Coevolution of Rumen Epithelial circRNAs with Their Microbiota and Metabolites in Response to Cold-Season Nutritional Stress in Tibetan Sheep
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Design and Sample Collection
2.3. Determination of VFAs, Microbiota Structure, and Differential Metabolites in Rumen
2.4. RNA Isolation, Library Preparation, and circRNA Sequencing
2.5. Identification of circRNAs
2.6. Identification of Differentially Expressed circRNAs
2.7. KEGG and GO Analysis of the Source Genes of DE circRNAs
2.8. CircRNA Targeted miRNA Loci Analysis
2.9. RT-qPCR Analysis
2.10. Statistical Analysis
3. Results
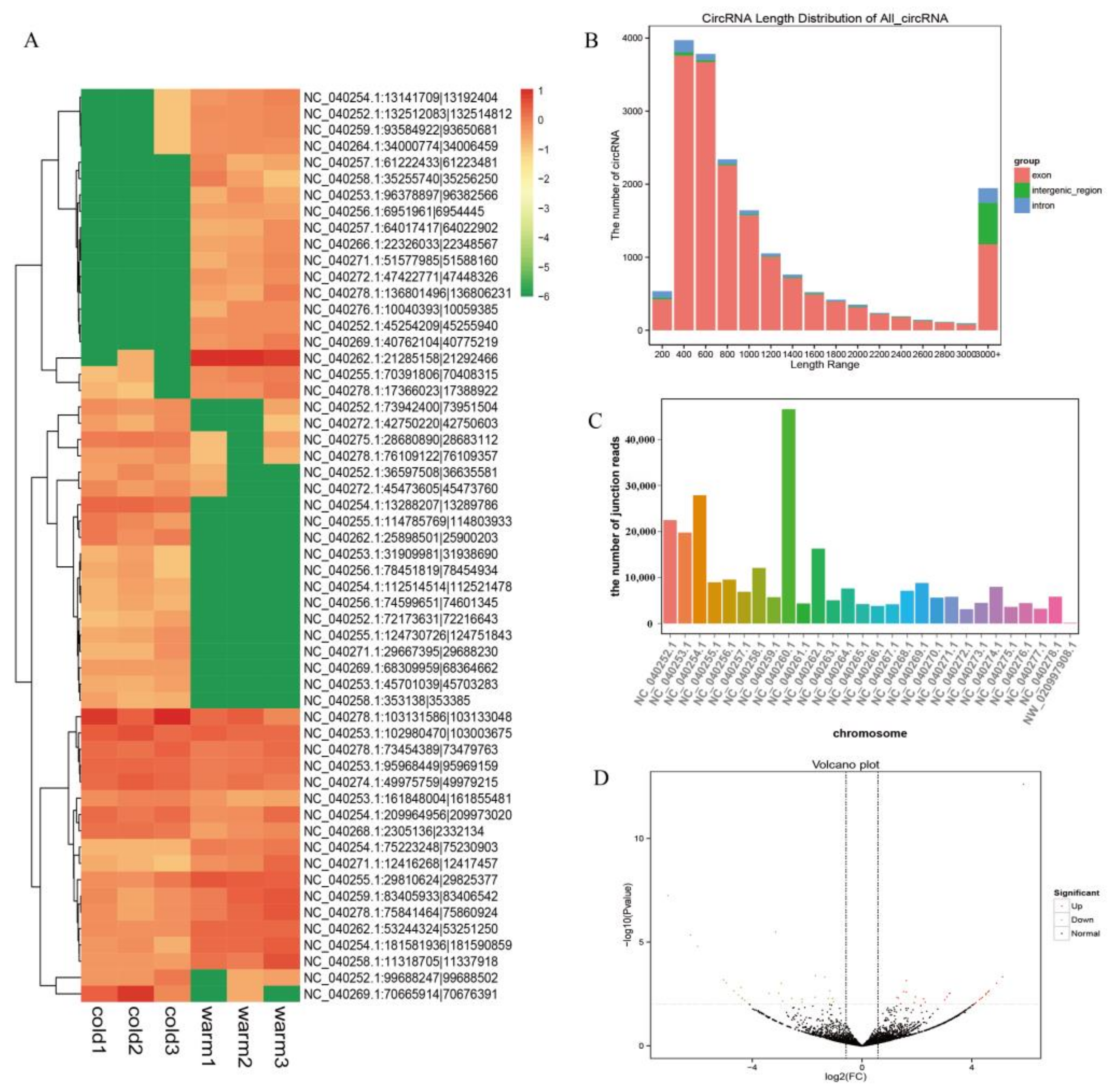
3.1. Identification of Rumen Epithelium circRNAs
3.2. Analysis and Validation of DE circRNAs
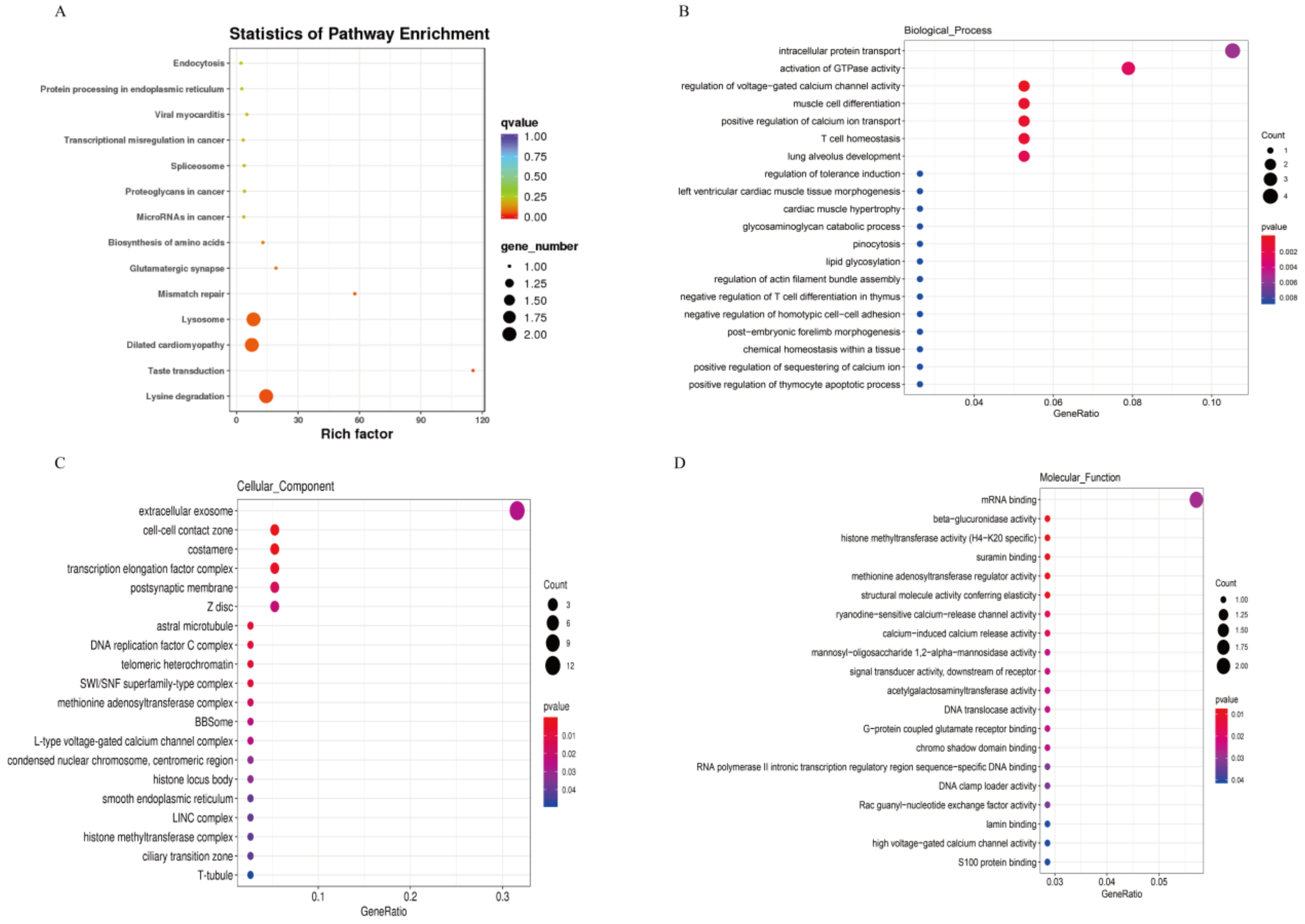
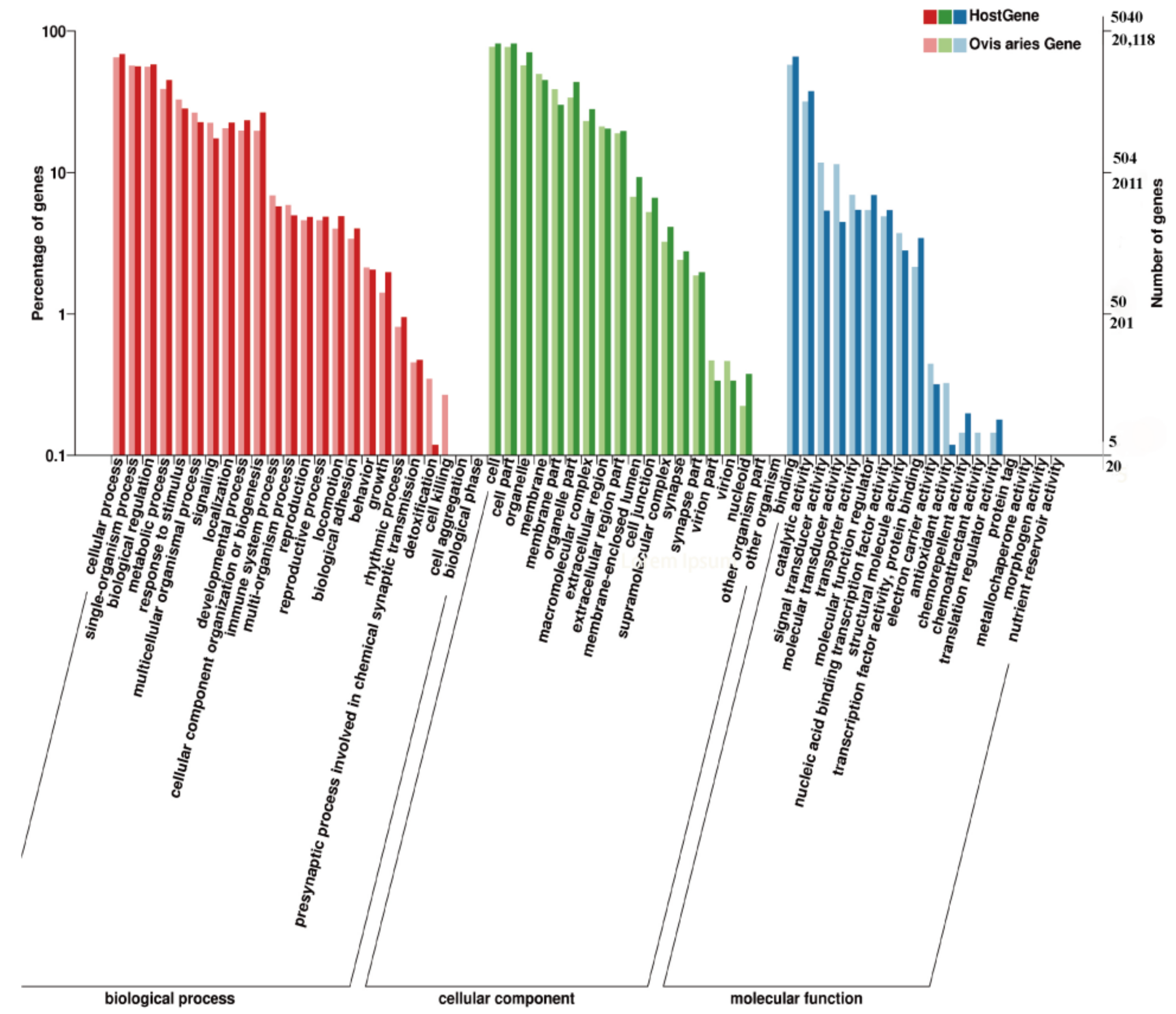
3.3. KEGG Pathway and GO Enrichment for Source Genes of DE circRNAs
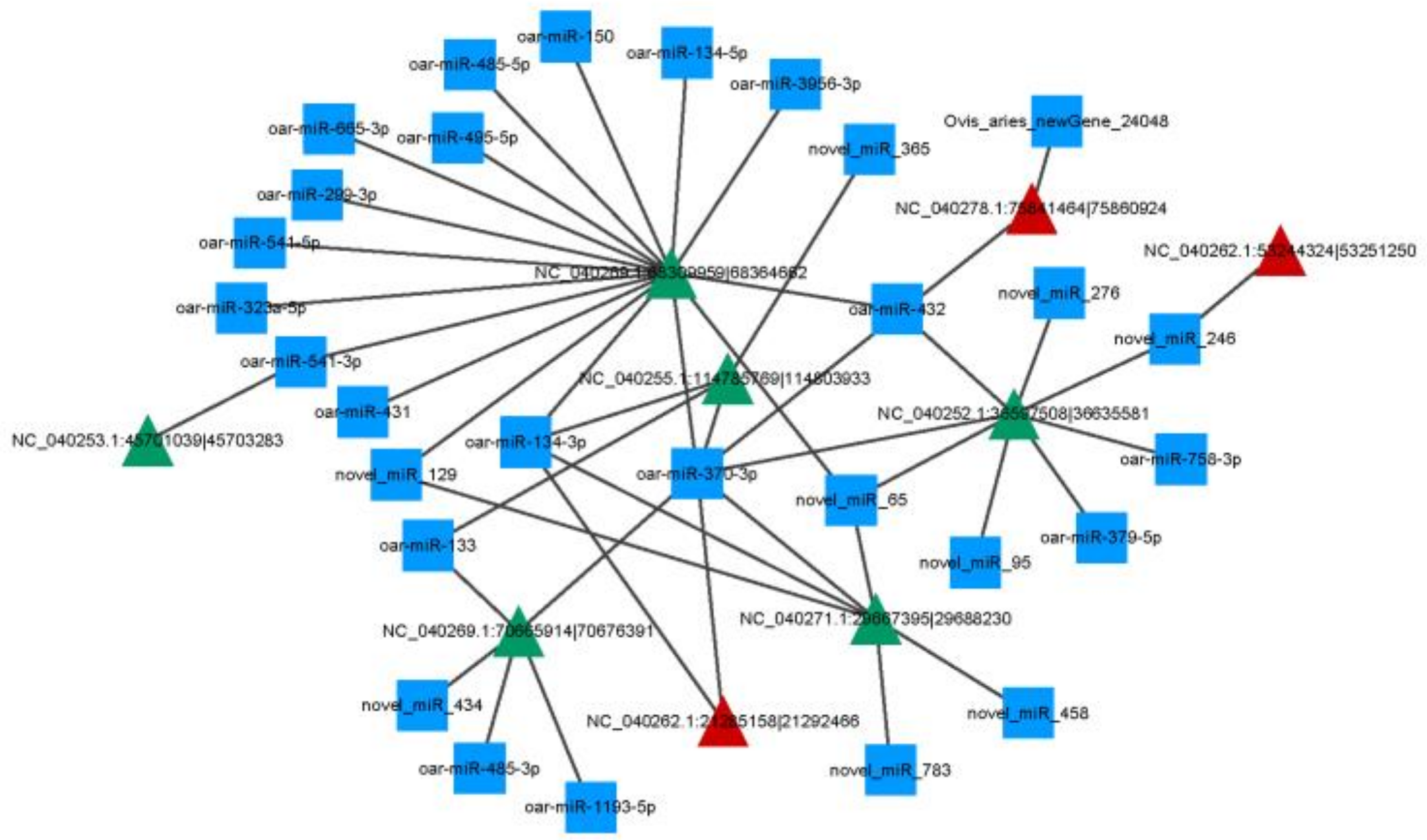
3.4. Construction of circRNA-miRNA Regulatory Network
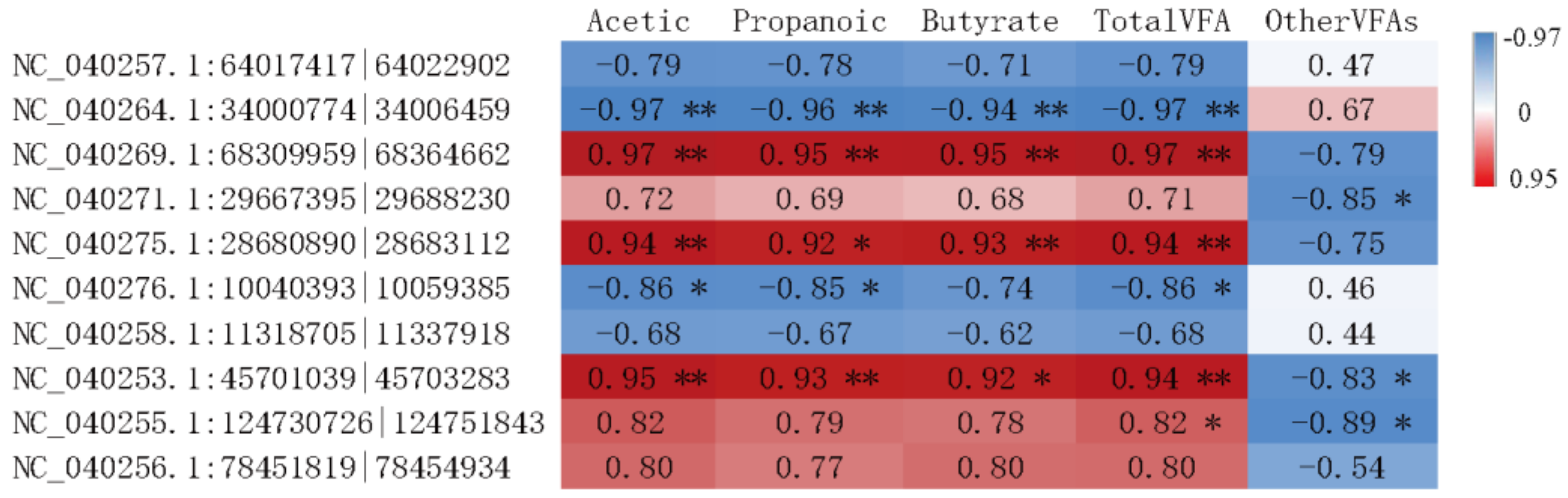
3.5. Interaction Analysis of circRNAs in the Rumen Epithelium and VFAs
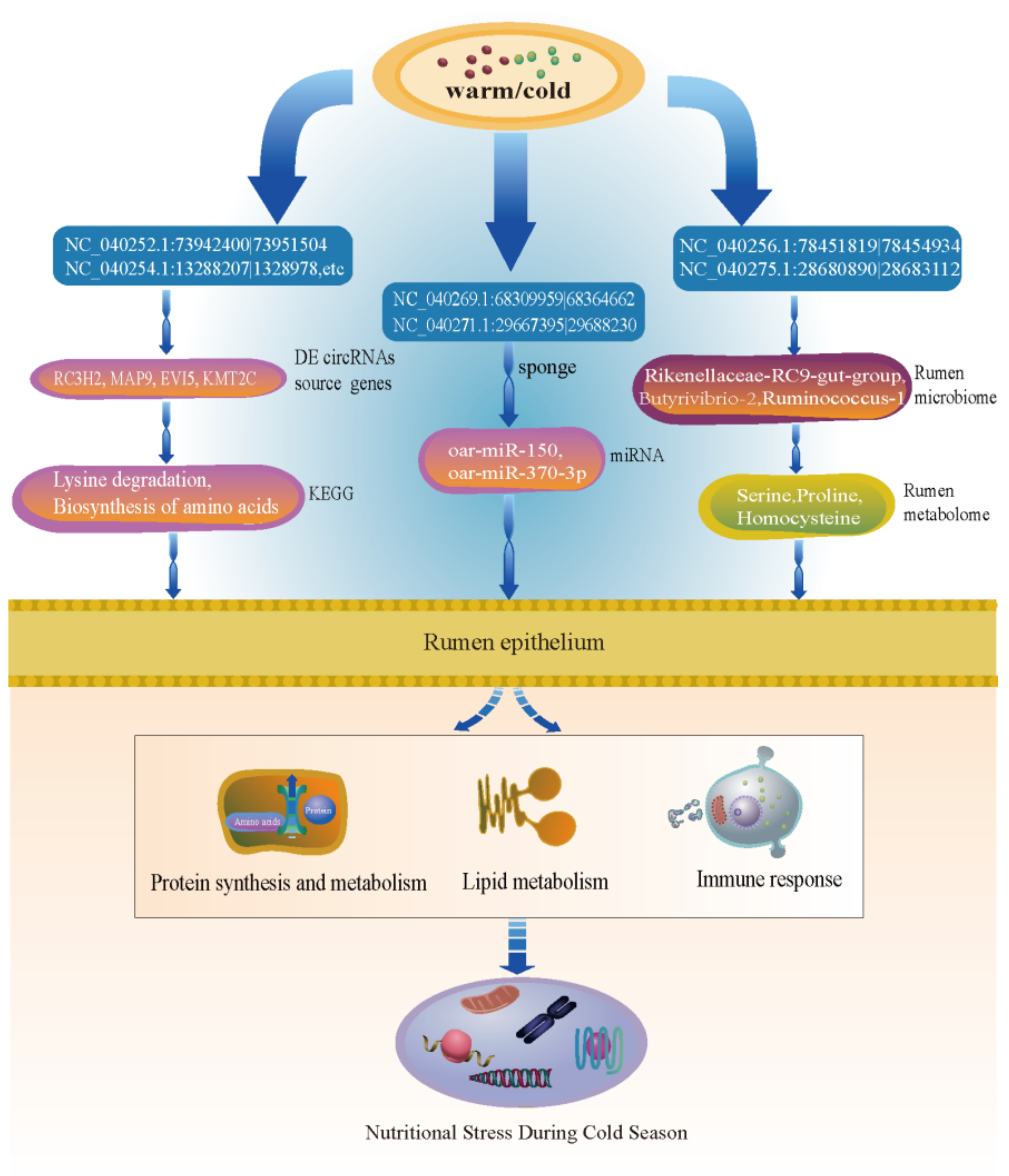
3.6. Combined Analysis of the Rumen Epithelium circRNA-Microbiome-Metabolome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, T.; Zhao, N.; Hu, L.; Xu, S.; Liu, H.; Ma, L.; Zhao, X. Characterizing CH4, CO2 and N2O emission from barn feeding Tibetan sheep in Tibetan alpine pastoral area in cold season. Atmos. Environ. 2017, 157, 84–90. [Google Scholar] [CrossRef]
- Guo, P.; Gao, P.; Li, F.; Chang, S.; Wang, Z.; Yan, T.; Hou, F. Prediction of Metabolizable Energy Concentrations of Herbage in the Qinghai–Tibetan Plateau Using Tibetan Sheep Digestibility Data. Animals 2020, 10, 376. [Google Scholar] [CrossRef]
