Role of Circular RNAs in Pulmonary Fibrosis
Abstract
:1. Introduction
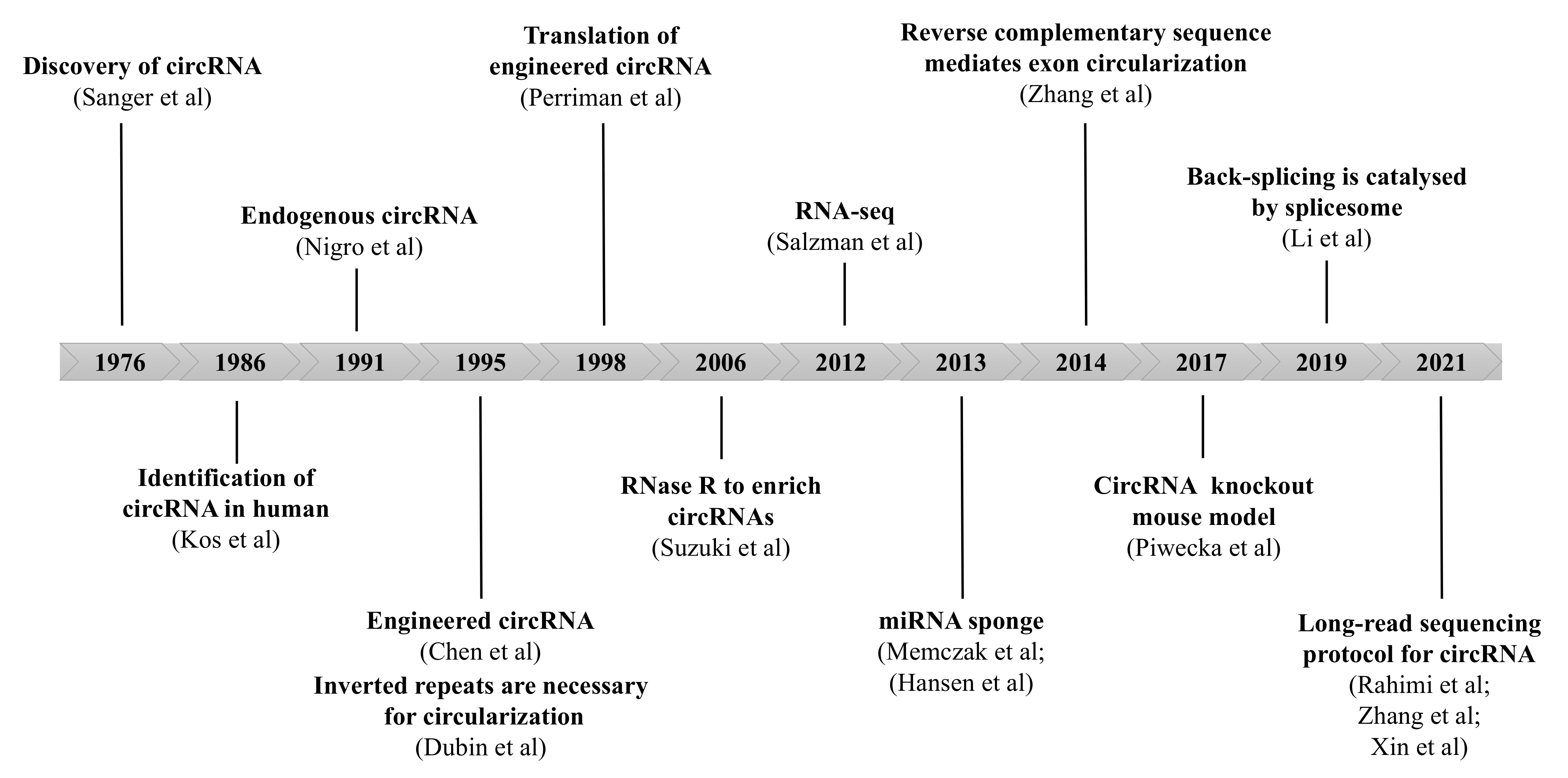
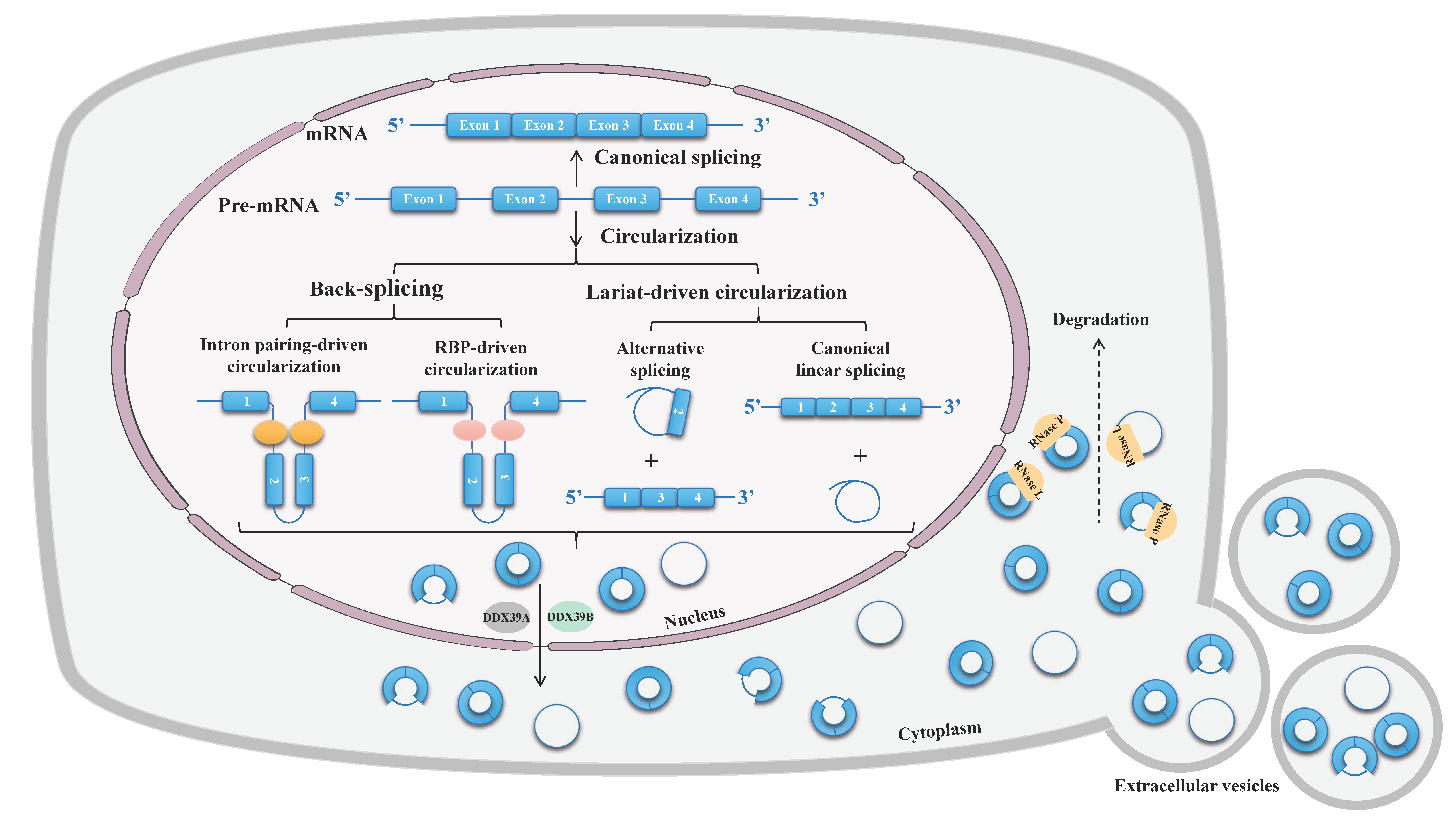
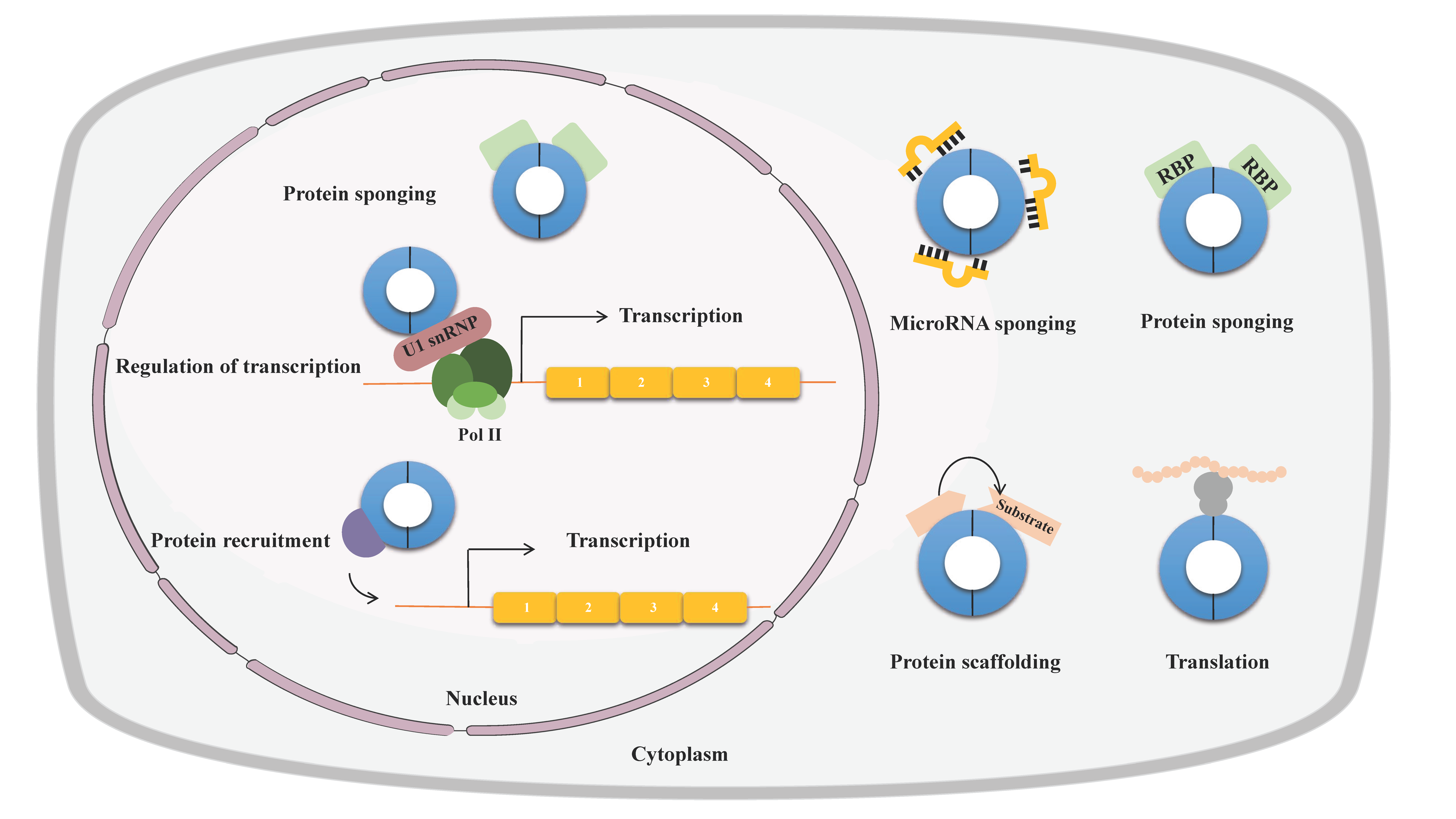
2. Properties of circRNAs
3. The circRNA Expression Profiling and Integrative Analysis in Pulmonary Fibrosis
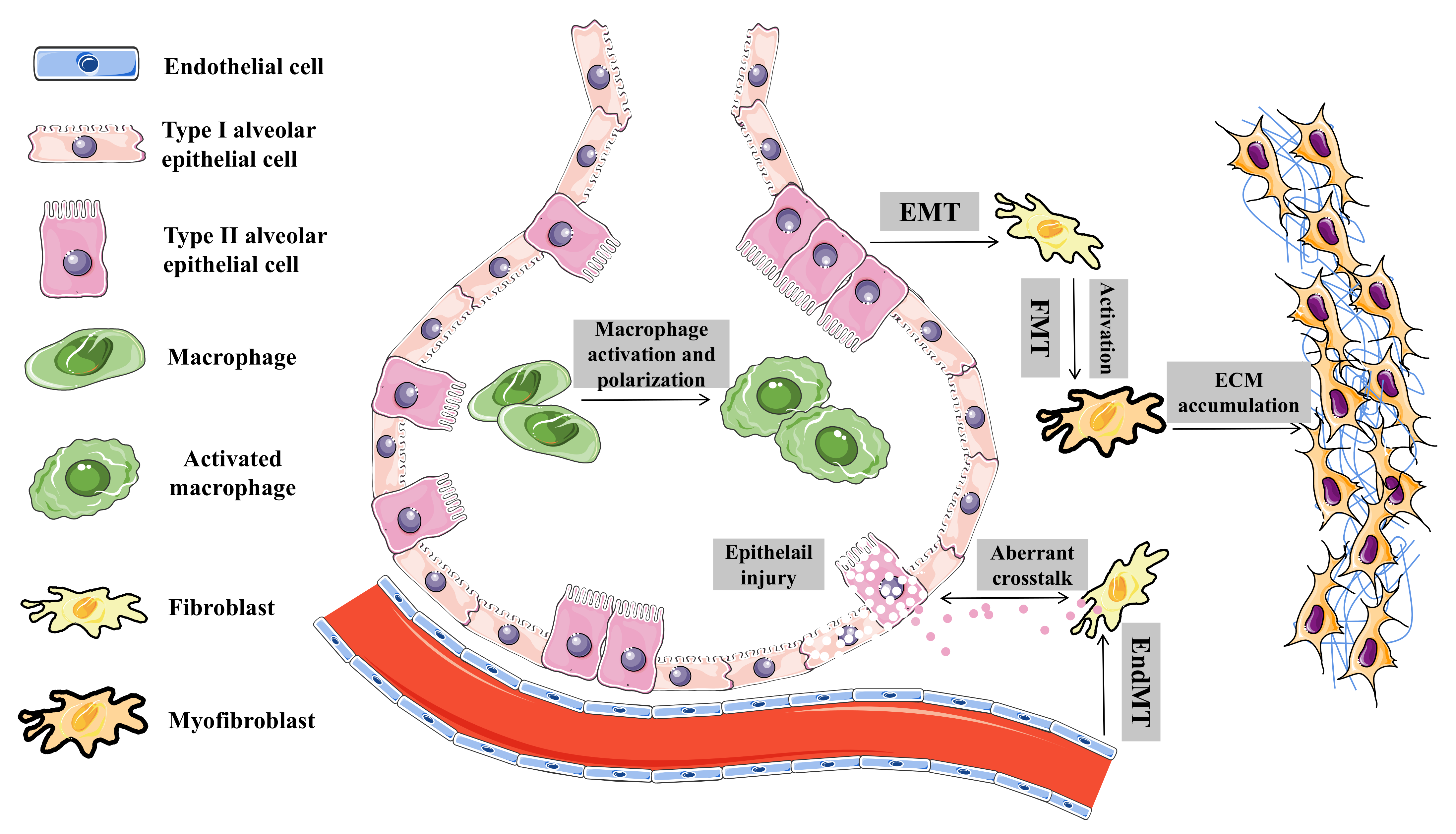
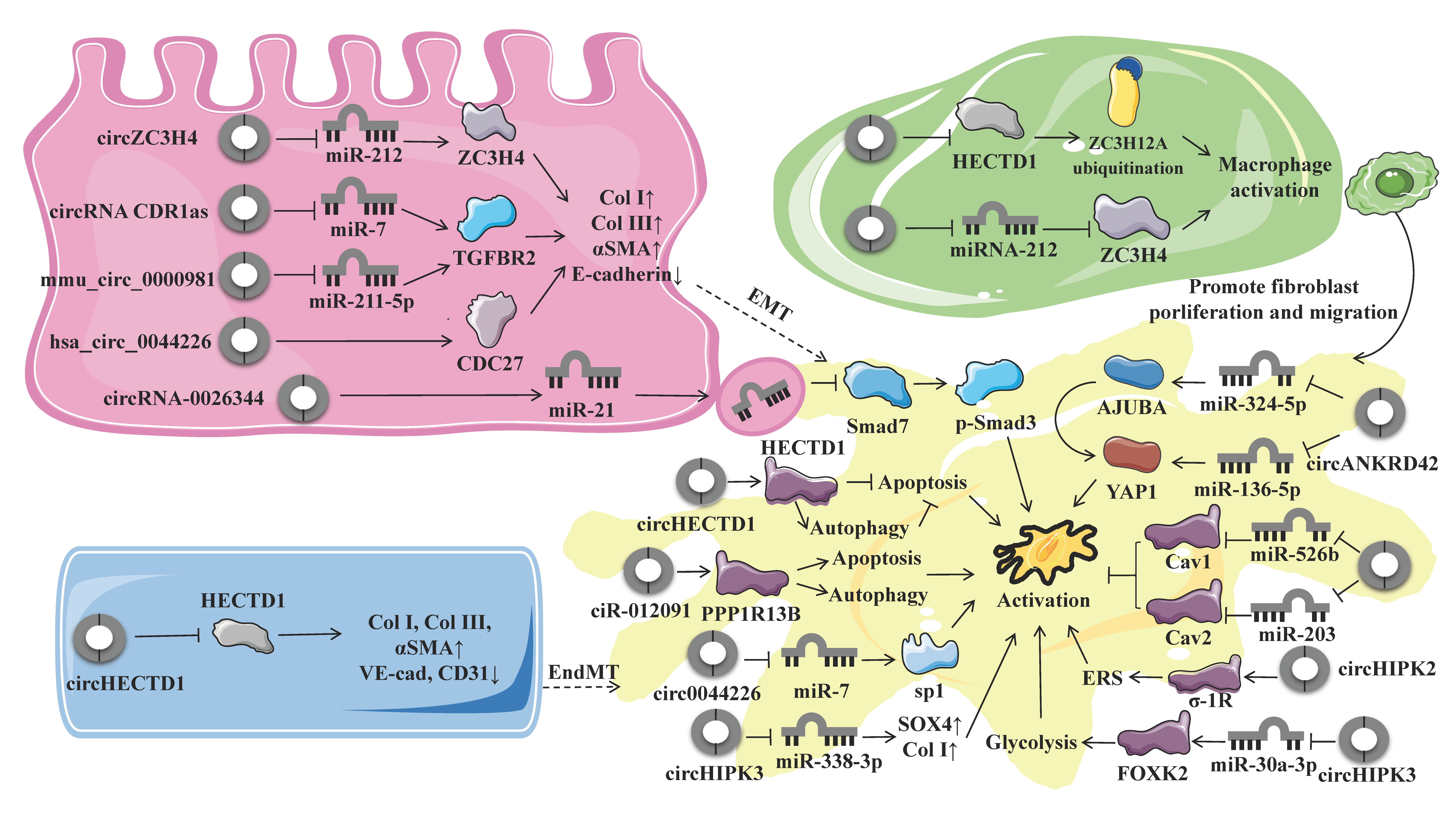
4. Biological Roles and Regulatory Mechanisms of circRNAs in Pulmonary Fibrosis
4.1. Endothelial–Mesenchymal Transition
4.2. Epithelial-To-Mesenchymal Transition
4.3. Macrophage Activation and Polarization
4.4. Fibroblast Activation and Fibroblast-To-Myofibroblast Transition
4.5. Other Biological Processes
5. Diagnostic and Therapeutic Potential of Targeting circRNAs in Pulmonary Fibrosis
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wijsenbeek, M.; Cottin, V. Spectrum of Fibrotic Lung Diseases. N. Engl. J. Med. 2020, 383, 958–968. [Google Scholar] [CrossRef] [PubMed]
- Richeldi, L.; Collard, H.R.; Jones, M.G. Idiopathic pulmonary fibrosis. Lancet 2017, 389, 1941–1952. [Google Scholar] [CrossRef]
- Lederer, D.J.; Martinez, F.J. Idiopathic Pulmonary Fibrosis. N. Engl. J. Med. 2018, 378, 1811–1823. [Google Scholar] [CrossRef]
- Moss, B.J.; Ryter, S.W.; Rosas, I.O. Pathogenic Mechanisms Underlying Idiopathic Pulmonary Fibrosis. Annu. Rev. Pathol. Mech. Dis. 2022, 17, 515–546. [Google Scholar] [CrossRef] [PubMed]
- Spagnolo, P.; Kropski, J.A.; Jones, M.G.; Lee, J.S.; Rossi, G.; Karampitsakos, T.; Maher, T.M.; Tzouvelekis, A.; Ryerson, C.J. Idiopathic pulmonary fibrosis: Disease mechanisms and drug development. Pharmacol. Ther. 2021, 222, 107798. [Google Scholar] [CrossRef] [PubMed]
- Chanda, D.; Otoupalova, E.; Smith, S.R.; Volckaert, T.; De Langhe, S.P.; Thannickal, V.J. Developmental pathways in the pathogenesis of lung fibrosis. Mol. Asp. Med. 2019, 65, 56–69. [Google Scholar] [CrossRef]
- Martinez, F.J.; Collard, H.R.; Pardo, A.; Raghu, G.; Richeldi, L.; Selman, M.; Swigris, J.J.; Taniguchi, H.; Wells, A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Primers 2017, 3, 17074. [Google Scholar] [CrossRef]
- Johannson, K.A.; Chaudhuri, N.; Adegunsoye, A.; Wolters, P.J. Treatment of fibrotic interstitial lung disease: Current approaches and future directions. Lancet 2021, 398, 1450–1460. [Google Scholar] [CrossRef]
- Liu, C.-X.; Chen, L.-L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef] [Green Version]
- Kos, A.; Dijkema, R.; Arnberg, A.C.; Van Der Meide, P.H.; Schellekens, H. The hepatitis delta (δ) virus possesses a circular RNA. Nature 1986, 323, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Sarnow, P. Initiation of Protein Synthesis by the Eukaryotic Translational Apparatus on Circular RNAs. Science 1995, 268, 415–417. [Google Scholar] [CrossRef] [PubMed]
- Dubin, R.A.; Kazmi, M.A.; Ostrer, H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 1995, 167, 245–248. [Google Scholar] [CrossRef]
- Perriman, R.; Ares, M., Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 1998, 4, 1047–1054. [Google Scholar] [CrossRef]
