TRIB3 Modulates PPARγ-Mediated Growth Inhibition by Interfering with the MLL Complex in Breast Cancer Cells
Abstract
1. Introduction
2. Results
2.1. TRIB3 Regulates PPARγ Expression in Breast Cancer Cells
2.2. Phospho-Proteome of TRIB3-KD in MCF7 Cells Reveals Its Role as an Epigenetic Regulator
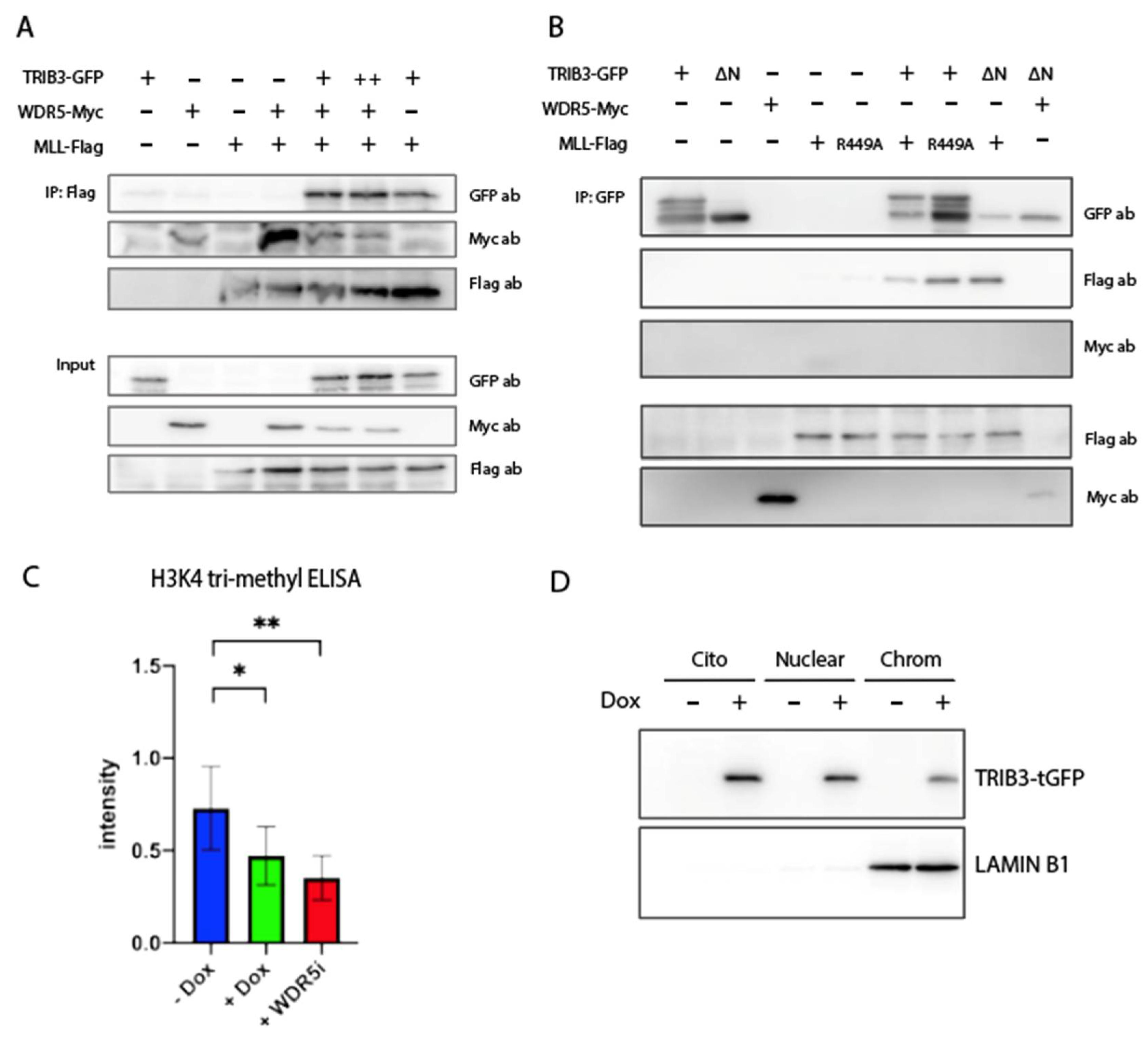
2.3. TRIB3 Interacts with WDR5 and ASHL2 Subunits of the WRAD Complex and with the SET Domain of MLL/SET1 Proteins
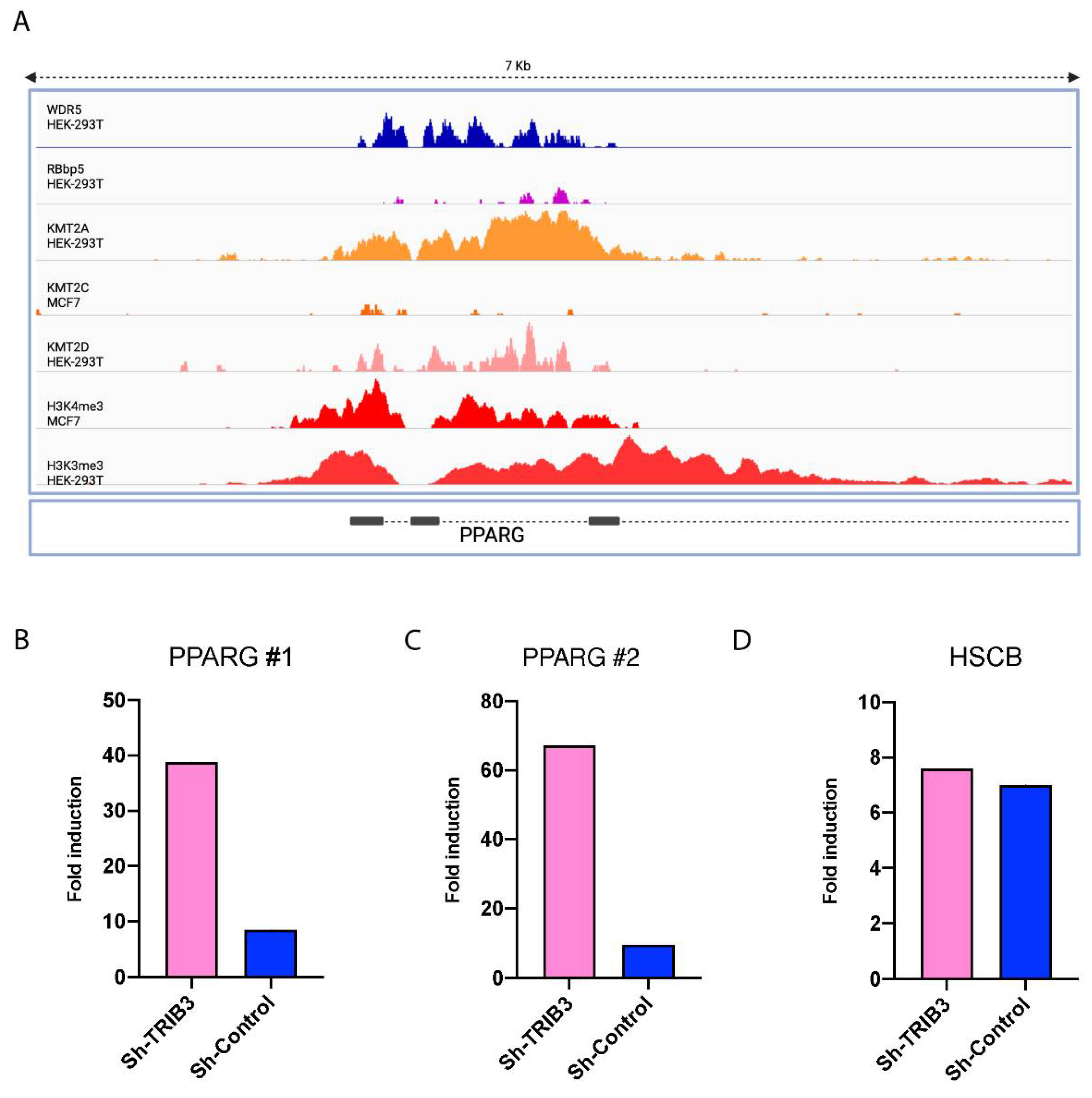
2.4. TRIB3 Interferes with H3K4me3 Levels on a Global and Local Level
2.5. TRIB3 Modulates PPARγ-Mediated Growth Inhibition in MCF7 Cells
3. Materials
3.1. Cell Culture
3.2. Western Blotting
3.3. RNA Isolation and RT-qPCR
3.4. RNA Sequencing
3.5. Immunoprecipitation
3.6. Phosphoproteomics
3.7. LC-MS/MS Analysis
3.8. Data Analysis
3.9. Cell Proliferation Assay
3.10. H3K4me3 ELISA
3.11. ChIP Followed by RT-qPCR
3.12. Chromatin Bond Protein Extraction
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiss-Toth, E.; Velasco, G.; Pear, W.S. Tribbles at the cross-roads. Biochem. Soc. Trans. 2015, 43, 1049–1050. [Google Scholar] [CrossRef] [PubMed]
- Dobens, L.L.; Nauman, C.; Fischer, Z.; Yao, X. Control of Cell Growth and Proliferation by the Tribbles Pseudokinase: Lessons from Drosophila. Cancers 2021, 13, 883. [Google Scholar] [CrossRef] [PubMed]
- Eyers, P.A.; Keeshan, K.; Kannan, N. Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol. 2017, 27, 284–298. [Google Scholar] [CrossRef] [PubMed]
- Richmond, L.; Keeshan, K. Pseudokinases: A tribble-edged sword. FEBS J. 2020, 287, 4170–4182. [Google Scholar] [CrossRef] [PubMed]
- Durzynska, I.; Xu, X.; Adelmant, G.; Ficarro, S.B.; Marto, J.A.; Sliz, P.; Uljon, S.; Blacklow, S.C. STK40 Is a Pseudokinase that Binds the E3 Ubiquitin Ligase COP1. Structure 2017, 25, 287–294. [Google Scholar] [CrossRef]
- Jamieson, S.A.; Ruan, Z.; Burgess, A.E.; Curry, J.R.; McMillan, H.D.; Brewster, J.L.; Dunbier, A.K.; Axtman, A.D.; Kannan, N.; Mace, P.D. Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci. Signal. 2018, 11, eaau0597. [Google Scholar] [CrossRef]
- Murphy, J.M.; Nakatani, Y.; Jamieson, S.A.; Dai, W.; Lucet, I.S.; Mace, P.D. Molecular Mechanism of CCAAT-Enhancer Binding Protein Recruitment by the TRIB1 Pseudokinase. Structure 2015, 23, 2111–2121. [Google Scholar] [CrossRef]
