Hepatocellular Carcinoma. Cancer Cell 2012, 22, 291–303. [Google Scholar] [CrossRef]
- Cermelli, S.; Ruggieri, A.; Marrero, J.A.; Ioannou, G.N.; Beretta, L. Circulating MicroRNAs in Patients with Chronic Hepatitis C and Non-Alcoholic Fatty Liver Disease. PLoS ONE 2011, 6, e23937. [Google Scholar] [CrossRef]
- Rossi, A.D.; Higa, L.M.; Herlinger, A.L.; Ribeiro-Alves, M.; de Menezes, M.T.; Giannini, A.L.M.; Cardoso, C.C.; Da Poian, A.T.; Tanuri, A.; Aguiar, R.S. Differential Expression of Human MicroRNAs During Dengue Virus Infection in THP-1 Monocytes. Front. Cell. Infect. Microbiol. 2021, 11, 714088. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.L.; Jeng, S.; McWeeney, S.K.; Hirsch, A.J. A MicroRNA Screen Identifies the Wnt Signaling Pathway as a Regulator of the Interferon Response during Flavivirus Infection. J. Virol. 2017, 91, e02388–e02404. [Google Scholar] [CrossRef]
- Fan, N.; Wang, J. MicroRNA 34a contributes to virus-mediated apoptosis through binding to its target gene Bax in influenza A virus infection. Biomed. Pharmacother. 2016, 83, 1464–1470. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Beezhold, D.H.; Umbright, C.M.; Noti, J.D. Influenza Virus-Induced Novel miRNAs Regulate the STAT Pathway. Viruses 2021, 13, 967. [Google Scholar] [CrossRef]
- Centa, A.; Fonseca, A.S.; Ferreira, S.G.D.S.; Azevedo, M.L.V.; de Paula, C.B.V.; Nagashima, S.; Machado-Souza, C.; Miggiolaro, A.F.R.D.S.; Baena, C.P.; de Noronha, L.; et al. Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients. Am. J. Physiol. Cell. Mol. Physiol. 2021, 320, L405–L412. [Google Scholar] [CrossRef] [PubMed]
- Bartoszewski, R.; Dabrowski, M.; Jakiela, B.; Matalon, S.; Harrod, K.S.; Sanak, M.; Collawn, J.F. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Cell. Mol. Physiol. 2020, 319, L444–L455. [Google Scholar] [CrossRef] [PubMed]
- Demirci, M.D.S.; Adan, A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ 2020, 8, e9369. [Google Scholar] [CrossRef]
- Verdonck, K.; González, E.; Van Dooren, S.; Vandamme, A.-M.; Vanham, G.; Gotuzzo, E. Human T-lymphotropic virus 1: Recent knowledge about an ancient infection. Lancet Infect. Dis. 2007, 7, 266–281. [Google Scholar] [CrossRef]
- Sharma, V.K.; Raimondi, V.; Ruggero, K.; Pise-Masison, C.A.; Cavallari, I.; Silic-Benussi, M.; Ciminale, V.; D’Agostino, D.M. Expression of miR-34a in T-Cells Infected by Human T-Lymphotropic Virus. Front. Microbiol. 2018, 9, 832. [Google Scholar] [CrossRef]
- Kumarswamy, R.; Volkmann, I.; Thum, T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011, 8, 706–713. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Sheetz, M. A Tale of Two States: Normal and Transformed, With and Without Rigidity Sensing. Annu. Rev. Cell Dev. Biol. 2019, 35, 169–190. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Shi, Z.; Zhang, M.; Dong, X.; Zheng, L.; Li, G.; Han, X.; Yao, Z.; Han, T.; Hong, W. Silencing lncRNA Lfar1 alleviates the classical activation and pyoptosis of macrophage in hepatic fibrosis. Cell Death Dis. 2020, 11, 132. [Google Scholar] [CrossRef] [PubMed]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Lee, Y.-H.; Yun, Y. HBx Protein of Hepatitis B Virus Activates Jak1-STAT Signaling. J. Biol. Chem. 1998, 273, 25510–25515. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.I.; Kang-Park, S.; Do, S.-I.; Lee, Y.I. The Hepatitis B Virus-X Protein Activates a Phosphatidylinositol 3-Kinase-dependent Survival Signaling Cascade. J. Biol. Chem. 2001, 276, 16969–16977. [Google Scholar] [CrossRef]
- Wu, H.-C.; Huang, C.-L.; Wang, H.-W.; Hsu, W.-F.; Tsai, T.-Y.; Chen, S.-H.; Peng, C.-Y. Serum miR-21 correlates with the histological stage of chronic hepatitis B-associated liver fibrosis. Int. J. Clin. Exp. Pathol. 2019, 12, 3819–3829. [Google Scholar]
- Li, W.; Yu, X.; Chen, X.; Wang, Z.; Yin, M.; Zhao, Z.; Zhu, C. HBV induces liver fibrosis via the TGF-β1/miR-21-5p pathway. Exp. Ther. Med. 2021, 21, 169. [Google Scholar] [CrossRef]
- Wei, J.; Feng, L.; Li, Z.; Xu, G.; Fan, X. MicroRNA-21 activates hepatic stellate cells via PTEN/Akt signaling. Biomed. Pharmacother. 2013, 67, 387–392. [Google Scholar] [CrossRef]
- Fu, R.-Q.; Hu, D.-P.; Hu, Y.-B.; Hong, L.; Sun, Q.-F.; Ding, J.-G. miR-21 promotes α-SMA and collagen I expression in hepatic stellate cells via the Smad7 signaling pathway. Mol. Med. Rep. 2017, 16, 4327–4333. [Google Scholar] [CrossRef]
- Qiu, X.; Dong, S.; Qiao, F.; Lu, S.; Song, Y.; Lao, Y.; Li, Y.; Zeng, T.; Hu, J.; Zhang, L.; et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene 2013, 32, 3296–3305. [Google Scholar] [CrossRef] [PubMed]
- Damania, P.; Sen, B.; Dar, S.B.; Kumar, S.; Kumari, A.; Gupta, E.; Sarin, S.K.; Venugopal, S.K. Hepatitis B Virus Induces Cell Proliferation via HBx-Induced microRNA-21 in Hepatocellular Carcinoma by Targeting Programmed Cell Death Protein4 (PDCD4) and Phosphatase and Tensin Homologue (PTEN). PLoS ONE 2014, 9, e91745. [Google Scholar] [CrossRef]
- Li, C.H.; Xu, F.; Chow, S.; Feng, L.; Yin, D.; Ng, T.B.; Chen, Y. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur. J. Cancer 2014, 50, 2560–2569. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Chow, S.C.; Yin, D.L.; Ng, T.B.; Chen, Y.C. Functional characterisation of hepatitis B viral X protein/microRNA-21 interaction in HBV-associated hepatocellular carcinoma. Hong Kong Med. J. 2016, 22, 37–42. [Google Scholar]
- Bandopadhyay, M.; Banerjee, A.; Sarkar, N.; Panigrahi, R.; Datta, S.; Pal, A.; Singh, S.P.; Biswas, A.; Chakrabarti, S.; Chakravarty, R. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by Hepatitis B virus X protein in malignant hepatocytes. BMC Cancer 2014, 14, 721. [Google Scholar] [CrossRef]
- Yin, D.; Wang, Y.; Sai, W.; Zhang, L.; Miao, Y.; Cao, L.; Zhai, X.; Feng, X.; Yang, L. HBx-induced miR-21 suppresses cell apoptosis in hepatocellular carcinoma by targeting interleukin-12. Oncol. Rep. 2016, 36, 2305–2312. [Google Scholar] [CrossRef]
- Otania, T.; Nakamuraa, S.; Tokia, M.; Motodaa, R.; Kurimotob, M.; Oritaa, K. Identification of IFN-γ-Producing Cells in IL-12/IL-18-Treated Mice. Cell. Immunol. 1999, 198, 111–119. [Google Scholar] [CrossRef]
- Zeh, H.J.; Hurd, S.; Storkus, W.J.; Lotze, M.T. Interleukin-12 Promotes the Proliferation and Cytolytic Maturation of Immune Effectors: Implications for the immunotherapy of cancer. J. Immunother. 1993, 14, 155–161. [Google Scholar] [CrossRef]
