TIAM2 Contributes to Osimertinib Resistance, Cell Motility, and Tumor-Associated Macrophage M2-like Polarization in Lung Adenocarcinoma
Abstract
:1. Introduction
2. Results
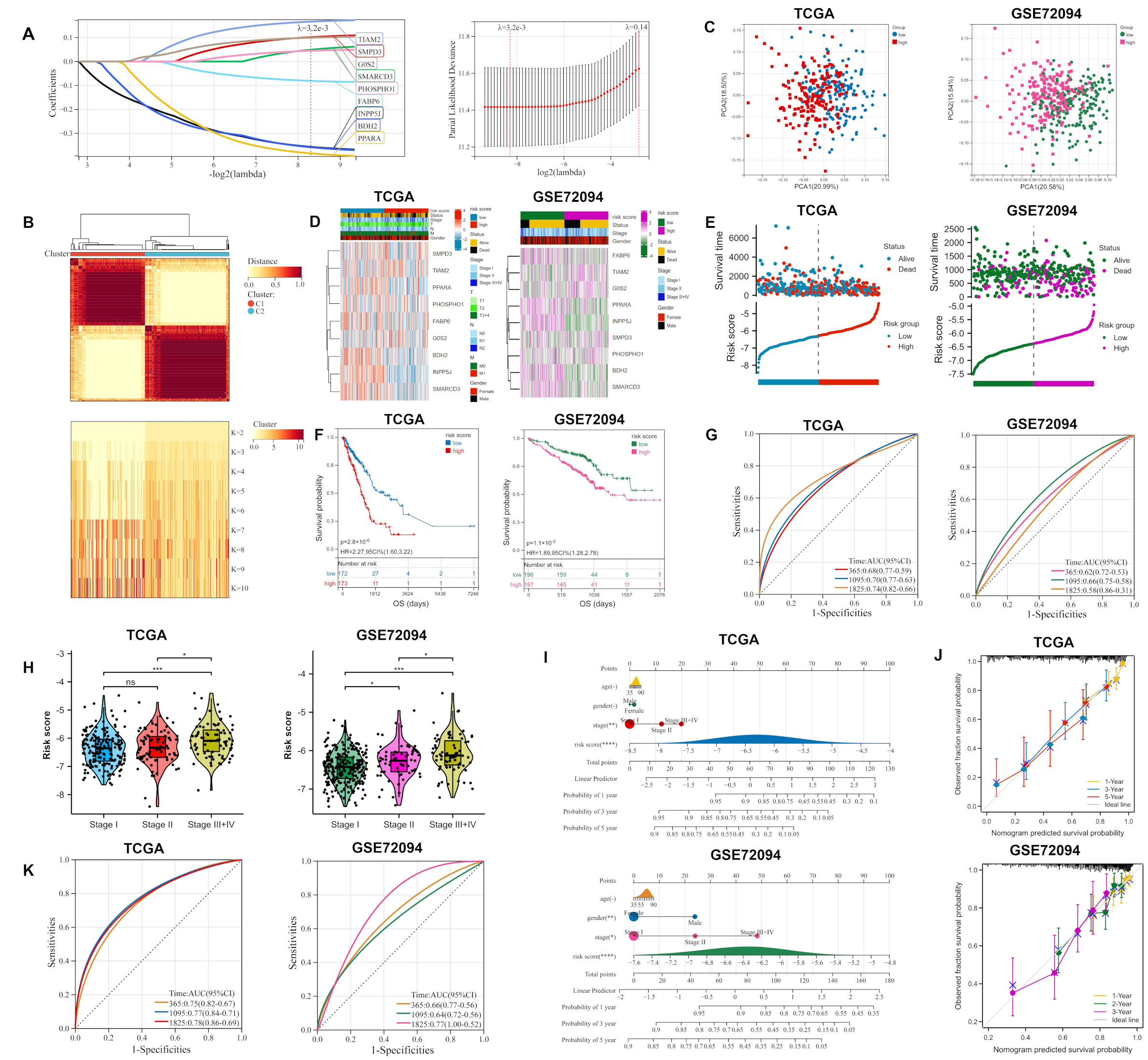
2.1. Lipid Metabolism-Related Genes Were Selected as Candidates for ORi
2.2. Construction and Validation of Ori
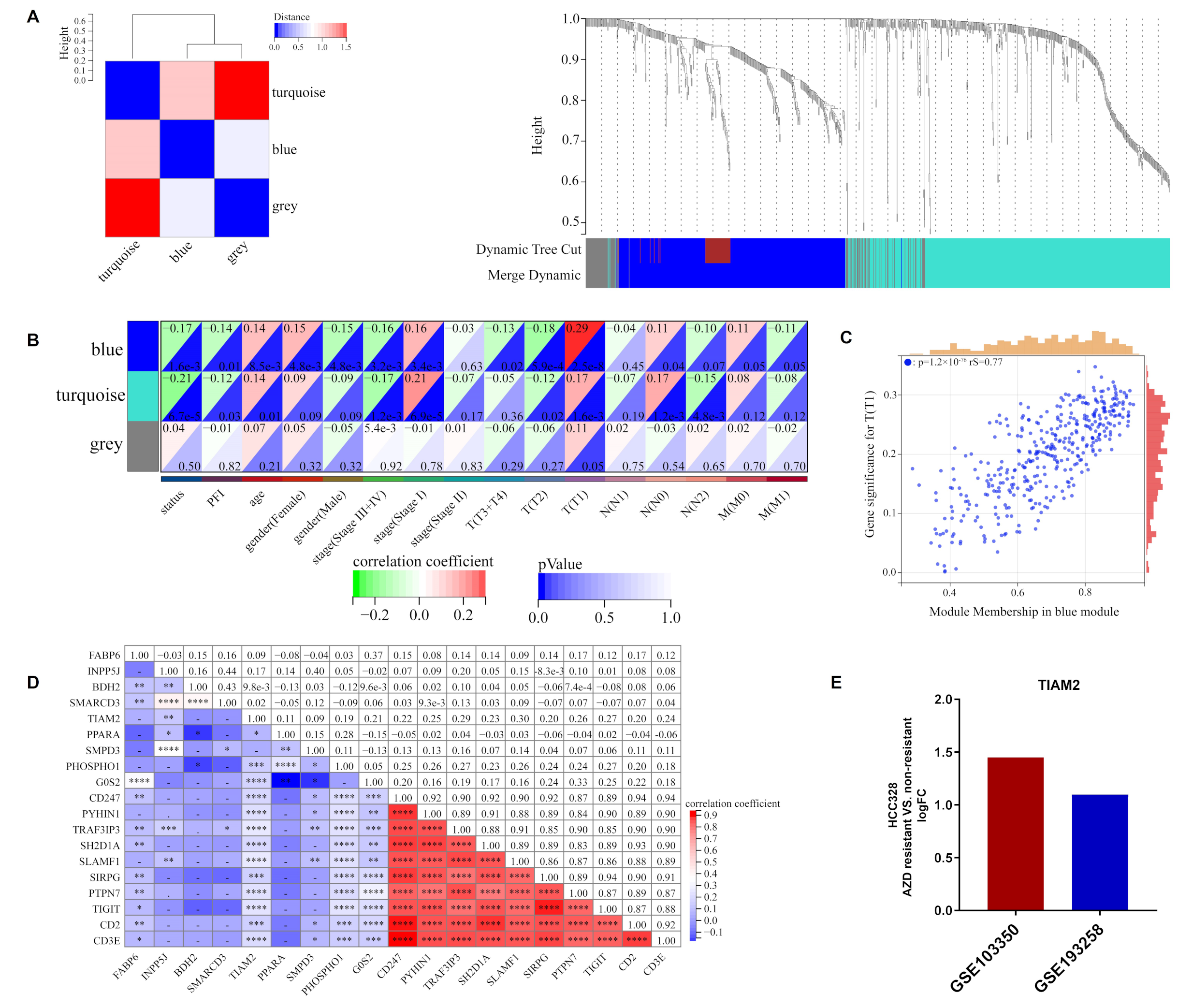
2.3. ORi Was Associated with M2-like TAM Infiltration
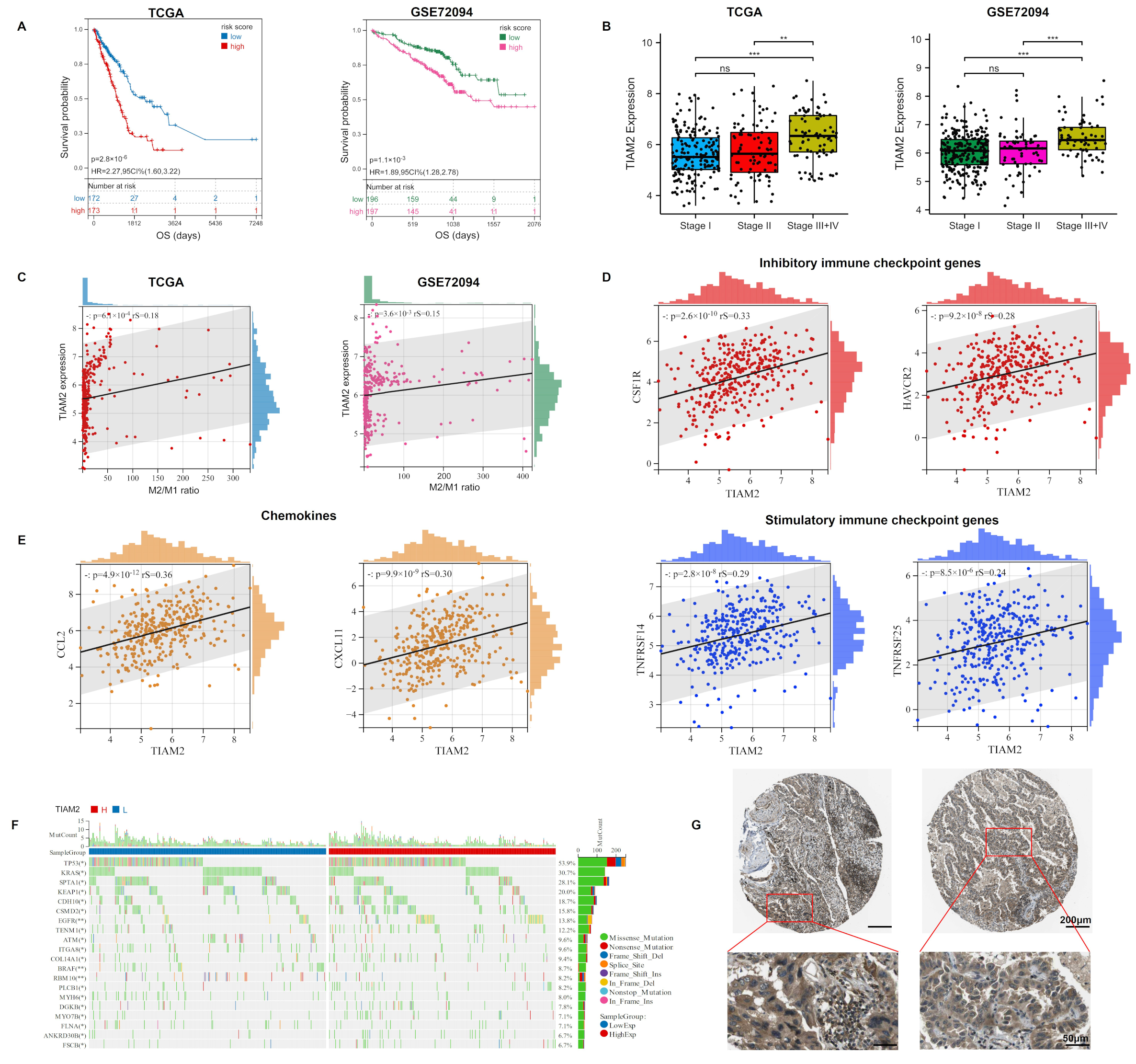
2.4. TIAM2 Involved in Immune Infiltration and Osimertinib Resistance
2.5. TIAM2 Correlated with Prognostic, Immunity, Mutational, and Pathological Changes in LUAD Patients
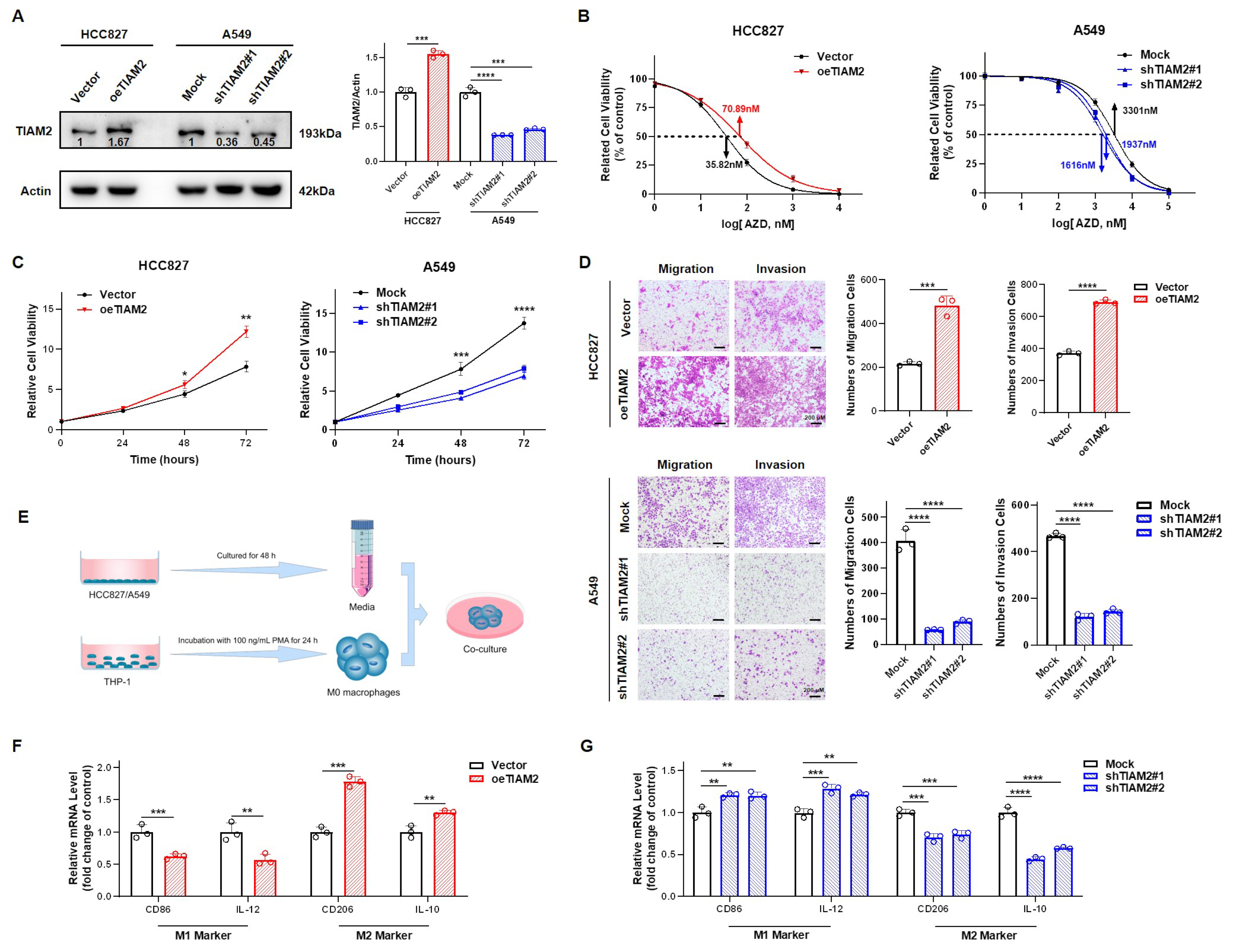
2.6. TIAM2 Contributed to Osimertinib Resistance, Cell Motility, and M2-like TAM Polarization in LUAD
3. Discussion
4. Materials and Methods
4.1. Collection of Public Data
4.2. Enrichment Analysis
4.3. Protein-Protein Interactions (PPI) Networks and Correlation Analysis
4.4. Construction and Validation of Prognostic Osimertinib Sensitive Risk Model
4.5. Immune Infiltration Analysis
4.6. Weighted Gene Correlation Network Analysis (WGCNA)
4.7. Genome Mutation and Human Protein Atlas (HPA)
4.8. Cell Culture, Plasmids, and shRNAs
4.9. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
4.10. Western Blot (WB)
4.11. Cell Counting Kit 8 (CCK-8) Assay
4.12. Transwell Assay
4.13. Differentiation of THP-1 and Conditioned Medium Treatment
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LC | Lung cancer |
| NSCLC | non-small cell lung cancer |
| LUAD | lung adenocarcinoma |
| ORi | osimertinib resistance index |
| EGFR | epidermal growth factor receptor |
| TKIs | tyrosine kinase inhibitors |
| EMT | epithelial–mesenchymal transition |
| TME | the tumor microenvironment |
| TAMs | tumor-associated macrophages |