- Suzuki, H.; Zuo, Y.; Wang, J.; Zhang, M.Q.; Malhotra, A.; Mayeda, A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006, 34, e63. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs Are the Predominant Transcript Isoform from Hundreds of Human Genes in Diverse Cell Types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary Sequence-Mediated Exon Circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [Green Version]
- Piwecka, M.; Glažar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Li, X.; Liu, S.; Zhang, L.; Issaian, A.; Hill, R.C.; Espinosa, S.; Shi, S.; Cui, Y.; Kappel, K.; Das, R.; et al. A unified mechanism for intron and exon definition and back-splicing. Nature 2019, 573, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, K.; Venø, M.T.; Dupont, D.M.; Kjems, J. Nanopore sequencing of brain-derived full-length circRNAs reveals circRNA-specific exon usage, intron retention and microexons. Nat. Commun. 2021, 12, 4825. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hou, L.; Zuo, Z.; Ji, P.; Zhang, X.; Xue, Y.; Zhao, F. Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat. Biotechnol. 2021, 39, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Xin, R.; Gao, Y.; Gao, Y.; Wang, R.; Kadash-Edmondson, K.E.; Liu, B.; Wang, Y.; Lin, L.; Xing, Y. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat. Commun. 2021, 12, 266. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef]
- Mehta, S.L.; Dempsey, R.J.; Vemuganti, R. Role of circular RNAs in brain development and CNS diseases. Prog. Neurobiol. 2020, 186, 101746. [Google Scholar] [CrossRef]
- Kim, E.; Kim, Y.K.; Lee, S.-J.V. Emerging functions of circular RNA in aging. Trends Genet. 2021, 37, 819–829. [Google Scholar] [CrossRef]
- Mei, X.; Chen, S.-Y. Circular RNAs in cardiovascular diseases. Pharmacol. Ther. 2022, 232, 107991. [Google Scholar] [CrossRef]
- van Zonneveld, A.J.; Kölling, M.; Bijkerk, R.; Lorenzen, J.M. Circular RNAs in kidney disease and cancer. Nat. Rev. Nephrol. 2021, 17, 814–826. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2022, 19, 188–206. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.S.; Ai, Y.; Wilusz, J.E. Biogenesis and Functions of Circular RNAs Come into Focus. Trends Cell Biol. 2020, 30, 226–240. [Google Scholar] [CrossRef] [PubMed]
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.R.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xue, W.; Li, X.; Zhang, J.; Chen, S.; Zhang, J.-L.; Yang, L.; Chen, L.-L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016, 15, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, X.-O.; Chen, T.; Xiang, J.-F.; Yin, Q.-F.; Xing, Y.-H.; Zhu, S.; Yang, L.; Chen, L.-L. Circular Intronic Long Noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Hsu, M.