- Clément, S.; Sobolewski, C.; Gomes, D.; Rojas, A.; Goossens, N.; Conzelmann, S.; Calo, N.; Negro, F.; Foti, M. Activation of the oncogenic miR-21-5p promotes HCV replication and steatosis induced by the viral core 3a protein. Liver Int. 2019, 39, 1226–1236. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Wang, H.; Shi, J.; Wu, K.; Liu, S.; Liu, Y.; Wu, J. HCV-Induced miR-21 Contributes to Evasion of Host Immune System by Targeting MyD88 and IRAK1. PLOS Pathog. 2013, 9, e1003248. [Google Scholar] [CrossRef]
- Marquez, R.T.; Bandyopadhyay, S.; Wendlandt, E.B.; Keck, K.; Hoffer, B.A.; Icardi, M.S.; Christensen, R.N.; Schmidt, W.N.; McCaffrey, A.P. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab. Investig. 2010, 90, 1727–1736. [Google Scholar] [CrossRef] [PubMed]
- Kanokudom, S.; Vilaivan, T.; Wikan, N.; Thepparit, C.; Smith, D.R.; Assavalapsakul, W. miR-21 promotes dengue virus serotype 2 replication in HepG2 cells. Antivir. Res. 2017, 142, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Duy, J.; Koehler, J.; Honko, A.; Schoepp, R.; Wauquier, N.; Gonzalez, J.-P.; Pitt, M.L.; Mucker, E.; Johnson, J.; O’Hearn, A.; et al. Circulating microRNA profiles of Ebola virus infection. Sci. Rep. 2016, 6, 24496. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Feng, P.; Gu, T. MicroRNA-21-3p modulates FGF2 to facilitate influenza A virus H5N1 replication by refraining type I interferon response. Biosci. Rep. 2020, 40, BSR20200158. [Google Scholar] [CrossRef]
- Xia, B.; Lu, J.; Wang, R.; Yang, Z.; Zhou, X.; Huang, P. miR-21-3p Regulates Influenza A Virus Replication by Targeting Histone Deacetylase-8. Front. Cell. Infect. Microbiol. 2018, 8, 175. [Google Scholar] [CrossRef]
- Chew, C.L.; Conos, S.A.; Unal, B.; Tergaonkar, V. Noncoding RNAs: Master Regulators of Inflammatory Signaling. Trends Mol. Med. 2018, 24, 66–84. [Google Scholar] [CrossRef]
- Lam, W.-Y.; Yeung, A.C.-M.; Ngai, K.L.-K.; Li, M.-S.; To, K.-F.; Tsui, S.K.-W.; Chan, P.K.-S. Effect of avian influenza A H5N1 infection on the expression of microRNA-141 in human respiratory epithelial cells. BMC Microbiol. 2013, 13, 104. [Google Scholar] [CrossRef]
- Tan, K.S.; Choi, H.; Jiang, X.; Yin, L.; Seet, J.E.; Patzel, V.; Engelward, B.P.; Chow, V.T. Micro-RNAs in regenerating lungs: An integrative systems biology analysis of murine influenza pneumonia. BMC Genom. 2014, 15, 587. [Google Scholar] [CrossRef]
- Zhong, Z.; Dong, Z.; Yang, L.; Gong, Z. miR-21 induces cell cycle at S phase and modulates cell proliferation by down-regulating hMSH2 in lung cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 1781–1788. [Google Scholar] [CrossRef]
- Vaporidi, K.; Vergadi, E.; Kaniaris, E.; Hatziapostolou, M.; Lagoudaki, E.; Georgopoulos, D.; Zapol, W.M.; Bloch, K.D.; Iliopoulos, D. Pulmonary microRNA profiling in a mouse model of ventilator-induced lung injury. Am. J. Physiol. Cell. Mol. Physiol. 2012, 303, L199–L207. [Google Scholar] [CrossRef]
- Nersisyan, S.; Engibaryan, N.; Gorbonos, A.; Kirdey, K.; Makhonin, A.; Tonevitsky, A. Potential role of cellular miRNAs in coronavirus-host interplay. PeerJ 2020, 8, e9994. [Google Scholar] [CrossRef] [PubMed]
- Jafarinejad-Farsangi, S.; Jazi, M.M.; Rostamzadeh, F.; Hadizadeh, M. High affinity of host human microRNAs to SARS-CoV-2 genome: An in silico analysis. Non-Coding RNA Res. 2020, 5, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Chahar, H.S.; Corsello, T.; Kudlicki, A.S.; Komaravelli, N.; Casola, A. Respiratory Syncytial Virus Infection Changes Cargo Composition of Exosome Released from Airway Epithelial Cells. Sci. Rep. 2018, 8, 387. [Google Scholar] [CrossRef] [PubMed]
- Boyapalle, S.; Wong, T.; Garay, J.; Teng, M.; Vergara, H.S.J.; Mohapatra, S.; Mohapatra, S. Respiratory Syncytial Virus NS1 Protein Colocalizes with Mitochondrial Antiviral Signaling Protein MAVS following Infection. PLoS ONE 2012, 7, e29386. [Google Scholar] [CrossRef]
- Kim, R.Y.; Horvat, J.C.; Pinkerton, J.W.; Starkey, M.R.; Essilfie, A.T.; Mayall, J.R.; Nair, P.M.; Hansbro, N.G.; Jones, B.; Haw, T.J.; et al. MicroRNA-21 drives severe, steroid-insensitive experimental asthma by amplifying phosphoinositide 3-kinase–mediated suppression of histone deacetylase 2. J. Allergy Clin. Immunol. 2017, 139, 519–532. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, P.; Chen, M.; Xu, Y.; Zhao, D. miRNAs and Leukotrienes in Respiratory Syncytial Virus Infection. Front. Pediatr. 2021, 9, 602195. [Google Scholar] [CrossRef]
- Ruiz-De-León, M.J.; Jiménez-Sousa, M.A.; Moreno, S.; García, M.; Gutiérrez-Rivas, M.; León, A.; Montero-Alonso, M.; González-García, J.; Resino, S.; Rallón, N.; et al. Lower expression of plasma-derived exosome miR-21 levels in HIV-1 elite controllers with decreasing CD4 T cell count. J. Microbiol. Immunol. Infect. 2019, 52, 667–671. [Google Scholar] [CrossRef]
- Cojo, M.S.-D.; López-Huertas, M.R.; Díez-Fuertes, F.; Rodríguez-Mora, S.; Bermejo, M.; López-Campos, G.; Mateos, E.; Jiménez-Tormo, L.; Gómez-Esquer, F.; Díaz-Gil, G.; et al. Changes in the cellular microRNA profile by the intracellular expression of HIV-1 Tat regulator: A potential mechanism for resistance to apoptosis and impaired proliferation in HIV-1 infected CD4+ T cells. PLoS ONE 2017, 12, e0185677. [Google Scholar] [CrossRef]
- Jiao, Y.; Zhang, T.; Wang, R.; Zhang, H.; Huang, X.; Yin, J.; Zhang, L.; Xu, X.; Wu, H. Plasma IP-10 Is Associated with Rapid Disease Progression in Early HIV-1 Infection. Viral Immunol. 2012, 25, 333–337. [Google Scholar] [CrossRef]
- Liu, P.P.; Mason, J.W. Advances in the Understanding of Myocarditis. Circulation 2001, 104, 1076–1082. [Google Scholar] [CrossRef]
- He, F.; Yao, H.; Xiao, Z.; Han, J.; Zou, J.; Liu, Z. Inhibition of IL-2 inducible T-cell kinase alleviates T-cell activation and murine myocardial inflammation associated with CVB3 infection. Mol. Immunol. 2014, 59, 30–38. [Google Scholar] [CrossRef]
- He, F.; Xiao, Z.; Yao, H.; Li, S.; Feng, M.; Wang, W.; Liu, Z.; Liu, Z.; Wu, J. The protective role of microRNA-21 against coxsackievirus B3 infection through targeting the MAP2K3/P38 MAPK signaling pathway. J. Transl. Med. 2019, 17, 335. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Zhang, H.M.; Qiu, Y.; Hanson, P.J.; Hemida, M.; Wei, W.; Hoodless, P.A.; Chu, F.; Yang, D. Coxsackievirus-Induced miR-21 Disrupts Cardiomyocyte Interactions via the Downregulation of Intercalated Disk Components. PLOS Pathog. 2014, 10, e1004070. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/rhabdoviridae/805/genus-vesiculovirus (accessed on 19 July 2022).