- Long, R.J.; Apori, S.O.; Castro, F.B.; Ørskov, E.R. Feed value of native forages of the Tibetan Plateau of China. Anim. Feed. Sci. Technol. 1999, 80, 101–113. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Dingkao, R.; Zhang, W.; Lv, W.; Wei, H.; Shi, H.; Hu, J.; Wang, J.; Li, S.; et al. Interactions Between Rumen Microbes, VFAs, and Host Genes Regulate Nutrient Absorption and Epithelial Barrier Function During Cold Season Nutritional Stress in Tibetan Sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent Evolution of Rumen Microbiomes in High-Altitude Mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Li, T.; Luo, R.; Wang, X.; Wang, H.; Zhao, X.; Guo, Y.; Jiang, H.; Ma, Y. Unraveling Stage-Dependent Expression Patterns of Circular RNAs and Their Related ceRNA Modulation in Ovine Postnatal Testis Development. Front. Cell Dev. Biol. 2021, 9, 627439. [Google Scholar] [CrossRef]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 1–11. [Google Scholar] [CrossRef]
- Li, Z.-X.; Chen, W.; Qin, M.; Wang, L.-X.; Zeng, Y.-Q. Characteristics of circRNAs expression profiles in the piglets intestine induced by oxidative stress. Genes Genom. 2021, 44, 425–433. [Google Scholar] [CrossRef]
- Zhang, X.; Zhan, S.; Yang, S.; Zhong, T.; Guo, J.; Cao, J.; Wang, Y.; Li, L.; Zhang, H.; Wang, L. Dynamic Expression Profiles of Circular RNAs during Brown to White Adipose Tissue Transformation in Goats (Capra hircus). Animals 2021, 11, 1351. [Google Scholar] [CrossRef]