- Salazar, M.; Lorente, M.; Garcia-Taboada, E.; Perez Gomez, E.; Davila, D.; Zuniga-Garcia, P.; Maria Flores, J.; Rodriguez, A.; Hegedus, Z.; Mosen-Ansorena, D.; et al. Loss of Tribbles pseudokinase-3 promotes Akt-driven tumorigenesis via FOXO inactivation. Cell Death Differ. 2015, 22, 131–144. [Google Scholar] [CrossRef]
- Mace, P.D.; Murphy, J.M. There’s more to death than life: Noncatalytic functions in kinase and pseudokinase signaling. J. Biol. Chem. 2021, 296, 100705. [Google Scholar] [CrossRef]
- Jousse, C.; Deval, C.; Maurin, A.C.; Parry, L.; Cherasse, Y.; Chaveroux, C.; Lefloch, R.; Lenormand, P.; Bruhat, A.; Fafournoux, P. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J. Biol. Chem. 2007, 282, 15851–15861. [Google Scholar] [CrossRef]
- Takahashi, Y.; Ohoka, N.; Hayashi, H.; Sato, R. TRB3 suppresses adipocyte differentiation by negatively regulating PPARgamma transcriptional activity. J. Lipid. Res. 2008, 49, 880–892. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Sun, W.; Wang, Z.H.; Liang, X.; Hua, F.; Li, K.; Lv, X.X.; Zhang, X.W.; Liu, Y.Y.; Yu, J.J.; et al. TRIB3 supports breast cancer stemness by suppressing FOXO1 degradation and enhancing SOX2 transcription. Nat. Commun. 2019, 10, 5720. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Quiles, M.; Baak, R.; Borgman, A.; den Haan, S.; Sobrevals Alcaraz, P.; van Es, R.; Kiss-Toth, E.; Vos, H.; Kalkhoven, E. Comprehensive Profiling of Mammalian Tribbles Interactomes Implicates TRIB3 in Gene Repression. Cancers 2021, 13, 6318. [Google Scholar] [CrossRef]
- Jiang, H. The complex activities of the SET1/MLL complex core subunits in development and disease. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194560. [Google Scholar] [CrossRef] [PubMed]
- Vedadi, M.; Blazer, L.; Eram, M.S.; Barsyte-Lovejoy, D.; Arrowsmith, C.H.; Hajian, T. Targeting human SET1/MLL family of proteins. Protein Sci. 2017, 26, 662–676. [Google Scholar] [CrossRef] [PubMed]
- Shilatifard, A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr. Opin. Cell Biol. 2008, 20, 341–348. [Google Scholar] [CrossRef]
- Ernst, P.; Vakoc, C.R. WRAD: Enabler of the SET1-family of H3K4 methyltransferases. Brief Funct. Genom. 2012, 11, 217–226. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef]