-T.; Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979, 280, 339–340. [Google Scholar] [CrossRef]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Liang, D.; Tatomer, D.C.; Wilusz, J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018, 32, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Molinie, B.; Daneshvar, K.; Pondick, J.V.; Wang, J.; Van Wittenberghe, N.; Xing, Y.; Giallourakis, C.C.; Mullen, A.C. Genome-Wide Maps of m6A circRNAs Identify Widespread and Cell-Type-Specific Methylation Patterns that Are Distinct from mRNAs. Cell Rep. 2017, 20, 2262–2276. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.-X.; Chen, X.; Xia, L.-P.; Zhang, J.-X.; Pan, Z.-Z.; Ma, X.-D.; Han, K.; Chen, J.-W.; Judde, J.-G.; Deas, O.; et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 2019, 10, 4695. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Enuka, Y.; Lauriola, M.; Feldman, M.E.; Sas-Chen, A.; Ulitsky, I.; Yarden, Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016, 44, 1370–1383. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Wiklund, E.D.; Bramsen, J.B.; Villadsen, S.B.; Statham, A.L.; Clark, S.J.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422. [Google Scholar] [CrossRef]
- Park, O.H.; Ha, H.; Lee, Y.; Boo, S.H.; Kwon, D.H.; Song, H.K.; Kim, Y.K. Endoribonucleolytic Cleavage of m6A-Containing RNAs by RNase P/MRP Complex. Mol. Cell 2019, 74, 494–507.e8. [Google Scholar] [CrossRef]
- Liu, C.-X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.-K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e21. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA Hypothesis with Quantitative Measurements of miRNA and Target Abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Das Mahapatra, K.; Pasquali, L.; Søndergaard, J.N.; Lapins, J.; Nemeth, I.B.; Baltás, E.; Kemény, L.; Homey, B.; Moldovan, L.-I.; Kjems, J.; et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci. Rep. 2020, 10, 3637. [Google Scholar] [CrossRef]
- Xia, P.; Wang, S.; Ye, B.; Du, Y.; Li, C.; Xiong, Z.; Qu, Y.; Fan, Z. A Circular RNA Protects Dormant Hematopoietic Stem Cells from DNA Sensor cGAS-Mediated Exhaustion. Immunity 2018, 48, 688–701.e7. [Google Scholar] [CrossRef] [PubMed]
- Essers, M.A.; Offner, S.; Blanco-Bose, W.E.; Waibler, Z.; Kalinke, U.; Duchosal, M.A.; Trumpp, A. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature 2009, 458, 904–908. [Google Scholar] [CrossRef] [PubMed]
- Suenkel, C.; Cavalli, D.; Massalini, S.; Calegari, F.; Rajewsky, N. A Highly Conserved Circular RNA Is Required to Keep Neural Cells in a Progenitor State in the Mammalian Brain. Cell Rep. 2020, 30, 2170–2179.e5. [Google Scholar] [CrossRef] [PubMed]
- Hollensen, A.K.; Thomsen, H.S.; Lloret-Llinares, M.; Kamstrup, A.B.; Jensen, J.M.; Luckmann, M.; Birkmose, N.; Palmfeldt, J.; Jensen, T.H.; Hansen, T.B.; et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. eLife 2020, 9, e58478. [Google Scholar] [CrossRef] [PubMed]
- Jakobsen, T.; Dahl, M.; Dimopoulos, K.; Grønbæk, K.; Kjems, J.; Kristensen, L.S. Genome-Wide Circular RNA Expression Patterns Reflect Resistance to Immunomodulatory Drugs in Multiple Myeloma Cells. Cancers 2021, 13, 365. [Google Scholar] [CrossRef] [PubMed]