- Pandey, N.; Rastogi, M.; Singh, S.K. Chandipura virus dysregulates the expression of hsa-miR-21-5p to activate NF-κB in human microglial cells. J. Biomed. Sci. 2021, 28, 52. [Google Scholar] [CrossRef]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2010, 17, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liang, H.; Deng, T.; Zhu, K.; Zhang, S.; Wang, N.; Jiang, X.; Wang, X.; Liu, R.; Zen, K.; et al. The identification of novel targets of miR-16 and characterization of their biological functions in cancer cells. Mol. Cancer 2013, 12, 92. [Google Scholar] [CrossRef]
- Yan, L.; Liang, M.; Hou, X.; Zhang, Y.; Zhang, H.; Guo, Z.; Jinyu, J.; Feng, Z.; Mei, Z. The role of microRNA-16 in the pathogenesis of autoimmune diseases: A comprehensive review. Biomed. Pharmacother. 2019, 112, 108583. [Google Scholar] [CrossRef]
- Wu, G.; Yu, F.; Xiao, Z.; Xu, K.; Xu, J.; Tang, W.; Wang, J.; Song, E. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br. J. Cancer 2011, 105, 146–153. [Google Scholar] [CrossRef]
- Zhu, B.; Wei, X.-X.; Wang, T.-B.; Zhou, Y.-C.; Liu, A.-M.; Zhang, G.-W. Increased miR-16 expression induced by hepatitis C virus infection promotes liver fibrosis through downregulation of hepatocyte growth factor and Smad7. Arch. Virol. 2015, 160, 2043–2050. [Google Scholar] [CrossRef]
- Wang, H.; Tian, Z.; Xu, Y.; Wang, Q.; Ding, S.-W.; Li, Y. Altering Intracellular Localization of the RNA Interference Factors by Influenza A Virus Non-structural Protein 1. Front. Microbiol. 2020, 11, 590904. [Google Scholar] [CrossRef]
- Sun, X.; Sit, A.; Feinberg, M.W. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc. Med. 2014, 24, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Teymoori-Rad, M.; Nakstad, B.; Salimi, V. Anti-Inflammatory MicroRNAs and Their Potential for Inflammatory Diseases Treatment. Front. Immunol. 2018, 9, 1377. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Henao-Mejia, J.; Harman, C.C.D.; Flavell, R.A. miR-181 and Metabolic Regulation in the Immune System. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-Y.; Zhang, F.-R.; Shang, J.-Y.; Liu, Y.-Y.; Lv, X.-F.; Yuan, J.-N.; Zhang, T.-T.; Li, K.; Lin, X.-C.; Liu, X.; et al. Renal inhibition of miR-181a ameliorates 5-fluorouracil-induced mesangial cell apoptosis and nephrotoxicity. Cell Death Dis. 2018, 9, 610. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhou, G.; Li, G.; Chen, B.; Dong, P.; Zheng, J. Serum miR-181b Is Correlated with Hepatitis B Virus Replication and Disease Progression in Chronic Hepatitis B Patients. Dig. Dis. Sci. 2015, 60, 2346–2352. [Google Scholar] [CrossRef]
- Wang, B.; Li, W.; Guo, K.; Xiao, Y.; Wang, Y.; Fan, J. miR-181b Promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem. Biophys. Res. Commun. 2012, 421, 4–8. [Google Scholar] [CrossRef]
- Zheng, J.; Wu, C.; Xu, Z.; Xia, P.; Dong, P.; Chen, B.; Yu, F. Hepatic stellate cell is activated by microRNA-181b via PTEN/Akt pathway. Mol. Cell. Biochem. 2015, 398, 1–9. [Google Scholar] [CrossRef]
- Zou, C.; Li, Y.; Cao, Y.; Zhang, J.; Jiang, J.; Sheng, Y.; Wang, S.; Huang, A.; Tang, H. Up-regulated MicroRNA-181a induces carcinogenesis in Hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer 2014, 14, 97. [Google Scholar] [CrossRef][Green Version]
- Tian, Y.; Xiao, X.; Gong, X.; Peng, F.; Xu, Y.; Jiang, Y.; Gong, G. HBx promotes cell proliferation by disturbing the cross-talk between miR-181a and PTEN. Sci. Rep. 2017, 7, 40089. [Google Scholar] [CrossRef]
- Zou, C.; Chen, J.; Chen, K.; Wang, S.; Cao, Y.; Zhang, J.; Sheng, Y.; Huang, A.; Tang, H. Functional analysis of miR-181a and Fas involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Exp. Cell Res. 2015, 331, 352–361. [Google Scholar] [CrossRef]
- Elhelw, D.S.; Mekky, R.Y.; El-Ekiaby, N.; Ahmed, R.; Eldin, M.A.M.; El-Sayed, M.; AbouElKhair, M.; Salah, A.; Zekri, A.R.; Esmat, G.; et al. Predictive prognostic role of miR-181a with discrepancy in the liver and serum of genotype 4 hepatitis C virus patients. Biomed. Rep. 2014, 2, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Boštjančič, E.; Bandelj, E.; Luzar, B.; Poljak, M.; Glavač, D. Hepatic expression of miR-122, miR-126, miR-136 and miR-181a and their correlation to histopathological and clinical characteristics of patients with hepatitis C. J. Viral Hepat. 2015, 22, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Zhou, Y.; Ying, R.S.; Shi, L.; Cheng, Y.Q.; Ren, J.P.; Griffin, J.W.; Jia, Z.S.; Li, C.F.; Moorman, J.P.; et al. Hepatitis C virus-induced reduction in miR-181a impairs CD4+T-cell responses through overexpression of DUSP6. Hepatology 2015, 61, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Shrivastava, S.; Chowdhury, J.B.; Ray, R.; Ray, R.B. Transcriptional Suppression of miR-181c by Hepatitis C Virus Enhances Homeobox A1 Expression. J. Virol. 2014, 88, 7929–7940. [Google Scholar] [CrossRef]
- Patra, T.; Meyer, K.; Ray, R.B.; Ray, R. Hepatitis C Virus Mediated Inhibition of miR-181c Activates ATM Signaling and Promotes Hepatocyte Growth. Hepatology 2020, 71, 780–793. [Google Scholar] [CrossRef]
- Lim, J.; Byun, J.; Guk, K.; Hwang, S.G.; Bae, P.K.; Jung, J.; Kang, T.; Lim, E.-K. Highly Sensitive in Vitro Diagnostic System of Pandemic Influenza A (H1N1) Virus Infection with Specific MicroRNA as a Biomarker. ACS Omega 2019, 4, 14560–14568. [Google Scholar] [CrossRef]
- Peng, F.; He, J.; Loo, J.F.C.; Yao, J.; Shi, L.; Liu, C.; Zhao, C.; Xie, W.; Shao, Y.; Kong, S.K.; et al. Identification of microRNAs in Throat Swab as the Biomarkers for Diagnosis of Influenza. Int. J. Med. Sci. 2016, 13, 77–84. [Google Scholar] [CrossRef]
- Atherton, L.J.; Jorquera, P.A.; Bakre, A.A.; Tripp, R.A. Determining Immune and miRNA Biomarkers Related to Respiratory Syncytial Virus (RSV) Vaccine Types. Front. Immunol. 2019, 10, 2323. [Google Scholar] [CrossRef]
- Su, J.-L.; Chen, P.-S.; Johansson, G.; Kuo, M.-L. Function and Regulation of Let-7 Family microRNAs. MicroRNA 2012, 1, 34–39. [Google Scholar] [CrossRef]
- Thammaiah, C.K.; Jayaram, S. Role of let-7 family microRNA in breast cancer. Non-Coding RNA Res. 2016, 1, 77–82. [Google Scholar] [CrossRef]
- Jiang, S. Recent findings regarding let-7 in immunity. Cancer Lett. 2018, 434, 130–131. [Google Scholar] [CrossRef] [PubMed]
- Letafati, A.; Najafi, S.; Mottahedi, M.; Karimzadeh, M.; Shahini, A.; Garousi, S.; Abbasi-Kolli, M.; Nahand, J.S.; Zadeh, S.S.T.; Hamblin, M.R.; et al. MicroRNA let-7 and viral infections: Focus on mechanisms of action. Cell. Mol. Biol. Lett. 2022, 27, 1–47. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, Y.; Toh, S.T.; Sung, W.-K.; Tan, P.; Chow, P.; Chung, A.Y.; Jooi, L.L.; Lee, C.G.L. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription. J. Hepatol. 2010, 53, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Huang, P.; Ju, X.; Li, Z.; Wang, Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int. J. Clin. Exp. Med. 2015, 8, 15108–15116. [Google Scholar]
- Al Zaid Siddiquee, K.; Turkson, J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008, 18, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Chen, J.; Liu, J.; Luo, Z.; Jiang, W.; Huang, J.; Qiu, Z.; Yue, W.; Wu, L. Expression of microRNA let-7a positively correlates with hepatitis B virus replication in hepatocellular carcinoma tissues. Exp. Biol. Med. 2017, 242, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Takata, A.; Otsuka, M.; Ohno, M.; Kishikawa, T.; Yoshikawa, T.; Koike, K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci. Rep. 2016, 6, 23237. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lowey, B.; Sodroski, C.; Krishnamurthy, S.; Alao, H.; Cha, H.; Chiu, S.; El-Diwany, R.; Ghany, M.G.; Liang, T.J. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017, 8, 1789. [Google Scholar] [CrossRef]
- Cheng, J.-C.; Yeh, Y.-J.; Tseng, C.-P.; Hsu, S.-D.; Chang, Y.-L.; Sakamoto, N.; Huang, H.-D. Let-7b is a novel regulator of hepatitis C virus replication. Cell. Mol. Life Sci. 2012, 69, 2621–2633. [Google Scholar] [CrossRef]
- Yeh, Y.-J.; Tseng, C.-P.; Hsu, S.-D.; Huang, H.-Y.; Lai, M.M.C.; Huang, H.-D.; Cheng, J.-C. Dual Effects of Let-7b in the Early Stage of Hepatitis C Virus Infection. J. Virol. 2021, 95, e01800–e01820. [Google Scholar] [CrossRef]
- Cheng, M.; Si, Y.; Niu, Y.; Liu, X.; Li, X.; Zhao, J.; Jin, Q.; Yang, W. High-Throughput Profiling of Alpha Interferon- and Interleukin-28B-Regulated MicroRNAs and Identification of let-7s with Anti-Hepatitis C Virus Activity by Targeting IGF2BP. J. Virol. 2013, 87, 9707–9718. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; Aizawa, N.; Enomoto, H.; Nishiguchi, S.; Toyoda, H.; Kumada, T.; Iio, E.; Ito, K.; Ogawa, S.; Isogawa, M.; et al. Circulating let-7 Levels in Serum Correlate with the Severity of Hepatic Fibrosis in Chronic Hepatitis C. Open Forum Infect. Dis. 2018, 5, ofy268. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, K.; De Giorgi, V.; Schechterly, C.; Wang, R.Y.; Farci, P.; Tanaka, Y.; Alter, H.J. Circulating let-7 levels in plasma and extracellular vesicles correlate with hepatic fibrosis progression in chronic hepatitis C. Hepatology 2016, 64, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Escalera-Cueto, M.; Medina-Martínez, I.; del Angel, R.; Berumen, J.; Gutiérrez-Escolano, A.L.; Yocupicio-Monroy, M. Let-7c overexpression inhibits dengue virus replication in human hepatoma Huh-7 cells. Virus Res. 2015, 196, 105–112. [Google Scholar] [CrossRef]