| DEGs | differentially expressed genes |
| WGCNA | weighted gene co-expression network analysis |
| GDSC | Drug Sensitivity in Cancer |
| IC50 | half-maximal inhibitory concentration |
| GEO | Gene Expression Omnibus database |
| GO | Gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | protein–protein interactions |
| LASSO | logistic least absolute shrinkage and selection operator |
| TCGA | The Cancer Genome Atlas |
| PCA | principal component analysis |
| t-SNE | T-distribution stochastic neighbor embedding |
| ROC | The receiver operating characteristic |
| AUC | area under the curve |
| KM | Kaplan–Meier |
| OS | overall survival |
| HR | hazard rate |
| CI | confidence interval |
| BP | biological processes |
| GSEA | Gene Set Enrichment Analysis |
| NF-kb | nuclear factor-Kb |
| CC | cellular composition |
| GEFs | guanine nucleotide exchange factors |
| HPA | Human Protein Atlas |
| WB | Western blot |
| RT-qPCR | Real-Time Quantitative PCR |
| shRNA | short hairpin RNA |
| ATCC | American Type Culture Collection |
| cDNA | complement DNA |
| CCK-8 | Cell Counting Kit 8 |
| PMA | phorbol 12-myristate 13-acetate |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, T.; Sun, Y.; Liang, X. The long noncoding RNA LINC00483 promotes lung adenocarcinoma progression by sponging miR-204-3p. Cell. Mol. Biol. Lett. 2019, 24, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Yang, F.; Wang, Y.; Zhao, H.; Wei, F.; Zhang, P.; Ren, X. Significantly different immunoscores in lung adenocarcinoma and squamous cell carcinoma and a proposal for a new immune staging system. Oncoimmunology 2020, 9, 1828538. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Wang, X.; Zang, R.; Liu, C.; Zheng, S.; Li, H.; Sun, N.; He, J. Desmoglein-2 modulates tumor progression and osimertinib drug resistance through the EGFR/Src/PAK1 pathway in lung adenocarcinoma. Cancer Lett. 2020, 483, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zheng, R.; Baade, P.D.; Zhang, S.; Zeng, H.; Bray, F.; Jemal, A.; Yu, X.Q.; He, J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016, 66, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.A.; Hughes, B.G. Targeted therapy for non-small cell lung cancer: Current standards and the promise of the future. Transl. Lung Cancer Res. 2015, 4, 36–54. [Google Scholar] [CrossRef]
- Li, F.; Huang, Q.; Luster, T.A.; Hu, H.; Zhang, H.; Ng, W.-L.; Khodadadi-Jamayran, A.; Wang, W.; Chen, T.; Deng, J.; et al. In Vivo Epigenetic CRISPR Screen Identifies Asf1a as an Immunotherapeutic Target in Kras-Mutant Lung Adenocarcinoma. Cancer Discov. 2020, 10, 270–287. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, R.; Zeng, Y.; Li, Y.; Chen, Y.; Zhou, J.; Huang, J.A. ALCAP2 inhibits lung adenocarcinoma cell proliferation, migration and invasion via the ubiquitination of beta-catenin by upregulating the E3 ligase NEDD4L. Cell Death Dis. 2021, 12, 755. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Yang, J.C.; Lee, C.K.; Kurata, T.; Kim, D.W.; John, T.; Jänne, P.A. Osimertinib as First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 841–849. [Google Scholar] [CrossRef] [Green Version]
- Finlay, M.R.V.; Anderton, M.; Ashton, S.; Ballard, P.; Bethel, P.A.; Box, M.R.; Bradbury, R.H.; Brown, S.J.; Butterworth, S.; Campbell, A.; et al. Discovery of a Potent and Selective EGFR Inhibitor (AZD9291) of Both Sensitizing and T790M Resistance Mutations That Spares the Wild Type Form of the Receptor. J. Med. Chem. 2014, 57, 8249–8267. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Ramalingam, S.S. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Taniguchi, H.; Yamada, T.; Wang, R.; Tanimura, K.; Adachi, Y.; Nishiyama, A.; Tanimoto, A.; Takeuchi, S.; Araujo, L.H.; Boroni, M.; et al. AXL confers intrinsic resistance to osimertinib and advances the emergence of tolerant cells. Nat. Commun. 2019, 10, 259. [Google Scholar] [CrossRef]
- Ramalingam, S.S.; Vansteenkiste, J.; Planchard, D.; Cho, B.C.; Gray, J.E.; Ohe, Y.; Zhou, C.; Reungwetwattana, T.; Cheng, Y.; Chewaskulyong, B.; et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N. Engl. J. Med. 2020, 382, 41–50. [Google Scholar] [CrossRef]
- Song, H.; Liu, D.; Wang, L.; Liu, K.; Chen, C.; Wang, L.; Ren, Y.; Ju, B.; Zhong, F.; Jiang, X.; et al. Methyltransferase like 7B is a potential therapeutic target for reversing EGFR-TKIs resistance in lung adenocarcinoma. Mol. Cancer 2022, 21, 1–20. [Google Scholar] [CrossRef]
- Janne, P.A.; Yang, J.C.H.; Kim, D.W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ranson, M. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-H.; Lu, J.-J. Osimertinib resistance in non-small cell lung cancer: Mechanisms and therapeutic strategies. Cancer Lett. 2018, 420, 242–246. [Google Scholar] [CrossRef]
- Bian, X.; Liu, R.; Meng, Y.; Xing, D.; Xu, D.; Lu, Z. Lipid metabolism and cancer. J. Exp. Med. 2021, 218, e20201606. [Google Scholar] [CrossRef]
- Pan, Z.; Wang, K.; Wang, X.; Jia, Z.; Yang, Y.; Duan, Y.; Ding, X. Cholesterol promotes EGFR-TKIs resistance in NSCLC by inducing EGFR/Src/Erk/SP1 signaling-mediated ERRalpha re-expression. Mol. Cancer 2022, 21, 77. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, D.; Owonikoko, T.K.; Ramalingam, S.S.; Sun, S.-Y. Induction of SREBP1 degradation coupled with suppression of SREBP1-mediated lipogenesis impacts the response of EGFR mutant NSCLC cells to osimertinib. Oncogene 2021, 40, 6653–6665. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wang, Q.; Li, D.; Wei, X.; Jia, Y.; Zhang, Z.; Ai, B.; Cao, X.; Guo, T.; Liao, Y. Co-administration of 20(S)-protopanaxatriol (g-PPT) and EGFR-TKI overcomes EGFR-TKI resistance by decreasing SCD1 induced lipid accumulation in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Jin, H.; He, Y.; Zhao, P.; Hu, Y.; Tao, J.; Chen, J.; Huang, Y. Targeting lipid metabolism to overcome EMT-associated drug resistance via integrin beta3/FAK pathway and tumor-associated macrophage repolarization using legumain-activatable delivery. Theranostics 2019, 9, 265–278. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. 2016, 73, 377–392. [Google Scholar] [CrossRef]
- Zheng, X.; Weigert, A.; Reu, S.; Guenther, S.; Mansouri, S.; Bassaly, B.; Gattenlöhner, S.; Grimminger, F.; Pullamsetti, S.S.; Seeger, W.; et al. Spatial Density and Distribution of Tumor-Associated Macrophages Predict Survival in Non–Small Cell Lung Carcinoma. Cancer Res. 2020, 80, 4414–4425. [Google Scholar] [CrossRef]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Mehla, K.; Singh, P.K. Metabolic Regulation of Macrophage Polarization in Cancer. Trends Cancer 2019, 5, 822–834. [Google Scholar] [CrossRef]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreño, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, H.; Li, Y.; Zhang, Y.; Bian, Y.; Zeng, Y.; Qin, Z. S100A4 enhances protumor macrophage polarization by control of PPAR-gamma-dependent induction of fatty acid oxidation. J. Immunother. Cancer 2021, 9, e002548. [Google Scholar] [CrossRef]