- Aho, E.R.; Weissmiller, A.M.; Fesik, S.W.; Tansey, W.P. Targeting WDR5: A WINning Anti-Cancer Strategy? Epigenet. Insights 2019, 12, 2516865719865282. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; Troy, A.E.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Gimple, R.C.; Zhou, N.; Zhao, L.; Gustafsson, J.A.; Zhou, S. Targeting Nuclear Receptors for Cancer Therapy: Premises, Promises, and Challenges. Trends Cancer 2021, 7, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Q.; Dong, L.; Liu, C.; Sun, Z.; Gao, L.; Wang, X. Morusin suppresses breast cancer cell growth in vitro and in vivo through C/EBPbeta and PPARgamma mediated lipoapoptosis. J. Exp. Clin. Cancer Res. 2015, 34, 137. [Google Scholar] [CrossRef] [PubMed]
- Vella, V.; Nicolosi, M.L.; Giuliano, S.; Bellomo, M.; Belfiore, A.; Malaguarnera, R. PPAR-gamma Agonists As Antineoplastic Agents in Cancers with Dysregulated IGF Axis. Front. Endocrinol. 2017, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Ishay-Ronen, D.; Diepenbruck, M.; Kalathur, R.K.R.; Sugiyama, N.; Tiede, S.; Ivanek, R.; Bantug, G.; Morini, M.F.; Wang, J.; Hess, C.; et al. Gain Fat-Lose Metastasis: Converting Invasive Breast Cancer Cells into Adipocytes Inhibits Cancer Metastasis. Cancer Cell 2019, 35, 17–32.e6. [Google Scholar] [CrossRef]
- Dalamaga, M.; Christodoulatos, G.S.; Liu, J. Diverting epithelial-to-mesenchymal transition to transform cancer cells to adipocytes: A promising strategy to stop metastasis. Metabol. Open 2019, 3, 100012. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef]
- Martin, A.M.; Weber, B.L. Genetic and hormonal risk factors in breast cancer. J. Natl. Cancer Inst. 2000, 92, 1126–1135. [Google Scholar] [CrossRef]
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, C.; Wang, X.; Sun, Y.; Zhang, J.; Chen, J.; Shi, Y. An Epigenetic Role of Mitochondria in Cancer. Cells 2022, 11, 2518. [Google Scholar] [CrossRef]
- Bahat, A.; Gross, A. Mitochondrial plasticity in cell fate regulation. J. Biol. Chem. 2019, 294, 13852–13863. [Google Scholar] [CrossRef] [PubMed]
- Reid, M.A.; Dai, Z.; Locasale, J.W. The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat. Cell Biol. 2017, 19, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Beeraka, N.M.; Tang, C.; Babayeva, Y.V.; Sinelnikov, M.Y.; Zhang, X.; Zhang, J.; Liu, J.; Reshetov, I.V.; et al. Advances in the Prevention and Treatment of Obesity-Driven Effects in Breast Cancers. Front. Oncol. 2022, 12, 820968. [Google Scholar] [CrossRef]