- Pamudurti, N.R.; Bartok, O.; Jens, M.; Ashwal-Fluss, R.; Stottmeister, C.; Ruhe, L.; Hanan, M.; Wyler, E.; Perez-Hernandez, D.; Ramberger, E.; et al. Translation of CircRNAs. Mol. Cell 2017, 66, 9–21.e7. [Google Scholar] [CrossRef]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e9. [Google Scholar] [CrossRef]
- Yang, Y.; Fan, X.; Mao, M.; Song, X.; Wu, P.; Zhang, Y.; Jin, Y.; Yang, Y.; Chen, L.-L.; Wang, Y.; et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017, 27, 626–641. [Google Scholar] [CrossRef]
- Li, R.; Wang, Y.; Song, X.; Sun, W.; Zhang, J.; Liu, Y.; Li, H.; Meng, C.; Zhang, J.; Zheng, Q.; et al. Potential regulatory role of circular RNA in idiopathic pulmonary fibrosis. Int. J. Mol. Med. 2018, 42, 3256–3268. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Wang, Z.; Zhang, J.; Zhao, X.; Xu, P.; Liu, X.; Li, M.; Lv, C.; Song, X. Crosstalk of mRNA, miRNA, lncRNA, and circRNA and Their Regulatory Pattern in Pulmonary Fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 204–218. [Google Scholar] [CrossRef]
- Yang, L.; Liu, X.; Zhang, N.; Chen, L.; Xu, J.; Tang, W. Investigation of circular RNAs and related genes in pulmonary fibrosis based on bioinformatics analysis. J. Cell. Biochem. 2019, 120, 11022–11032. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Jia, X.; He, R.; Zhang, X.; Zhang, W. Changing Expression Profiles of Messenger RNA, MicroRNA, Long Non-coding RNA, and Circular RNA Reveal the Key Regulators and Interaction Networks of Competing Endogenous RNA in Pulmonary Fibrosis. Front. Genet. 2020, 11, 558095. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, Y.; Wu, S.; Zhao, R.; Yu, Y.; Zhou, Y.; Zhou, Z.; Dong, Y.; Qiu, A.; Xu, H.; et al. Peripheral blood circular RNA hsa_circ_0058493 as a potential novel biomarker for silicosis and idiopathic pulmonary fibrosis. Ecotoxicol. Environ. Saf. 2022, 236, 113451. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Jiang, R.; Yang, X.; Guo, H.; Fang, S.; Zhang, Y.; Cheng, Y.; Wang, J.; Yao, H.; Chao, J. circRNA Mediates Silica-Induced Macrophage Activation Via HECTD1/ZC3H12A-Dependent Ubiquitination. Theranostics 2018, 8, 575–592. [Google Scholar] [CrossRef] [PubMed]
- Piera-Velazquez, S.; Li, Z.; Jimenez, S.A. Role of Endothelial-Mesenchymal Transition (EndoMT) in the Pathogenesis of Fibrotic Disorders. Am. J. Pathol. 2011, 179, 1074–1080. [Google Scholar] [CrossRef]
- Qi, F.; Li, Y.; Yang, X.; Wu, Y.; Lin, L.; Liu, X. Hsa_circ_0044226 knockdown attenuates progression of pulmonary fibrosis by inhibiting CDC27. Aging 2020, 12, 14808–14818. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, J.; Wang, M.; Liu, B.; Li, R.; Li, H.; Zhai, N.; Liu, W.; Lv, C.; Song, X. hnRNPL-activated circANKRD42 back-splicing and circANKRD42-mediated crosstalk of mechanical stiffness and biochemical signal in lung fibrosis. Mol. Ther. 2022, 30, 2370–2387. [Google Scholar] [CrossRef]
- Fang, S.; Guo, H.; Cheng, Y.; Zhou, Z.; Zhang, W.; Han, B.; Luo, W.; Wang, J.; Xie, W.; Chao, J. circHECTD1 promotes the silica-induced pulmonary endothelial–mesenchymal transition via HECTD1. Cell Death Dis. 2018, 9, 396. [Google Scholar] [CrossRef]