- Zhang, S.; Gu, D.; Ouyang, X.; Xie, W. Proinflammatory effects of the hemagglutinin protein of the avian influenza A (H7N9) virus and microRNA-mediated homeostasis response in THP-1 cells. Mol. Med. Rep. 2015, 12, 6241–6246. [Google Scholar] [CrossRef][Green Version]
- Zhu, Z.; Qi, Y.; Ge, A.; Zhu, Y.; Xu, K.; Ji, H.; Shi, Z.; Cui, L.; Zhou, M. Comprehensive Characterization of Serum MicroRNA Profile in Response to the Emerging Avian Influenza A (H7N9) Virus Infection in Humans. Viruses 2014, 6, 1525–1539. [Google Scholar] [CrossRef]
- Ma, Y.-J.; Yang, J.; Fan, X.-L.; Zhao, H.-B.; Hu, W.; Li, Z.-P.; Yu, G.; Ding, X.-R.; Wang, J.-Z.; Bo, X.-C.; et al. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 2012, 16, 2539–2546. [Google Scholar] [CrossRef]
- Bakre, A.; Mitchell, P.; Coleman, J.K.; Jones, L.P.; Saavedra, G.; Teng, M.; Tompkins, S.; Tripp, R.A. Respiratory syncytial virus modifies microRNAs regulating host genes that affect virus replication. J. Gen. Virol. 2012, 93, 2346–2356. [Google Scholar] [CrossRef]
- Thornburg, N.; Hayward, S.L.; Crowe, J.E. Respiratory Syncytial Virus Regulates Human MicroRNAs by Using Mechanisms Involving Beta Interferon and NF-κB. mBio 2012, 3, e00220–e00232. [Google Scholar] [CrossRef]
- Bakre, A.A.; Harcourt, J.L.; Haynes, L.M.; Anderson, L.J.; Tripp, R.A. The Central Conserved Region (CCR) of Respiratory Syncytial Virus (RSV) G Protein Modulates Host miRNA Expression and Alters the Cellular Response to Infection. Vaccines 2017, 5, 16. [Google Scholar] [CrossRef]
- Chen, B.; Han, J.; Chen, S.; Xie, R.; Yang, J.; Zhou, T.; Zhang, Q.; Xia, R. MicroLet-7b Regulates Neutrophil Function and Dampens Neutrophilic Inflammation by Suppressing the Canonical TLR4/NF-κB Pathway. Front. Immunol. 2021, 12, 653344. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Chen, Y.; Luo, D.; Zhuang, Z.; Jin, H.; Zhou, H.; Li, X.; Lin, H.; Zheng, X.; Zhang, J.; et al. Therapeutic potential of C1632 by inhibition of SARS-CoV-2 replication and viral-induced inflammation through upregulating let-7. Signal Transduct. Target. Ther. 2021, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, S.; Suzuki, K.; Seddiki, N.; Kaplan, W.; Cowley, M.J.; Hood, C.L.; Clancy, J.L.; Murray, D.D.; Méndez, C.; Gelgor, L.; et al. Differential Regulation of the Let-7 Family of MicroRNAs in CD4+ T Cells Alters IL-10 Expression. J. Immunol. 2012, 188, 6238–6246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, Y.; Zhang, S.; Luo, H.; Zhang, H. HIV-1 Infection-Induced Suppression of the Let-7i/IL-2 Axis Contributes to CD4+ T Cell Death. Sci. Rep. 2016, 6, 25341. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Akbar, I.; Kumari, B.; Vrati, S.; Basu, A.; Banerjee, A. Japanese Encephalitis Virus-induced let-7a/b interacted with the NOTCH—TLR 7 pathway in microglia and facilitated neuronal death via caspase activation. J. Neurochem. 2019, 149, 518–534. [Google Scholar] [CrossRef]
- Bin, S.; Xin, L.; Lin, Z.; Jinhua, Z.; Rui, G.; Xiang, Z. Targeting miR-10a-5p/IL-6R axis for reducing IL-6-induced cartilage cell ferroptosis. Exp. Mol. Pathol. 2021, 118, 104570. [Google Scholar] [CrossRef]
- Tan, Y.; Ge, G.; Pan, T.; Wen, D.; Gan, J. Serum MiRNA panel as potential biomarkers for chronic hepatitis B with persistently normal alanine aminotransferase. Clin. Chim. Acta 2015, 451, 232–239. [Google Scholar] [CrossRef]
- Horii, R.; Honda, M.; Shirasaki, T.; Shimakami, T.; Shimizu, R.; Yamanaka, S.; Murai, K.; Kawaguchi, K.; Arai, K.; Yamashita, T.; et al. MicroRNA-10a Impairs Liver Metabolism in Hepatitis C Virus-Related Cirrhosis Through Deregulation of the Circadian Clock Gene Brain and Muscle Aryl Hydrocarbon Receptor Nuclear Translocator-Like 1. Hepatol. Commun. 2019, 3, 1687–1703. [Google Scholar] [CrossRef]
- Lawrence, T. The Nuclear Factor NF-kappa B Pathway in Inflammation. Cold Spring Harb. Perspect. Biol. 2009, 1, a001651. [Google Scholar] [CrossRef]
- Arthur, J.S.C.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef]
- Li, Q.; Verma, I.M. NF-κB regulation in the immune system. Nat. Rev. Immunol. 2002, 2, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Lin, L.; Wu, S.; Guo, Z.; Wang, T.; Qin, Y.; Wang, R.; Zhong, X.; Wu, X.; Wang, Y.; et al. MiR-10a* up-regulates coxsackievirus B3 biosynthesis by targeting the 3D-coding sequence. Nucleic Acids Res. 2013, 41, 3760–3771. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J.; Abrantes, J.; Bertagnoli, S.; Guitton, J.-S.; Le Gall-Reculé, G.; Lopes, A.M.; Marchandeau, S.; Alda, F.; Almeida, T.; Célio, A.P.; et al. Proposal for a unified classification system and nomenclature of lagoviruses. J. Gen. Virol. 2017, 98, 1658–1666. [Google Scholar] [CrossRef] [PubMed]
- Abrantes, J.; Van Der Loo, W.; Le Pendu, J.; Esteves, P.J. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): A review. Vet. Res. 2012, 43, 12. [Google Scholar] [CrossRef] [PubMed]
- Hukowska-Szematowicz, B.; Maciejak-Jastrzębska, A.; Blatkiewicz, M.; Maciak, K.; Góra, M.; Janiszewska, J.; Burzyńska, B. Changes in MicroRNA Expression during Rabbit Hemorrhagic Disease Virus (RHDV) Infection. Viruses 2020, 12, 965. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brahmakshatriya, V.; Lupiani, B.; Reddy, S.M.; Soibam, B.; Benham, A.L.; Gunaratne, P.; Liu, H.-C.; Trakooljul, N.; Ing, N.; et al. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genom. 2012, 13, 278. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/dsdna-viruses/w/herpesviridae/1611/genus-mardivirus (accessed on 19 July 2022).
- Burnside, J.; Morgan, R. Emerging roles of chicken and viral microRNAs in avian disease. BMC Proc. 2011, 5, S2. [Google Scholar] [CrossRef]
- Burnside, J.; Morgan, R.W. Genomics and Marek’s disease virus. Cytogenet. Genome Res. 2007, 117, 376–387. [Google Scholar] [CrossRef]
- Yao, Y.; Zhao, Y.; Smith, L.P.; Lawrie, C.; Saunders, N.; Watson, M.; Nair, V. Differential expression of microRNAs in Marek’s disease virus-transformed T-lymphoma cell lines. J. Gen. Virol. 2009, 90, 1551–1559. [Google Scholar] [CrossRef]
- Skovgaard, K.; Cirera, S.; Vasby, D.; Podolska, A.; Breum, S.; Dürrwald, R.; Schlegel, M.; Heegaard, P.M. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2). Innate Immun. 2013, 19, 531–544. [Google Scholar] [CrossRef]
- Chen, L.; Song, Y.; He, L.; Wan, X.; Lai, L.; Dai, F.; Liu, Y.; Wang, Q. MicroRNA-223 Promotes Type I Interferon Production in Antiviral Innate Immunity by Targeting Forkhead Box Protein O3 (FOXO3). J. Biol. Chem. 2016, 291, 14706–14716. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Brahmakshatriya, V.; Zhu, H.; Lupiani, B.; Reddy, S.M.; Yoon, B.-J.; Gunaratne, P.H.; Kim, J.H.; Chen, R.; Wang, J.; et al. Identification of differentially expressed miRNAs in chicken lung and trachea with avian influenza virus infection by a deep sequencing approach. BMC Genom. 2009, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Xue, M.; Wang, C.; Wang, J.; Chen, P. Bursopentine as a Novel Immunoadjuvant Enhances both Humoral and Cell-Mediated Immune Responses to Inactivated H9N2 Avian Influenza Virus in Chickens. Clin. Vaccine Immunol. 2011, 18, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef] [PubMed]
- Murray, K.; Selleck, P.; Hooper, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Westbury, H.; Hiley, L.; Selvey, L.; Rodwell, B.; et al. A Morbillivirus that Caused Fatal Fisease in Horses and Humans. Science 1995, 268, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.R.; Marsh, G.A.; Jenkins, K.A.; Gantier, M.P.; Tizard, M.L.; Middleton, D.; Lowenthal, J.W.; Haining, J.; Izzard, L.; Gough, T.J.; et al. Promotion of Hendra Virus Replication by MicroRNA 146a. J. Virol. 2013, 87, 3782–3791. [Google Scholar] [CrossRef]
- Cowled, C.; Foo, C.-H.; Deffrasnes, C.; Rootes, C.L.; Williams, D.T.; Middleton, D.; Wang, L.-F.; Bean, A.G.D.; Stewart, C.R. Circulating microRNA profiles of Hendra virus infection in horses. Sci. Rep. 2017, 7, 7431. [Google Scholar] [CrossRef]
- Witter, R.L.S.; Schat, K. Diseases of Poultry. In Marek’s Disease; Agricultural Research Service US Department of Agriculture: Urbana, IL, USA, 2003; pp. 407–465. [Google Scholar]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/picornaviridae/707/genus-aphthovirus (accessed on 19 July 2022).