- Calle, E.E.; Kaaks, R. Overweight, obesity and cancer: Epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 2004, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Greaney, M.L.; Sprunck-Harrild, K.; Ruddy, K.J.; Ligibel, J.; Barry, W.T.; Baker, E.; Meyer, M.; Emmons, K.M.; Partridge, A.H. Study protocol for Young & Strong: A cluster randomized design to increase attention to unique issues faced by young women with newly diagnosed breast cancer. BMC Public Health 2015, 15, 37. [Google Scholar] [CrossRef]
- Villarreal-Garza, C.; Aguila, C.; Magallanes-Hoyos, M.C.; Mohar, A.; Bargallo, E.; Meneses, A.; Cazap, E.; Gomez, H.; Lopez-Carrillo, L.; Chavarri-Guerra, Y.; et al. Breast cancer in young women in Latin America: An unmet, growing burden. Oncologist 2013, 18, 1298–1306. [Google Scholar] [CrossRef]
- Wennemers, M.; Bussink, J.; Scheijen, B.; Nagtegaal, I.D.; van Laarhoven, H.W.; Raleigh, J.A.; Varia, M.A.; Heuvel, J.J.; Rouschop, K.M.; Sweep, F.C.; et al. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 2011, 13, R82. [Google Scholar] [CrossRef]
- Wennemers, M.; Bussink, J.; van den Beucken, T.; Sweep, F.C.; Span, P.N. Regulation of TRIB3 mRNA and protein in breast cancer. PLoS ONE 2012, 7, e49439. [Google Scholar] [CrossRef]
- Orea-Soufi, A.; Castillo-Lluva, S.; Salvador-Tormo, N.; Martin-Cabrera, P.; Recuero, S.; Gabicagogeascoa, E.; Moreno-Valladares, M.; Mendiburu-Elicabe, M.; Blanco-Gomez, A.; Ramos-Pittol, J.M.; et al. The Pseudokinase TRIB3 Negatively Regulates the HER2 Receptor Pathway and Is a Biomarker of Good Prognosis in Luminal Breast Cancer. Cancers 2021, 13, 5307. [Google Scholar] [CrossRef]
- Carrier, M.; Joint, M.; Lutzing, R.; Page, A.; Rochette-Egly, C. Phosphoproteome and Transcriptome of RA-Responsive and RA-Resistant Breast Cancer Cell Lines. PLoS ONE 2016, 11, e0157290. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef] [PubMed]
- Guarnaccia, A.D.; Rose, K.L.; Wang, J.; Zhao, B.; Popay, T.M.; Wang, C.E.; Guerrazzi, K.; Hill, S.; Woodley, C.M.; Hansen, T.J.; et al. Impact of WIN site inhibitor on the WDR5 interactome. Cell Rep. 2021, 34, 108636. [Google Scholar] [CrossRef]