- Yang, X.; Wang, J.; Zhou, Z.; Jiang, R.; Huang, J.; Chen, L.; Cao, Z.; Chu, H.; Han, B.; Cheng, Y.; et al. Silica-induced initiation of circular ZC3H4 RNA/ZC3H4 pathway promotes the pulmonary macrophage activation. FASEB J. 2018, 32, 3264–3277. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Zhou, Z.; Liao, Y.; Yang, F.; Cheng, Y.; Huang, J.; Wang, J.; Chen, H.; Zhu, T.; Chao, J. The emerging roles of a novel CCCH-type zinc finger protein, ZC3H4, in silica-induced epithelial to mesenchymal transition. Toxicol. Lett. 2019, 307, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Li, Y.; Han, L.; Ji, X.; Pan, H.; Liu, Y.; Yuan, J.; Yan, W.; Ni, C. The CDR1as/miR-7/TGFBR2 Axis Modulates EMT in Silica-Induced Pulmonary Fibrosis. Toxicol. Sci. 2018, 166, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Gao, H.; Zhang, M.; Wang, J.; Gu, Y.; Wang, Y.; Zhang, H.; Liu, P.; Zhang, X.; Zhao, L. Atractylon Treatment Attenuates Pulmonary Fibrosis via Regulation of the mmu_circ_0000981/miR-211-5p/TGFBR2 Axis in an Ovalbumin-Induced Asthma Mouse Model. Inflammation 2021, 44, 1856–1864. [Google Scholar] [CrossRef]
- Xu, Q.; Cheng, D.; Li, G.; Liu, Y.; Li, P.; Sun, W.; Ma, D.; Ni, C. CircHIPK3 regulates pulmonary fibrosis by facilitating glycolysis in miR-30a-3p/FOXK2-dependent manner. Int. J. Biol. Sci. 2021, 17, 2294–2307. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chi, X.; Luo, W.; Yu, S.; Zhang, J.; Guo, Y.; Ren, Q.; Zhang, W. Lung myofibroblast transition and fibrosis is regulated by circ0044226. Int. J. Biochem. Cell Biol. 2020, 118, 105660. [Google Scholar] [CrossRef]
- Chu, H.; Wang, W.; Luo, W.; Zhang, W.; Cheng, Y.; Huang, J.; Wang, J.; Dai, X.; Fang, S.; Chao, J. CircHECTD1 mediates pulmonary fibroblast activation via HECTD1. Ther. Adv. Chronic Dis. 2019, 10, 2040622319891558. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, W.; Li, Z.; Cao, M.; Zhu, Z.; Han, C.; Dai, X.; Zhang, W.; Wang, J.; Yao, H.; et al. CircRNA-012091/PPP1R13B-mediated Lung Fibrotic Response in Silicosis via Endoplasmic Reticulum Stress and Autophagy. Am. J. Respir. Cell Mol. Biol. 2019, 61, 380–391. [Google Scholar] [CrossRef]
- Li, J.; Li, P.; Zhang, G.; Qin, P.; Zhang, D.; Zhao, W. CircRNA TADA2A relieves idiopathic pulmonary fibrosis by inhibiting proliferation and activation of fibroblasts. Cell Death Dis. 2020, 11, 553. [Google Scholar] [CrossRef]
- Cao, Z.; Xiao, Q.; Dai, X.; Zhou, Z.; Jiang, R.; Cheng, Y.; Yang, X.; Guo, H.; Wang, J.; Xi, Z.; et al. circHIPK2-mediated sigma-1R promotes endoplasmic reticulum stress in human pulmonary fibroblasts exposed to silica. Cell Death Dis. 2017, 8, 3212. [Google Scholar] [CrossRef]
- Bai, J.; Deng, J.; Han, Z.; Cui, Y.; He, R.; Gu, Y.; Zhang, Q. CircRNA_0026344 via exosomal miR-21 regulation of Smad7 is involved in aberrant cross-talk of epithelium-fibroblasts during cigarette smoke-induced pulmonary fibrosis. Toxicol. Lett. 2021, 347, 58–66. [Google Scholar] [CrossRef]