- Mason, P.W.; Grubman, M.J.; Baxt, B. Molecular basis of pathogenesis of FMDV. Virus Res. 2003, 91, 9–32. [Google Scholar] [CrossRef]
- Mason, P.W.; Chinsangaram, J.; Moraes, M.P.; Mayr, G.A.; Grubman, M.J. Engineering better vaccines for foot-and-mouth disease. Dev. Biol. 2003, 114, 79–88. [Google Scholar]
- Stenfeldt, C.; Arzt, J.; Smoliga, G.; LaRocco, M.; Gutkoska, J.; Lawrence, P. Proof-of-concept study: Profile of circulating microRNAs in Bovine serum harvested during acute and persistent FMDV infection. Virol. J. 2017, 14, 71. [Google Scholar] [CrossRef][Green Version]
- Quinn, S.R.; O’Neill, L. A trio of microRNAs that control Toll-like receptor signalling. Int. Immunol. 2011, 23, 421–425. [Google Scholar] [CrossRef] [PubMed]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/positive-sense-rna-viruses/w/arteriviridae/1590/genus-betaarterivirus (accessed on 19 July 2022).
- Chand, R.J.; Trible, B.R.; Rowland, R.R. Pathogenesis of porcine reproductive and respiratory syndrome virus. Curr. Opin. Virol. 2012, 2, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.A.; Yoo, D.; Liu, H.-C. Characterization of the microRNAome in Porcine Reproductive and Respiratory Syndrome Virus Infected Macrophages. PLoS ONE 2013, 8, e82054. [Google Scholar] [CrossRef]
- Ye, D.; Shen, Z.; Zhou, S. Function of microRNA-145 and mechanisms underlying its role in malignant tumor diagnosis and treatment. Cancer Manag. Res. 2019, 11, 969–979. [Google Scholar] [CrossRef]
- He, T.; Feng, G.; Chen, H.; Wang, L.; Wang, Y. Identification of host encoded microRNAs interacting with novel swine-origin influenza A (H1N1) virus and swine influenza virus. Bioinformation 2009, 4, 112–118. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses (ICTV). Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/rhabdoviridae/795/genus-lyssavirus (accessed on 19 July 2022).
- Zhao, P.; Zhao, L.; Zhang, K.; Feng, H.; Wang, H.; Wang, T.; Xu, T.; Feng, N.; Wang, C.; Gao, Y.; et al. Infection with street strain rabies virus induces modulation of the microRNA profile of the mouse brain. Virol. J. 2012, 9, 159. [Google Scholar] [CrossRef]
- Booth, V.; Keizer, D.; Kamphuis, M.B.; Clark-Lewis, A.I.; Sykes, B.D. The CXCR3 Binding Chemokine IP-10/CXCL10: Structure and Receptor Interactions. Biochemistry 2002, 41, 10418–10425. [Google Scholar] [CrossRef]
- Shi, N.; Zhang, X.Y.; Dong, C.; Hou, J.L.; Zhang, M.L.; Guan, Z.H.; Li, Z.Y.; Duan, M. Alterations in microRNA expression profile in rabies virus-infected mouse neurons. Acta Virol. 2014, 58, 120–127. [Google Scholar] [CrossRef]
- Liu, T.; Xu, Z.; Ou, D.; Liu, J.; Zhang, J. The miR-15a/16 gene cluster in human cancer: A systematic review. J. Cell. Physiol. 2019, 234, 5496–5506. [Google Scholar] [CrossRef]
- Indrieri, A.; Carrella, S.; Carotenuto, P.; Banfi, S.; Franco, B. The Pervasive Role of the miR-181 Family in Development, Neurodegeneration, and Cancer. Int. J. Mol. Sci. 2020, 21, 2092. [Google Scholar] [CrossRef]
- Peng, X.; Gao, Q.S.; Zhou, L.; Chen, Z.H.; Lu, S.; Huang, H.J.; Zhan, C.Y.; Xiang, M. microRNAs in avian influenza virus H9N2-infected and non-infected chicken embryo fibroblasts. Genet. Mol. Res. 2015, 14, 9081–9091. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.-K.; Zhang, Q.; Gao, L.; Li, N.; Chen, X.-X.; Feng, W.-H. Increasing Expression of MicroRNA 181 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication and Has Implications for Controlling Virus Infection. J. Virol. 2013, 87, 1159–1171. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Luo, J.; Zhang, H.; Chang, S.; Song, J. MiRNA expression signatures induced by Marek’s disease virus infection in chickens. Genomics 2012, 99, 152–159. [Google Scholar] [CrossRef]
- Payne, L.N.; Nair, V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012, 41, 11–19. [Google Scholar] [CrossRef]
- Ji, J.; Shang, H.; Zhang, H.; Li, H.; Ma, J.; Bi, Y.; Xie, Q. Temporal changes of microRNA gga-let-7b and gga-let-7i expression in chickens challenged with subgroup J avian leukosis virus. Vet. Res. Commun. 2017, 41, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ji, J.; Xie, Q.; Shang, H.; Zhang, H.; Xin, X.; Chen, F.; Sun, B.; Xue, C.; Ma, J.; et al. Aberrant expression of liver microRNA in chickens infected with subgroup avian leukosis virus. Virus Res. 2012, 169, 268–271. [Google Scholar] [CrossRef] [PubMed]
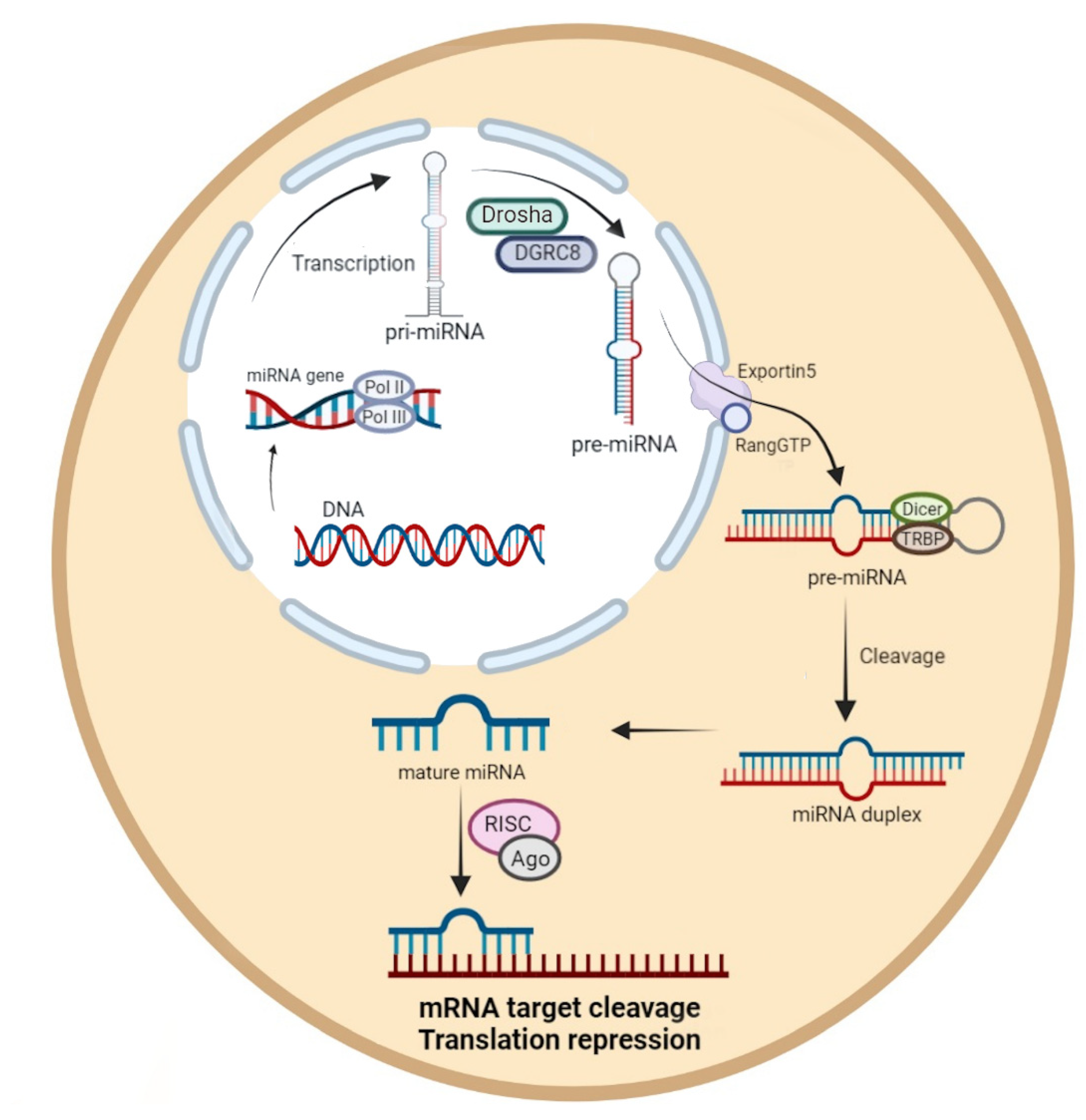
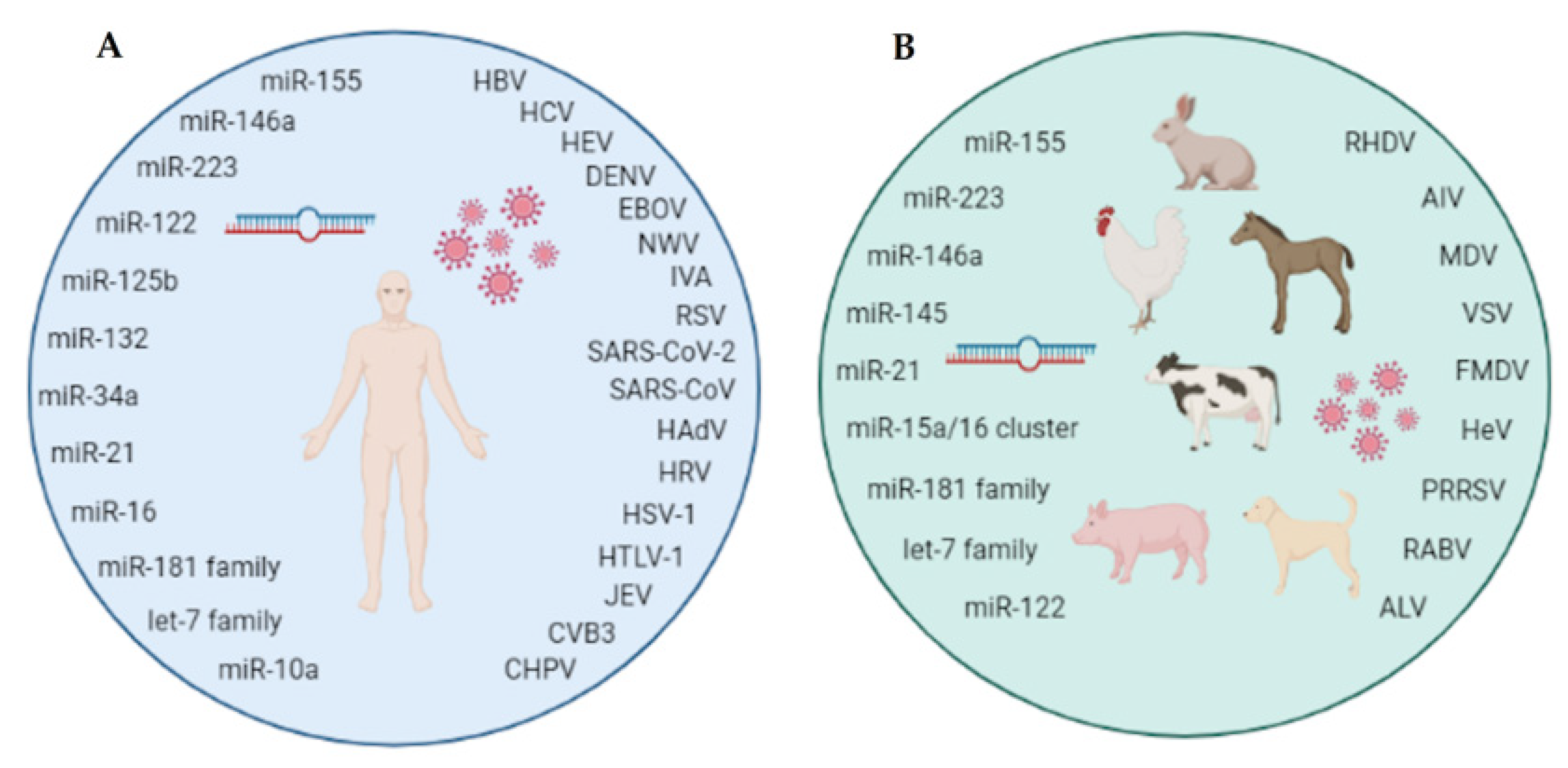
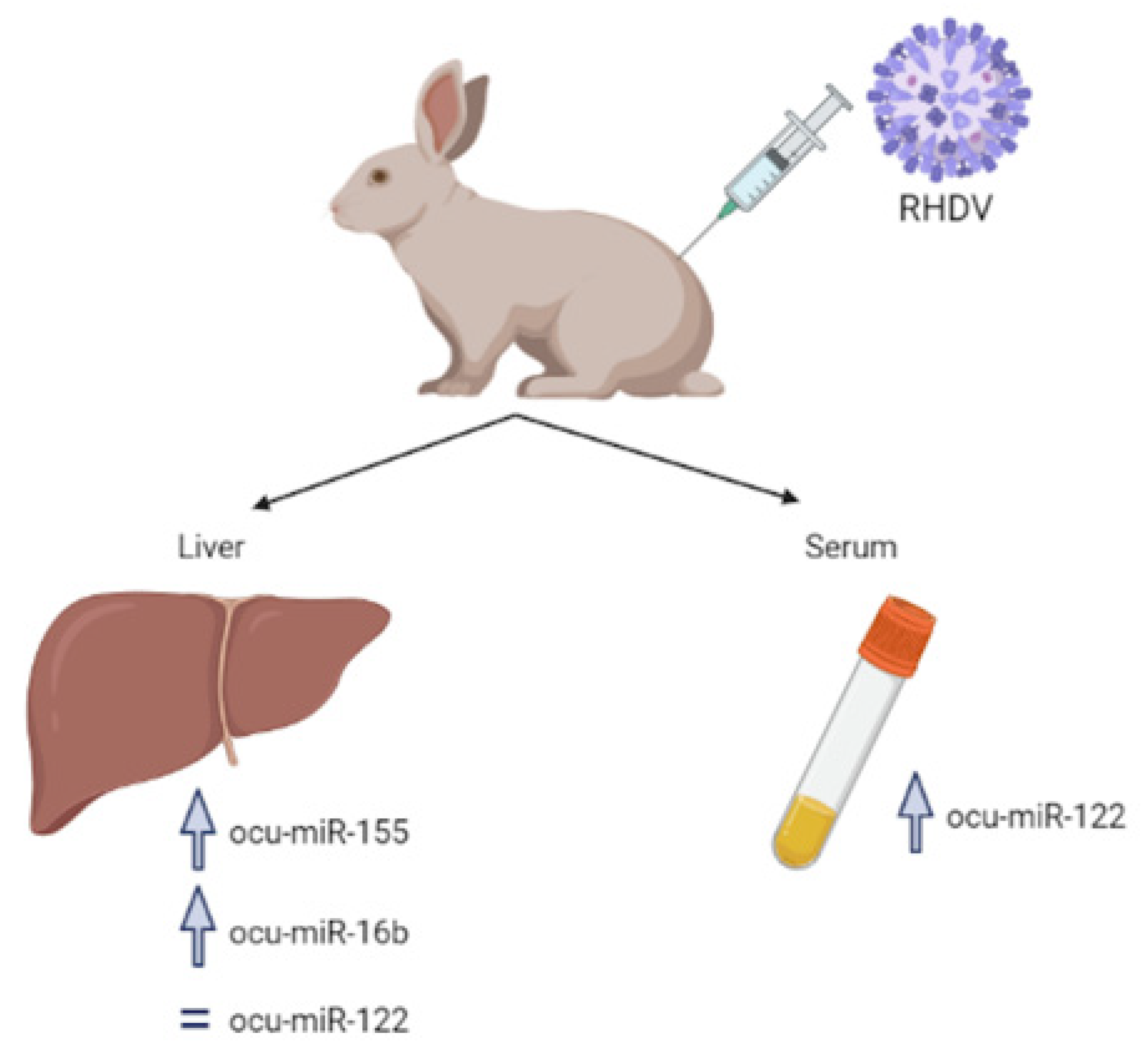
| MicroRNA | Virus/Disease | Target Genes | Biological Function/Modified Pathways | Reference |
|---|---|---|---|---|
| miR-155 | HBV/HB | SOCS1 | Enhances the phosphorylation of STAT1 and STAT3; enhances JAK/STAT signaling pathway; suppresses HBV infection in hepatocytes; regulates IFN-γ production; inhibits Akt/mTOR pathway | [23,26,29] |
| C/EBP-β | Modulates HBV replication | [24] | ||