- Song, J.J.; Kingston, R.E. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008, 283, 35258–35264. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.K.; Fields, A.T.; Cheng, K.; Lee, A.; Wagenaar, E.; Lagrois, R.; Schmidt, B.; Xia, B.; Ma, D. WD repeat-containing protein 5 (WDR5) localizes to the midbody and regulates abscission. J. Biol. Chem. 2015, 290, 8987–9001. [Google Scholar] [CrossRef] [PubMed]
- Grebien, F.; Vedadi, M.; Getlik, M.; Giambruno, R.; Grover, A.; Avellino, R.; Skucha, A.; Vittori, S.; Kuznetsova, E.; Smil, D.; et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol. 2015, 11, 571–578. [Google Scholar] [CrossRef]
- Lee, S.; Park, S.; Lee, H.; Jeong, D.; Ham, J.; Choi, E.H.; Kim, S.J. ChIP-seq analysis reveals alteration of H3K4 trimethylation occupancy in cancer-related genes by cold atmospheric plasma. Free Radic. Biol. Med. 2018, 126, 133–141. [Google Scholar] [CrossRef]
- Kim, K.Y.; Kim, S.S.; Cheon, H.G. Differential anti-proliferative actions of peroxisome proliferator-activated receptor-gamma agonists in MCF-7 breast cancer cells. Biochem. Pharmacol. 2006, 72, 530–540. [Google Scholar] [CrossRef]
- Jeninga, E.H.; van Beekum, O.; van Dijk, A.D.; Hamers, N.; Hendriks-Stegeman, B.I.; Bonvin, A.M.; Berger, R.; Kalkhoven, E. Impaired peroxisome proliferator-activated receptor gamma function through mutation of a conserved salt bridge (R425C) in familial partial lipodystrophy. Mol. Endocrinol. 2007, 21, 1049–1065. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Smyth, G.K. Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012, 40, e133. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Ramos Pittol, J.M.; Milona, A.; Morris, I.; Willemsen, E.C.L.; van der Veen, S.W.; Kalkhoven, E.; van Mil, S.W.C. FXR Isoforms Control Different Metabolic Functions in Liver Cells via Binding to Specific DNA Motifs. Gastroenterology 2020, 159, 1853–1865.e10. [Google Scholar] [CrossRef] [PubMed]
- van Nuland, R.; Smits, A.H.; Pallaki, P.; Jansen, P.W.; Vermeulen, M.; Timmers, H.T. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol. Cell Biol. 2013, 33, 2067–2077. [Google Scholar] [CrossRef]