- Li, B.; Li, Y.; Li, L.; Yu, Y.; Gu, X.; Liu, C.; Long, X.; Yu, Y.; Zuo, X. Hsa_circ_0001021 regulates intestinal epithelial barrier function via sponging miR-224-5p in ulcerative colitis. Epigenomics 2021, 13, 1385–1401. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Ma, X.-X.; Luo, J.; Chung, H.K.; Kwon, M.S.; Yu, T.-X.; Rao, J.N.; Kozar, R.; Gorospe, M.; Wang, J.-Y. Circular RNA CircHIPK3 Promotes Homeostasis of the Intestinal Epithelium by Reducing MicroRNA 29b Function. Gastroenterology 2021, 161, 1303–1317. [Google Scholar] [CrossRef]
- Okholm, T.L.H.; Sathe, S.; Park, S.S.; Kamstrup, A.B.; Rasmussen, A.M.; Shankar, A.; Chua, Z.M.; Fristrup, N.; Nielsen, M.M.; Vang, S.; et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liao, Y.; Lou, J.; Zhuang, M.; Yan, H.; Li, Q.; Deng, Y.; Xu, X.; Wen, D.; Sun, Y. CircRNA_Maml2 promotes the proliferation and migration of intestinal epithelial cells after severe burns by regulating the miR-93-3p/FZD7/Wnt/beta-catenin pathway. Burn. Trauma 2022, 10, c9. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Lv, W.; Cao, G.; Guo, X.; Pu, X.; Wang, J.; Li, S.; Hu, J.; Luo, Y. Multi-Omics Reveals That the Rumen Transcriptome, Microbiome, and Its Metabolome Co-regulate Cold Season Adaptability of Tibetan Sheep. Front. Microbiol. 2022, 13, 859601. [Google Scholar] [CrossRef] [PubMed]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H.; et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Liu, L.; Lin, L.; Tang, H.; Fan, X.; Lin, H.; Li, X. Cecal CircRNAs Are Associated With the Response to Salmonella Enterica Serovar Enteritidis Inoculation in the Chicken. Front. Immunol. 2019, 10, 1186. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Bao, G.; Zhao, F.; Wang, J.; Liu, X.; Hu, J.; Shi, B.; Wen, Y.; Zhao, L.; Luo, Y.; Li, S. Characterization of the circRNA–miRNA–mRNA Network to Reveal the Potential Functional ceRNAs Associated With Dynamic Changes in the Meat Quality of the Longissimus Thoracis Muscle in Tibetan Sheep at Different Growth Stages. Front. Veter. Sci. 2022, 9, 803758. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Betel, D.; Wilson, M.; Gabow, A.; Marks, D.S.; Sander, C. The microRNA.org resource: Targets and expression. Nucleic Acids Res. 2008, 36, D149–D153. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, R.L.; Connor, E.E. Rumen Function and Development. Vet. Clin. Food Anim. Pract. 2017, 33, 427–439. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 2018, 9, e1478. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chen, J.; Qin, Y.; Wang, J.; Zhou, L. Mutations in voltage-gated L-type calcium channel: Implications in cardiac arrhythmia. Channels 2018, 12, 201–218. [Google Scholar] [CrossRef]
- Schröder, B.; Vössing, S.; Breves, G. In vitro studies on active calcium absorption from ovine rumen. J. Comp. Physiol. B 1999, 169, 487–494. [Google Scholar] [CrossRef]
- Lai, N.; Lu, W.; Wang, J. Ca(2+) and ion channels in hypoxia-mediated pulmonary hypertension. Int. J. Clin. Exp. Pathol. 2015, 8, 1081–1092. [Google Scholar]
- Del Rizzo, P.A.; Trievel, R.C. Molecular basis for substrate recognition by lysine methyltransferases and demethylases. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 1404–1415. [Google Scholar] [CrossRef]
- Assiry, A.A.; AlBalawi, A.M.; Zafar, M.S.; Khan, S.D.; Ullah, A.; AlMatrafi, A.; Ramzan, K.; Basit, S. KMT2C, a histone methyltransferase, is mutated in a family segregating non-syndromic primary failure of tooth eruption. Sci. Rep. 2019, 9, 16410–16469. [Google Scholar] [CrossRef]
- Corvalan, A.Z.; Coller, H.A. Methylation of histone 4’s lysine 20: A critical analysis of the state of the field. Physiol. Genom. 2021, 53, 22–32. [Google Scholar] [CrossRef]