- Jia, Y.; Li, X.; Nan, A.; Zhang, N.; Chen, L.; Zhou, H.; Zhang, H.; Qiu, M.; Zhu, J.; Ling, Y.; et al. Circular RNA 406961 interacts with ILF2 to regulate PM2.5-induced inflammatory responses in human bronchial epithelial cells via activation of STAT3/JNK pathways. Environ. Int. 2020, 141, 105755. [Google Scholar] [CrossRef]
- Weiskirchen, R.; Weiskirchen, S.; Tacke, F. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol. Asp. Med. 2018, 65, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Lovisa, S. Epithelial-to-Mesenchymal Transition in Fibrosis: Concepts and Targeting Strategies. Front. Pharmacol. 2021, 12, 737570. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef]
- Kishore, A.; Petrek, M. Roles of Macrophage Polarization and Macrophage-Derived miRNAs in Pulmonary Fibrosis. Front. Immunol. 2021, 12, 678457. [Google Scholar] [CrossRef]
- Liu, G.; Philp, A.M.; Corte, T.; Travis, M.A.; Schilter, H.; Hansbro, N.G.; Burns, C.J.; Eapen, M.S.; Sohal, S.S.; Burgess, J.K.; et al. Therapeutic targets in lung tissue remodelling and fibrosis. Pharmacol. Ther. 2021, 225, 107839. [Google Scholar] [CrossRef]
- Tschumperlin, D.J.; Lagares, D. Mechano-therapeutics: Targeting Mechanical Signaling in Fibrosis and Tumor Stroma. Pharmacol. Ther. 2020, 212, 107575. [Google Scholar] [CrossRef]
- Jaffar, J.; Yang, S.-H.; Kim, S.Y.; Kim, H.-W.; Faiz, A.; Chrzanowski, W.; Burgess, J.K. Greater cellular stiffness in fibroblasts from patients with idiopathic pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 315, L59–L65. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-X.; Lu, J.; Xie, H.; Wang, D.-P.; Ni, H.E.; Zhu, Y.; Ren, L.-H.; Meng, X.-X.; Wang, R.-L. circHIPK3 regulates lung fibroblast-to-myofibroblast transition by functioning as a competing endogenous RNA. Cell Death Dis. 2019, 10, 182. [Google Scholar] [CrossRef]
- Xie, L.; Zeng, Y. Therapeutic Potential of Exosomes in Pulmonary Fibrosis. Front. Pharmacol. 2020, 11, 590972. [Google Scholar] [CrossRef] [PubMed]
- Hewlett, J.C.; Kropski, J.A.; Blackwell, T.S. Idiopathic pulmonary fibrosis: Epithelial-mesenchymal interactions and emerging therapeutic targets. Matrix Biol. 2018, 71–72, 112–127. [Google Scholar] [CrossRef] [PubMed]
- Katzen, J.; Beers, M.F. Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J. Clin. Investig. 2020, 130, 5088–5099. [Google Scholar] [CrossRef] [PubMed]
- Sesé, L.; Nunes, H.; Cottin, V.; Sanyal, S.; Didier, M.; Carton, Z.; Israel-Biet, D.; Crestani, B.; Cadranel, J.; Wallaert, B.; et al. Role of atmospheric pollution on the natural history of idiopathic pulmonary fibrosis. Thorax 2017, 73, 145–150. [Google Scholar] [CrossRef]
- Spagnolo, P.; Ryerson, C.J.; Putman, R.; Oldham, J.; Salisbury, M.; Sverzellati, N.; Valenzuela, C.; Guler, S.; Jones, S.; Wijsenbeek, M.; et al. Early diagnosis of fibrotic interstitial lung disease: Challenges and opportunities. Lancet Respir. Med. 2021, 9, 1065–1076. [Google Scholar] [CrossRef]