- Wu, H.; Han, Y.; Sillke, Y.R.; Deng, H.; Siddiqui, S.; Treese, C.; Schmidt, F.; Friedrich, M.; Keye, J.; Wan, J.; et al. Lipid droplet-dependent fatty acid metabolism controls the immune suppressive phenotype of tumor-associated macrophages. EMBO Mol. Med. 2019, 11, e10698. [Google Scholar] [CrossRef]
- Kishore, U.; Bulla, R.; Madan, T. Editorial: Odyssey of Surfactant Proteins SP-A and SP-D: Innate Immune Surveillance Molecules. Front. Immunol. 2020, 11, 394. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, J.; Lu, D.; Xu, X. Targeting tumor-associated macrophages to synergize tumor immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 1–12. [Google Scholar] [CrossRef]
- Bianco, A.; Malapelle, U.; Rocco, D.; Perrotta, F.; Mazzarella, G. Targeting immune checkpoints in non small cell lung cancer. Curr. Opin. Pharmacol. 2018, 40, 46–50. [Google Scholar] [CrossRef]
- Martin, T.D.; Patel, R.S.; Cook, D.R.; Choi, M.Y.; Patil, A.; Liang, A.C.; Li, M.Z.; Haigis, K.M.; Elledge, S.J. The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 2021, 373, 1327–1335. [Google Scholar] [CrossRef]
- Ren, S.; Tian, Q.; Amar, N.; Yu, H.; Rivard, C.J.; Caldwell, C.; Ng, T.L.; Tu, M.; Liu, Y.; Gao, D.; et al. The immune checkpoint, HVEM may contribute to immune escape in non-small cell lung cancer lacking PD-L1 expression. Lung Cancer 2018, 125, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Qin, T.; Huang, D.; Liu, Z.; Zhang, X.; Jia, Y.; Xian, C.J.; Li, K. Tumor necrosis factor superfamily 15 promotes lymphatic metastasis via upregulation of vascular endothelial growth factor-C in a mouse model of lung cancer. Cancer Sci. 2018, 109, 2469–2478. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, R.; Tokunaga, R.; Zhang, W.U.; Naseem, M.; Puccini, A.; Berger, M.D.; Soni, S.; Lenz, H.J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—A target for novel cancer therapy. Cancer Treat Rev. 2018, 63, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef]