- Xie, K.; Peng, Y.; Zhong, W.; Liu, X. KMT2C is a Potential Biomarker of Anti-PD-1 Treatment Response in Metastatic Melanoma. Front. Biosci. 2022, 27, 103. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, R.; Russell, R.G.; Goldman, I. Localization of the murine reduced folate carrier as assessed by immunohistochemical analysis. Biochim. Biophys. Acta Biomembr. 2001, 1513, 49–54. [Google Scholar] [CrossRef]
- Sirotnak, F.M.; Tolner, B. Carrier-mediated membrane transport of folates in mammalian cells. Annu. Rev. Nutr. 1999, 19, 91–122. [Google Scholar] [CrossRef]
- Large, P. Folates and pterins: Volume 3 nutritional, pharmacological and physiological aspects. FEBS Lett. 1988, 228, 203. [Google Scholar] [CrossRef]
- Hamid, A.; Wani, N.A.; Rana, S.; Vaiphei, K.; Mahmood, A.; Kaur, J. Down-regulation of reduced folate carrier may result in folate malabsorption across intestinal brush border membrane during experimental alcoholism. FEBS J. 2007, 274, 6317–6328. [Google Scholar] [CrossRef] [PubMed]
- Ragaller, V.; Hüther, L.; Lebzien, P. Folic acid in ruminant nutrition: A review. Br. J. Nutr. 2008, 101, 153–164. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ramkumar, A.; Jong, B.Y.; Ori-McKenney, K.M. ReMAPping the microtubule landscape: How phosphorylation dictates the activities of microtubule-associated proteins. Dev. Dyn. 2017, 247, 138–155. [Google Scholar] [CrossRef] [PubMed]
- Faitar, S.L.; Dabbeekeh, J.T.; Ranalli, T.A.; Cowell, J.K. EVI5 is a novel centrosomal protein that binds to α- and γ-tubulin. Genomics 2005, 86, 594–605. [Google Scholar] [CrossRef]