- Molyneaux, P.L.; Smith, J.J.; Saunders, P.; Chua, F.; Wells, A.U.; Renzoni, E.A.; Nicholson, A.G.; Fahy, W.A.; Jenkins, R.G.; Maher, T.M. BAL Is Safe and Well Tolerated in Individuals with Idiopathic Pulmonary Fibrosis: An Analysis of the PROFILE Study. Am. J. Respir. Crit. Care Med. 2021, 203, 136–139. [Google Scholar] [CrossRef]
- Tomassetti, S.; Ravaglia, C.; Wells, A.U.; Cavazza, A.; Colby, T.V.; Rossi, G.; Ley, B.; Ryu, J.H.; Puglisi, S.; Arcadu, A.; et al. Prognostic value of transbronchial lung cryobiopsy for the multidisciplinary diagnosis of idiopathic pulmonary fibrosis: A retrospective validation study. Lancet Respir. Med. 2020, 8, 786–794. [Google Scholar] [CrossRef]
- Renzoni, E.A.; Poletti, V.; Mackintosh, J.A. Disease pathology in fibrotic interstitial lung disease: Is it all about usual interstitial pneumonia? Lancet 2021, 398, 1437–1449. [Google Scholar] [CrossRef]
- He, A.T.; Liu, J.; Li, F.; Yang, B.B. Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target. Ther. 2021, 6, 185. [Google Scholar] [CrossRef]
- Meganck, R.M.; Borchardt, E.K.; Rivera, R.M.; Scalabrino, M.L.; Wilusz, J.E.; Marzluff, W.F.; Asokan, A. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol. Ther.-Nucleic Acids 2018, 13, 89–98. [Google Scholar] [CrossRef]
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215. [Google Scholar] [CrossRef]
- Li, S.; Li, X.; Xue, W.; Zhang, L.; Yang, L.-Z.; Cao, S.-M.; Lei, Y.-N.; Liu, C.-X.; Guo, S.-K.; Shan, L.; et al. Screening for functional circular RNAs using the CRISPR–Cas13 system. Nat. Methods 2021, 18, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.-O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021, 22, 41. [Google Scholar] [CrossRef] [PubMed]
- Ligresti, G.; Pham, T.X.; Sanders, Y.Y. Circular RNA Methylation: A New Twist in Lung Fibrosis. Am. J. Respir. Cell Mol. Biol. 2022, 66, 471–472. [Google Scholar] [CrossRef]
- Wang, S.; Luo, W.; Huang, J.; Chen, M.; Ding, J.; Cheng, Y.; Zhang, W.; Fang, S.; Wang, J.; Chao, J. The Combined Effects of Circular RNA Methylation Promote Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2022, 66, 510–523. [Google Scholar] [CrossRef] [PubMed]
| Species | Model | Method | Criteria | Total circRNA | Upregulated | Downregulated | Reference |
|---|---|---|---|---|---|---|---|
| Mice | Mouse model of SiO2-induced silicosis | Microarray analysis | Fold change > 2.0; p < 0.05 | 120 | 73 | 47 | [65] |
| Mice | Mice model of BIPF | RNA sequencing | Fold-change ≥ 3.0 | 74 | - | - | [61] |
| Human | IPF Patients (GEO database [GSE102660]) | Bioinformatics analysis | |Fold change| > 1.5; −log10 p-value > 1.3 | 45 | 22 | 23 | [67] |
| Human | Silicosis patients | RNA sequencing | Fold change > 1.5; p < 0.05 | 243 | 139 | 104 | [64] |
| Human | IPF Patients (GEO database [GSE102660]) | Bioinformatics analysis | p < 0.05 | 316 | 200 | 116 | |
| Human | Peripheral blood samples of IPF patients | Microarray hybridization | |Fold change| ≥ 1.5; p ≤ 0.05 | 67 | 38 | 29 | [60,68] |
| Rats | Rat model of BIPF | RNA sequencing | Fold change > 2.0; p < 0.05 | 10 | 2 | 8 | [62] |