- Badenhorst, P.; Voas, M.; Rebay, I.; Wu, C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002, 16, 3186–3198. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Deng, L.; Liao, L.; Yang, S.Y.; Hu, S.Y.; Ning, Y.; Zhang, F.L.; Li, D.Q. Chromatin complexes subunit BAP18 promotes triple-negative breast cancer progression through transcriptional activation of oncogene S100A9. Cell Death Dis. 2022, 13, 408. [Google Scholar] [CrossRef]
- Ord, T.; Ord, D.; Adler, P.; Vilo, J.; Ord, T. TRIB3 enhances cell viability during glucose deprivation in HEK293-derived cells by upregulating IGFBP2, a novel nutrient deficiency survival factor. Biochim. Biophys. Acta 2015, 1853, 2492–2505. [Google Scholar] [CrossRef]
- Carraro, V.; Maurin, A.C.; Lambert-Langlais, S.; Averous, J.; Chaveroux, C.; Parry, L.; Jousse, C.; Ord, D.; Ord, T.; Fafournoux, P.; et al. Amino acid availability controls TRB3 transcription in liver through the GCN2/eIF2alpha/ATF4 pathway. PLoS ONE 2010, 5, e15716. [Google Scholar] [CrossRef]
- Kubota, T.; Koshizuka, K.; Williamson, E.A.; Asou, H.; Said, J.W.; Holden, S.; Miyoshi, I.; Koeffler, H.P. Ligand for peroxisome proliferator-activated receptor gamma (troglitazone) has potent antitumor effect against human prostate cancer both in vitro and in vivo. Cancer Res. 1998, 58, 3344–3352. [Google Scholar] [PubMed]
- Sarraf, P.; Mueller, E.; Jones, D.; King, F.J.; DeAngelo, D.J.; Partridge, J.B.; Holden, S.A.; Chen, L.B.; Singer, S.; Fletcher, C.; et al. Differentiation and reversal of malignant changes in colon cancer through PPARgamma. Nat. Med. 1998, 4, 1046–1052. [Google Scholar] [CrossRef] [PubMed]
- Motomura, W.; Okumura, T.; Takahashi, N.; Obara, T.; Kohgo, Y. Activation of peroxisome proliferator-activated receptor gamma by troglitazone inhibits cell growth through the increase of p27KiP1 in human. Pancreatic carcinoma cells. Cancer Res. 2000, 60, 5558–5564. [Google Scholar]
- Tsubouchi, Y.; Sano, H.; Kawahito, Y.; Mukai, S.; Yamada, R.; Kohno, M.; Inoue, K.; Hla, T.; Kondo, M. Inhibition of human lung cancer cell growth by the peroxisome proliferator-activated receptor-gamma agonists through induction of apoptosis. Biochem. Biophys. Res. Commun. 2000, 270, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Bonofiglio, D.; Cione, E.; Qi, H.; Pingitore, A.; Perri, M.; Catalano, S.; Vizza, D.; Panno, M.L.; Genchi, G.; Fuqua, S.A.; et al. Combined low doses of PPARgamma and RXR ligands trigger an intrinsic apoptotic pathway in human breast cancer cells. Am. J. Pathol. 2009, 175, 1270–1280. [Google Scholar] [CrossRef]