- Stathatos, G.G.; Dunleavy, J.E.; Zenker, J.; O’Bryan, M.K. Delta and epsilon tubulin in mammalian development. Trends Cell Biol. 2021, 31, 774–787. [Google Scholar] [CrossRef]
- Athanasopoulos, V.; Ramiscal, R.R.; Vinuesa, C.G. ROQUIN signalling pathways in innate and adaptive immunity. Eur. J. Immunol. 2016, 46, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Dou, C.; Cao, Z.; Yang, B.; Ding, N.; Hou, T.; Luo, F.; Kang, F.; Li, J.; Yang, X.; Jiang, H.; et al. Changing expression profiles of lncRNAs, mRNAs, circRNAs and miRNAs during osteoclastogenesis. Sci. Rep. 2016, 6, 21499. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Ding, Y.; Tian, Y.; Lei, X. Circular RNA circ_0010283 regulates the viability and migration of oxidized low-density lipoprotein-induced vascular smooth muscle cells via an miR-370-3p/HMGB1 axis in atherosclerosis. Int. J. Mol. Med. 2020, 46, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Ren, K.; Li, B.; Jiang, L.; Liu, Z.; Wu, F.; Zhang, Y.; Liu, J.; Duan, W. circ_0023461 Silencing Protects Cardiomyocytes from Hypoxia-Induced Dysfunction through Targeting miR-370-3p/PDE4D Signaling. Oxid. Med. Cell. Longev. 2021, 2021, 8379962. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.; Zhang, S.; Wu, S.; Xiao, Q.; Gu, Y.; Guo, X.; Lin, X.; Chen, L.; Zhao, Y.; et al. miR-370-3p Regulates Adipogenesis through Targeting Mknk1. Molecules 2021, 26, 6926. [Google Scholar] [CrossRef]
- Garvey, W.T.; Luo, N.; Wang, D.-Z.; Fu, Y. MicroRNA-150 Regulates Lipid Metabolism and Inflammatory Response. J. Metab. Syndr. 2014, 3, 1–7. [Google Scholar] [CrossRef]
- Koskella, B.; Bergelson, J. The study of host–microbiome (co)evolution across levels of selection. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190604. [Google Scholar] [CrossRef]