| BCL-6, SHIP-1, SOCS-1 | Induces inflammatory cytokine production | [28] | ||
| HCV/HC | TNF-α | Pro-inflammatory response | [33] | |
| Tim3/T-bet | Increases IFN-γ production; regulates Tim-3/T-bet/STAT-5 signaling and cytokine expression in NK cells; immune injury during chronic viral disease | [36] | ||
| ISG15, TLR3 | Innate and adaptive immune response | [34] | ||
| APC | Wnt pathway; apoptosis; cell proliferation | [39] | ||
| DENV/dengue | Bach1 | Replication of DEVN; enhances anti-viral IFN responses | [43] | |
| SOCS1 | Regulates cytokine signal transduction | [41] | ||
| WNV/ West Nile fever | IL-13, BDNF, CCR9 | Cell survival | [45] | |
| IL-1β, IL-12, IL-6, IL-15, GM-CSF | Anti-viral response | [47] | ||
| IAV/influenza | S1PR1 | Inflammatory response; activates S1PR1/NF-κB/pro-inflammatory cytokine pathway | [51] | |
| IFN type I | Anti-viral response | [53] | ||
| RSV/upper respiratory tract infections | IFN type I | Anti-viral response | [53] | |
| TNF-α, IL-1β, IL-6, IL-8 | Pro-inflammatory response | [59] | ||
| SHIP1, Kif1 | Antigen presentation | [61,62] | ||
| SOCS1 | Enhances activation of STAT1 and up-regulation of ISGs gene | [63] | ||
| SARS-CoV-2/ COVID-19 | STAT1, STAT3, TGFB1, SMAD3, IRF1, AKT1, MYB, BCL6, TP6, HIF1A, FOXP3, JUNB, NFKB1 | Immune response; apoptosis | [71] | |
| SOCS1, IL-6, IL-1β, CSF1R CD274, TLF6, TNF | Regulates the host immune response; miR-155-5p–IL-6/TNF/IL-1β axis | [75] | ||
| IL-1α, G-CSF, IL-9, MIP-2, IL-12-P70 VEGF, IP-10, IFN-γ, MCP-1, MIG, MIP-1α, M−CSF, TNF-α, MIP-1β | Alleviates inflammation and lung cytokine storm | [73] | ||
| HAdV/respiratory infections (colds) | IFN-β | Anti-viral response | [80,81] | |
| SOCS1 | Enhances type I IFN Anti-viral response | [82] | ||
| SHIP1 | Enhances IFN type I signaling | [20] | ||
| HRV/COPD | HRV1B | Inhibits viral replication | [84] | |
| miR-223 | HCV/CHC | NF-κB | Chronic liver inflammation | [94] |
| DENV/dengue | STMN1 | DENV replication | [95] | |
| IAV/influenza | TNF-α | Pro-inflammatory response | [97,101] | |
| PI3K, IGF1R, GPCR, PP2A, PKA and Ca2+ channel | Represses the activity of CREB; T-cell development and cell survival | [96] | ||
| IL1RN, MDA5, STAT1 | Cell death; apoptosis | [100] | ||
| SARS-CoV/SARS | NF-κB | Activated CCR1, the inflammatory chemokine receptor for CCL3 and CCL5 enhances lung fibrosis | [105] | |
| SARS-CoV-2/ COVID-19 | TRAF6, FOXO1, TLR4, STMN1, PI3K/AKT, CXCL2, CCL3, IL-6, IFN-I, IL-1β, Caspase-1 and mainly NLRP3, IKKα, NF-κB | Regulates inflammatory processes; anti-oxidant and anti-viral role | [108] | |
| HIV-1/AIDS | RhoB Sp3, LIF | Inhibits HIV-1 production in resting primary CD4+ T-cells Activates the AKT–NF-κB pathway Viral replication | [110] [112] [112] | |
| miR-146a | HBV/CHB | NF-κB | Production of pro-inflammatory cytokines (TNF-α, IL-6, IL-8, IL-12, and IL-18) | [118] |
| STAT1 | Viral persistence Decreases cytotoxicity of T-cells | [117] | ||
| CFH | Regulation of the complement alternative pathway | [121] | ||
| XIAP | Regulation of HBV replication; regulation of XIAP-MDM2/p53 pathway | [118] | ||
| ZEB2 | Regulation of HBV transcription and replication | [119] | ||
| TRAF6, IRAK1 | Regulation of FEN1 by NF-κB activity; promotes HBV DNA replication | [120,124] | ||
| HCV/HC | SOCS1 | STAT3 inhibition; increases IL-23, IL-10, and TGF-β expression | [125] | |
| DENV/dengue | TRAF6 LC3 | Inhibits IFN-β and IL-28A/B; increases DENV2 replication Autophagy; viral replication | [129] [130] | |
| IAV/influenza | TRAF6 | Production of type I IFN; anti-viral response | [133] | |
| IRAK1 | IL-7, VEGF and, JAK-STAT signaling pathway | [134] | ||
| RSV/upper respiratory tract infections | TRAF6, IRAK1 | Viral replication | [137,138] | |
| SARS-CoV-2/ COVID-19 | TRAF6, IRAK1, IRAK2 | NF-κB pro-inflammatory signaling pathway | [139] | |
| STAT1 | Regulates JAK/STAT pathway | [139] | ||
| miR-122 | HBV/HB | cyclin G1 | Replication of HBV | [149] |
| HO-1 | Replication of HBV; oxidative stress | [153,154,155] | ||
| IFN type 1 | Replication of HBV; anti-viral response | [156] | ||
| SOCS3 | JAK/STAT pathway signaling; cytokine signaling | [157] | ||
| HCV/HC | viral 5′-NCR | Replication of HCV | [166] | |
| Bach1 | HO-1 gene regulation; HCV replication | [170] | ||
| IFN | Anti-viral response | [171] | ||
| TGFBRAP1 | Promotes HCC progression induced by HCV | [172] | ||
| DENV/dengue | CYP7A1, IGFR1, SFR, RAC1, RHOA, cyclin G1 | Immune response | [131] | |
| ZEBOV/RESTV/ Ebola | viral vp40 gene | Regulation of virus replication | [176,177] | |
| RSV/upper respiratory tract infections | Wnt | Inflammatory and immune response | [178] | |
| IL1R1 | NF-κB activation, inflammatory response | [179,180] | ||
| TLR4 | Innate immune response: NF-κB activation; cytokine secretion; inflammatory response | [180] | ||
| iNOS | Anti-viral response | [184] | ||
| HRV/COPD | CXCL2 | Chemotaxis | [185] | |
| SOCS1 | Regulates cytokine signal transduction | [185] | ||