| Rats | Rat model of BIPF | Whole transcriptome sequencing | |Fold change| > 2; p ≤ 0.05 | 605 | 287 | 318 | [63] |
| CircRNA | Dysregulation | Role in PF | Target Gene; Related Molecular | Function | Model | Cell | Reference |
|---|---|---|---|---|---|---|---|
| Endothelial–mesenchymal transition | |||||||
| circHECTD1 | Up | Pro | HECTD1 | EndMT | Murine model of silicosis | MML1 | [69] |
| Macrophage activation | |||||||
| circHECTD1 | Down | Anti | HECTD1; ZC3H12A | MA | Murine model of silicosis | RAW264.7 | [65] |
| circZC3H4 | Up | Pro | ZC3H4 | MA | Silicosis patients | RAW264.7 | [70] |
| Epithelial–mesenchymal transition | |||||||
| hsa_circ_0044226 | Up | Pro | CDC27 | EMT | Murine model of BIPF | RLE-6TN | [67] |
| circZC3H4 | Up | Pro | miR-212; ZC3H4 | EMT | Murine model of silicosis | MLE-12, A549, BEAS-2B | [71] |
| ciRS-7 | Up | Pro | miR-7; TGFBR2 | EMT | Murine model of silicosis | HBE, A549; MRC-5, NIH/3T3 | [72] |
| mmu_circ_0000981 | Up | Pro | miR-211-5p; TGFBR2 | EMT | Murine model of asthma | TC-1 | [73] |
| Fibroblast-to-myofibroblast transition | |||||||
| circHIPK3 | Up | Pro | miR-338-3p; SOX4 and COL1A1 | FMT | Murine model of BIPF | WI-38 | [74] |
| circ0044226 | Up | Pro | miR-7; sp1 | FMT | Murine model of BIPF | WI-38 | [75] |
| circANKRD42 | Up | Pro | miR-324-5p, miR-136-5p; AJUBA, YAP1 | FMT | Murine model of BIPF and IPF patients | MRC-5 | [68] |
| Fibroblast activation | |||||||
| circHECTD1 | Down | Anti | HECTD1 | FA | Murine model of silicosis | HPF-α | [76] |
| ciR-012091 | Down | Anti | PPP1R13B | FA | Murine model of silicosis | L929 and HPF-α | [77] |
| circ949 and circ057 | Up | Pro | miR-29b-2-5p; STAT3 phosphorylation | FA | Murine model of BIPF | L929 | [61] |
| circTADA2A | Down | Anti | miR-526b, miR-203; Caveolin-1, Caveolin-2 | FA | Murine model of BIPF | Fibroblasts | [78] |
| circHIPK3 | Up | Pro | miR-30a-3p; FOXK2 | FA | Murine model of silicosis | MRC-5 | [74] |
| circHIPK2 | Up | Pro | σ-1R | FA | NA | HPF-α | [79] |
| circRNA 0026344 | Down | Anti | miR-21; Smad7 | FA | Murine model of cigarette smoke-induced PF | HBE, MRC-5 | [80] |
| circ_406961 | Down | Anti | ILF2; STAT3, MAPK8, JNK | Airway inflammation | NA | BEAS-2B | [81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Chen, Y.; He, M.; Li, X.; Wang, R. Role of Circular RNAs in Pulmonary Fibrosis. Int. J. Mol. Sci. 2022, 23, 10493. https://doi.org/10.3390/ijms231810493
Zhou J, Chen Y, He M, Li X, Wang R. Role of Circular RNAs in Pulmonary Fibrosis. International Journal of Molecular Sciences. 2022; 23(18):10493. https://doi.org/10.3390/ijms231810493
Chicago/Turabian StyleZhou, Jian, Yali Chen, Menglin He, Xuehan Li, and Rurong Wang. 2022. "Role of Circular RNAs in Pulmonary Fibrosis" International Journal of Molecular Sciences 23, no. 18: 10493. https://doi.org/10.3390/ijms231810493
APA StyleZhou, J., Chen, Y., He, M., Li, X., & Wang, R. (2022). Role of Circular RNAs in Pulmonary Fibrosis. International Journal of Molecular Sciences, 23(18), 10493. https://doi.org/10.3390/ijms231810493