- Wilson, A.C.C.; Duncan, R.P. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. USA 2015, 112, 10255–10261. [Google Scholar] [CrossRef]
- Hassan, M.I.; Waheed, A.; Grubb, J.H.; Klei, H.E.; Korolev, S.; William, S.S. High Resolution Crystal Structure of Human β-Glucuronidase Reveals Structural Basis of Lysosome Targeting. PLoS ONE 2013, 8, e79687. [Google Scholar] [CrossRef]
- Gloux, K.; Berteau, O.; El Oumami, H.; Béguet, F.; Leclerc, M.; Doré, J. A metagenomic -glucuronidase uncovers a core adaptive function of the human intestinal microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4539–4546. [Google Scholar] [CrossRef]
- Sicard, J.-F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic Analysis of the Human Distal Gut Microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Chen, X.; Wu, W.; Wang, W.; Pang, W.; Yang, G. MAT2B promotes adipogenesis by modulating SAMe levels and activating AKT/ERK pathway during porcine intramuscular preadipocyte differentiation. Exp. Cell Res. 2016, 344, 11–21. [Google Scholar] [CrossRef]
- Grillo, M.A.; Colombatto, S. S-adenosylmethionine and its products. Amino Acids 2008, 34, 187–193. [Google Scholar] [CrossRef]
- Zhou, X.; He, L.; Wu, C.; Zhang, Y.; Wu, X.; Yin, Y. Serine alleviates oxidative stress via supporting glutathione synthesis and methionine cycle in mice. Mol. Nutr. Food Res. 2017, 61, 1700262. [Google Scholar] [CrossRef] [PubMed]
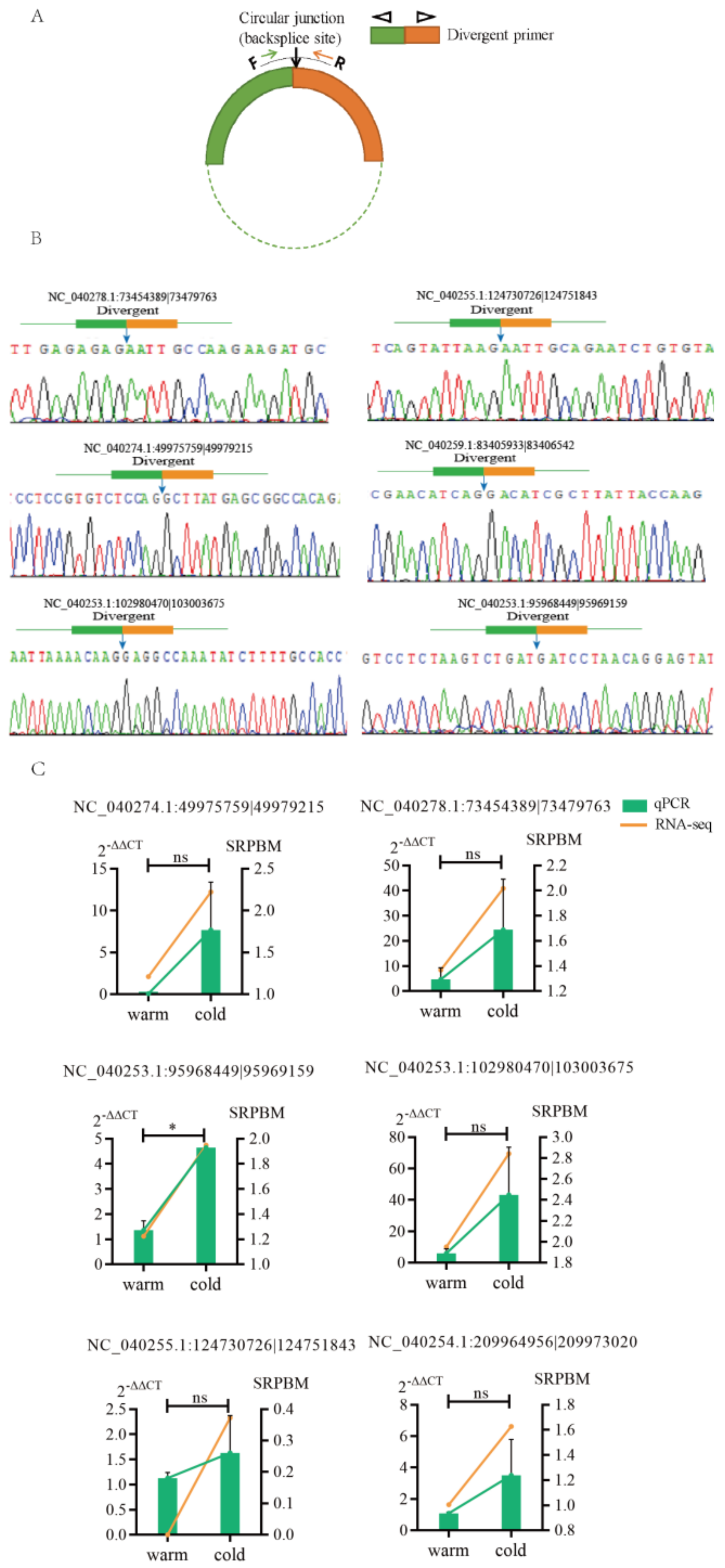

| CircRNA | Forward (5′→3′) | Reverse (5′→3′) |
|---|---|---|
| NC_040278.1:73454389|73479763 | TAAGAACAGAAACACAAAATGCTC | CTTCCTCATCTCCAGGGTTT |
| NC_040255.1:124730726|124751843 | AAGTTCACTCTGCATCAGG | ACTTCCCACAGAAAGGAC |
| NC_040274.1:49975759|49979215 | CAGTGATGTGAGTGCGAGTA | GTGTCACTGGTGAAATCCTGT |