| miR-125b | HBV/HB | LIN28B | Regulates let-7 and stimulates HBV replication | [194] |
| SCNN1A | Inhibits HBV core protein expression | [197] | ||
| HCV/HC | TLR2 | TLR2/MyD88 signaling pathway; phosphorylation of NF-κBp65, ERK, and P38 | [198] | |
| HuR | Viral replication | [200] | ||
| RSV/upper respiratory tract infections | NF-κB, MAPK | Immune response | [205] | |
| IAV/influenza | MAPK | Cell proliferation; apoptosis | [206] | |
| SARS-CoV-2/ COVID-19 | ACE2 | Activation of immune system | [207] | |
| HIV-1/AIDS | CPSF6 | Regulation of HIV-1 nuclear entry and viral replication | [208] | |
| JEV/Japanese encephalitis | STAT3, Map2k7, Triap1 | Reduces genome replication and virus titers | [212] | |
| miR-132 | HBV/HB | Akt | Development of HCC induced by HBV | [217] |
| IAV/influenza | IRF1 | Regulation of IFN-α and IFN-β production | [219] | |
| MAPK3 | Regulates innate immune signaling pathways | [134] | ||
| HSV/orofacial and genital disease | Ras | Inflammation | [224] | |
| miR-34a | HBV/HB | Wnt1 | Regulation of Wnt/β-catenin pathway; contributes to HCC induced by HBV | [229] |
| HMGB1 | Innate immune response; apoptosis | [227] | ||
| CCL22 | Chemotactic for monocytes, dendritic cells, natural killer cells, and T lymphocytes | [230] | ||
| Smad4 | Regulation of TGF-β/Smad3 pathway activity; liver fibrosis | [228] | ||
| DENV/dengue WNV/West Nile fever | Wnt | Activates type I IFN response; regulation of WNT/β-catenin signaling; anti-viral state | [233] | |
| IAV/influenza | Bax | Apoptosis | [234] | |
| STAT3 | Anti-inflammatory function Inhibits NF-κB gene reporters | [235] | ||
| SARS-CoV-2/ COVID-19 | Bcl-2, Bax, KLF4 IL-6R | Apoptosis Inflammation | [236] | |
| HTLV-1/leukemia | SIRT1 Bax | Apoptosis | [240] | |
| miR-21 | HBV/CHB | PTEN | PTEN/Akt signaling; activation of HSCs; apoptosis | [250,251] |
| Smad7 | Promotes α-SMA and collagen I expression in HSCs; liver fibrosis | [250,251] | ||
| TGF-β1 | Positive feedback loop; liver fibrosis | [249] | ||
| PDCD4 | Apoptosis | [252] | ||
| IL-12 | Regulates proliferation of NK and T-cells | [257] | ||
| HCV/HC | PTEN | Apoptosis | [260] | |
| MyD88 IRAK1 | Production of IFN type I; anti-viral response | [261] | ||
| Smad7 | Fibrosis | [262] | ||
| IAV/influenza | CCL1, CCL17, CCL19, IL-22, C2orf28 | Inflammation; apoptosis | [268] | |
| TGF-β | Fibrosis | [271] | ||
| Ad/respiratory infection | p53 TGF-β | Apoptosis | [80] | |
| SARS-CoV-2/ COVID-19 | ORF1ab ORF3a S protein gene | Regulation of viral entry | [273] | |
| CCL20 | Chemotactic response TGF-β and Akt signaling | [139] | ||
| IRAK1 | Participates in NF-κB pro-inflammatory signaling pathway | [139,273] | ||
| CXCL-10 | Pro-inflammatory response; chemotaxis | [273] | ||
| HIV-1/AIDS | PTEN PDCD4 CDKN1B | Apoptosis | [279] | |
| IP-10 | Inflammation | [280] | ||
| CVB3 | YOD1 | Metabolism of proteins; protein ubiquitination | [284] | |
| CHPV | PTEN | Regulation of phosphorylation of AKT and subunit p65 of NF-ĸB; production of pro-inflammatory cytokines (IL-6, TNF-α) | [286] | |
| miR-16 | HBV/HB | CCND1 | Cell proliferation | [290] |
| c-Myc | Cellular metabolism and proliferation | [290] | ||
| Bcl-2 | Apoptosis | [290] | ||
| HCV/HC | HGF | Hepatocyte proliferation | [291] | |
| Smad7 | Fibrosis | [291] | ||
| Human CoV (MERS, SARS, SARS-CoV-2) | polyprotein 1ab coding region | Regulation of viral replication | [272] | |
| SARS-CoV-2/ COVID-19 | MK2 CCND1 | Cell proliferation | [273] | |
| RSV/upper respiratory tract infections | TLR4 RIG-1 | Activation of NF-κB; pro-inflammatory | [58] | |
| miR-181a | HBV/HB | PTEN | Cell proliferation; apoptosis; development of hepatocellular carcinoma | [300] |
| CLDN1 | Blocks HCV entry into cells | [306] | ||
| E2F5 | Development of HCC induced by HBV | [301] | ||
| Fas | Apoptosis and regulation of HCC cell proliferation | [302] | ||
| HCV/CHC | DUSP6 | Regulation of T-cell response | [305] | |
| miR-181b | HBV/HB | p27 PTEN | Regulation of hepatic stellate cell proliferation; pro-fibrotic role | [298,299] |
| miR-181c | HCV/HC | HOXA1 | Cell growth regulator; activates STAT3 and STAT5 | [306] |
| ATM | Apoptosis | [307] | ||
| CDK-2 cyclin-A | Cell cycle | [307] | ||
| IAV/influenza | Bcl-2 | Apoptosis | [206] | |
| IL-2, TNF-α | Immune response | [206] | ||
| let-7a | HBV/HB | STAT3 | Immune suppression | [315,317] |
| HCV/HC | CHUK, IKBKE, XPNPEP1 | Regulation of HCV assembly or secretion | [320] | |
| JEV/Japanese encephalitis | cPARP | Apoptosis | [337] | |
| let-7b | HCV/HC | IFN type I SOCS1 IKKα | Immune response; anti-viral response; stimulation of JAK/STAT signaling pathway | [322] |
| GF2BP1 | Regulation of HCV replication | [323] | ||
| SARS-CoV-2/ COVID-19 | RELB, IL-6, KHSRP, EHMT2, KDM2B, CMYC, LIN28B | Immune response; repair function | [71] | |
| TLR4 | Regulation of inflammation via NF-κB | [333] | ||