- Saez, E.; Tontonoz, P.; Nelson, M.C.; Alvarez, J.G.; Ming, U.T.; Baird, S.M.; Thomazy, V.A.; Evans, R.M. Activators of the nuclear receptor PPARgamma enhance colon polyp formation. Nat. Med. 1998, 4, 1058–1061. [Google Scholar] [CrossRef]
- Lefebvre, A.M.; Chen, I.; Desreumaux, P.; Najib, J.; Fruchart, J.C.; Geboes, K.; Briggs, M.; Heyman, R.; Auwerx, J. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nat. Med. 1998, 4, 1053–1057. [Google Scholar] [CrossRef]
- Wang, X.; Southard, R.C.; Kilgore, M.W. The increased expression of peroxisome proliferator-activated receptor-gamma1 in human breast cancer is mediated by selective promoter usage. Cancer Res. 2004, 64, 5592–5596. [Google Scholar] [CrossRef][Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández-Quiles, M.; Baak, R.; Orea-Soufi, A.; Borgman, A.; den Haan, S.; Sobrevals Alcaraz, P.; Jongejan, A.; van Es, R.; Velasco, G.; Vos, H.; et al. TRIB3 Modulates PPARγ-Mediated Growth Inhibition by Interfering with the MLL Complex in Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 10535. https://doi.org/10.3390/ijms231810535
Hernández-Quiles M, Baak R, Orea-Soufi A, Borgman A, den Haan S, Sobrevals Alcaraz P, Jongejan A, van Es R, Velasco G, Vos H, et al. TRIB3 Modulates PPARγ-Mediated Growth Inhibition by Interfering with the MLL Complex in Breast Cancer Cells. International Journal of Molecular Sciences. 2022; 23(18):10535. https://doi.org/10.3390/ijms231810535
Chicago/Turabian StyleHernández-Quiles, Miguel, Rosalie Baak, Alba Orea-Soufi, Anouska Borgman, Suzanne den Haan, Paula Sobrevals Alcaraz, Aldo Jongejan, Robert van Es, Guillermo Velasco, Harmjan Vos, and et al. 2022. "TRIB3 Modulates PPARγ-Mediated Growth Inhibition by Interfering with the MLL Complex in Breast Cancer Cells" International Journal of Molecular Sciences 23, no. 18: 10535. https://doi.org/10.3390/ijms231810535
APA StyleHernández-Quiles, M., Baak, R., Orea-Soufi, A., Borgman, A., den Haan, S., Sobrevals Alcaraz, P., Jongejan, A., van Es, R., Velasco, G., Vos, H., & Kalkhoven, E. (2022). TRIB3 Modulates PPARγ-Mediated Growth Inhibition by Interfering with the MLL Complex in Breast Cancer Cells. International Journal of Molecular Sciences, 23(18), 10535. https://doi.org/10.3390/ijms231810535