| NC_040259.1:83405933|83406542 | ATGTGATCAGCGAGAAGCAG | CAGCATCGGTCTGTAACCTC |
| NC_040253.1:102980470|103003675 | AAACGCAGGAACAGCTAGAG | CGCCACAGTCAGGTCTAATC |
| NC_040253.1:95968449|95969159 | AGGTCAAAATTGAAAGTGAGGC | TCATTAATTTGTGAGGCTTGGAAT |
| β-actin | AGCCTTCCTTCCTGGGCATGGA | GGACAGCACCGTGTTGGCGTAGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Sha, Y.; Pu, X.; Xu, Y.; Yao, L.; Liu, X.; He, Y.; Hu, J.; Wang, J.; Li, S.; et al. Coevolution of Rumen Epithelial circRNAs with Their Microbiota and Metabolites in Response to Cold-Season Nutritional Stress in Tibetan Sheep. Int. J. Mol. Sci. 2022, 23, 10488. https://doi.org/10.3390/ijms231810488
Guo X, Sha Y, Pu X, Xu Y, Yao L, Liu X, He Y, Hu J, Wang J, Li S, et al. Coevolution of Rumen Epithelial circRNAs with Their Microbiota and Metabolites in Response to Cold-Season Nutritional Stress in Tibetan Sheep. International Journal of Molecular Sciences. 2022; 23(18):10488. https://doi.org/10.3390/ijms231810488
Chicago/Turabian StyleGuo, Xinyu, Yuzhu Sha, Xiaoning Pu, Ying Xu, Liangwei Yao, Xiu Liu, Yanyu He, Jiang Hu, Jiqing Wang, Shaobin Li, and et al. 2022. "Coevolution of Rumen Epithelial circRNAs with Their Microbiota and Metabolites in Response to Cold-Season Nutritional Stress in Tibetan Sheep" International Journal of Molecular Sciences 23, no. 18: 10488. https://doi.org/10.3390/ijms231810488
APA StyleGuo, X., Sha, Y., Pu, X., Xu, Y., Yao, L., Liu, X., He, Y., Hu, J., Wang, J., Li, S., & Chen, G. (2022). Coevolution of Rumen Epithelial circRNAs with Their Microbiota and Metabolites in Response to Cold-Season Nutritional Stress in Tibetan Sheep. International Journal of Molecular Sciences, 23(18), 10488. https://doi.org/10.3390/ijms231810488