| IL-6, IL-8, TNF-α, IL-10 | Regulation of pro- and anti-inflammatory cytokine production | [333] | ||
| ORF1ab (virus gene) | Regulation of viral replication | [238] | ||
| HIV-1/AIDS | IL-10 | Anti-viral response | [335] | |
| JEV/Japanese encephalitis | cPARP | Apoptosis | [337] | |
| let-7c | DENV/dengue | Bach1 | Regulation of HO-1; regulation of viral replication; oxidative stress; anti-inflammatory response | [326] |
| let-7f | RSV/upper respiratory tract infections | CCL7 SOCS3 | Anti-viral cytokine response | [330] |
| let-7i | HIV-1/AIDS | IL-2 | Th-cell death | [336] |
| miR-10a | HCV/HC | RORA | Immune response | [340] |
| SREBP1 FANS SREBP2 PGC1α | Lipid homeostasis; cholesterol biosynthesis; fatty acid synthesis; regulation of fatty acid metabolism | [340] | ||
| Bmal1 | Liver damage | [340] | ||
| RSV/upper respiratory tract infections | NF-κB MAPK | Immune response | [341,342,343] |
| MicroRNA | Virus/Disease | Target Genes | Biological Function/Modified Pathways | Reference |
|---|---|---|---|---|
| miR-155 | Lagovirus europaeus/RHDV/ RHD | JUN, IGFR1, KRAS, TNF-α, TGF-β, IL-1β, IL-6, IFN | Immune response | [347] |
| AIV/influenza | MX1 | Anti-viral response | [348] | |
| JNK pathway | Apoptosis | [348] | ||
| miR-223 | VSV/vesicular stomatitis | FOXO3 | Regulation of IFN type I production | [354] |
| miR-146a | H1N2/influenza | IRAK1 STAT2 TLR2 | Innate immune response; regulation of IFN type I production | [353] |
| HeV | RIG-I | Innate immune response | [359] | |
| RFN11 | Regulation of viral replication | [359] | ||
| VSV/vesicular stomatitis | IRAK1, IRAK2 | Regulation of IFN type I production | [116] | |
| FMDV/FMD | TRAF6 IRAK1 | Regulation of NF-κB activation; innate immune response | [365] | |
| PRRSV/PRRS | C1QTNF3 MAFB | Regulation of immune response | [369] | |
| miR-145 | AIV/influenza | HA | Regulation of pathogenicity of influenza virus and immunosuppression | [371] |
| RABV/rabies | MYC, STAM, STAT4, SOCS7 | JAK/STAT signaling pathway | [373] | |
| IL17RB TGFBR2 | Receptor for the pro-inflammatory cytokines IL17B and IL17E, controlling the growth and/or differentiation of hematopoietic cells Regulation of cell cycle arrest in epithelial and hematopoietic cells; control of mesenchymal cell proliferation and differentiation; wound healing; extracellular matrix production; immunosuppression; and carcinogenesis | [373] | ||
| MYC, SMAD3, INHBB, TGFBR2, SMAD5 | TGF-beta signaling pathway | [373] | ||
| ARF6, CFL2, LAT, ARPC5, CRKL | Fc gamma R-mediated phagocytosis | [373] | ||
| miR-21 | H1N2/influenza | CXXL10 | Chemoattractant for monocytes/macrophages, T-cells, NK cells, and dendritic cells; may also promote T-cell adhesion to endothelial cells | [353,355] |
| miR-16 miR-15a | Lagovirus europaeus/RHDV/RHD | HGF Bcl-2 | Cell proliferation during the liver regeneration process Apoptosis | [347] |
| AIV/influenza | CXCL10 | Chemoattractant for monocytes/macrophages, T-cells, NK cells, and dendritic cells; may also promote T-cell adhesion to endothelial cells | [353] | |
| TLR7 | Activation of innate immunity | [353] | ||
| miR-181a | PRRSV/PRRS | ORF7 (virus gene) | Inhibits viral gene expression and virus protein production | [379] |
| miR-181b | FMDV/FMD | RASSF1A NF-κB | Regulates cellular proliferation and inflammatory response | [365] |
| AC9 | Immunomodulatory effect | [365] | ||
| IFNα | Anti-viral response | [365] | ||
| miR-181c | PRRSV/PRRS | ORF7 (virus gene) | Inhibits viral gene expression and virus protein production | [379] |
| let-7g | FMDV/FMD | LOX | Cell proliferation | [365] |
| caspase-3 | Apoptosis | [365] | ||
| let-7i | MDV/MD | ATF2 | Proliferation, apoptosis | [380] |
| miR-122 | Lagovirus europaeus/RHDV/RHD | HFG, c-Met STAT1, STAT3 | Hepatic homeostasis Apoptosis | [347] |
| AIV/influenza | MX1, IL-8, IRF-7, TNFRS19 | Immune response | [348] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ostrycharz, E.; Hukowska-Szematowicz, B. Micro-Players of Great Significance—Host microRNA Signature in Viral Infections in Humans and Animals. Int. J. Mol. Sci. 2022, 23, 10536. https://doi.org/10.3390/ijms231810536
Ostrycharz E, Hukowska-Szematowicz B. Micro-Players of Great Significance—Host microRNA Signature in Viral Infections in Humans and Animals. International Journal of Molecular Sciences. 2022; 23(18):10536. https://doi.org/10.3390/ijms231810536
Chicago/Turabian StyleOstrycharz, Ewa, and Beata Hukowska-Szematowicz. 2022. "Micro-Players of Great Significance—Host microRNA Signature in Viral Infections in Humans and Animals" International Journal of Molecular Sciences 23, no. 18: 10536. https://doi.org/10.3390/ijms231810536
APA StyleOstrycharz, E., & Hukowska-Szematowicz, B. (2022). Micro-Players of Great Significance—Host microRNA Signature in Viral Infections in Humans and Animals. International Journal of Molecular Sciences, 23(18), 10536. https://doi.org/10.3390/ijms231810536