- Wu, S.-G.; Shih, J.-Y. Management of acquired resistance to EGFR TKI–targeted therapy in advanced non-small cell lung cancer. Mol. Cancer 2018, 17, 1–14. [Google Scholar] [CrossRef]
- Piotrowska, Z.; Isozaki, H.; Lennerz, J.K.; Gainor, J.F.; Lennes, I.T.; Zhu, V.W.; Marcoux, N.; Banwait, M.K.; Digumarthy, S.R.; Su, W.; et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer Discov. 2018, 8, 1529–1539. [Google Scholar] [CrossRef]
- Lin, C.-C.; Shih, J.-Y.; Yu, C.-J.; Ho, C.-C.; Liao, W.-Y.; Lee, J.-H.; Tsai, T.-H.; Su, K.-Y.; Hsieh, M.-S.; Chang, Y.-L.; et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: A genomic study. Lancet Respir. Med. 2018, 6, 107–116. [Google Scholar] [CrossRef]
- Weng, C.-H.; Chen, L.-Y.; Lin, Y.-C.; Shih, J.-Y.; Lin, Y.-C.; Tseng, R.-Y.; Chiu, A.-C.; Yeh, Y.-H.; Liu, C.; Lin, Y.-T.; et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene 2019, 38, 455–468. [Google Scholar] [CrossRef]
- Mok, T.S.; Mok, T.S.; Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Papadimitrakopoulou, V.A. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef]
- Snaebjornsson, M.T.; Janaki-Raman, S.; Schulze, A. Greasing the Wheels of the Cancer Machine: The Role of Lipid Metabolism in Cancer. Cell Metab. 2020, 31, 62–76. [Google Scholar] [CrossRef]
- Corn, K.C.; Windham, M.A.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef]
- Nomura, M.; Liu, J.; Rovira, I.I.; Gonzalez-Hurtado, E.; Lee, J.; Wolfgang, M.J.; Finkel, T. Fatty acid oxidation in macrophage polarization. Nat. Immunol. 2016, 17, 216–217. [Google Scholar] [CrossRef]
- Goossens, P.; Rodriguez-Vita, J.; Etzerodt, A.; Masse, M.; Rastoin, O.; Gouirand, V.; Ulas, T.; Papantonopoulou, O.; Van Eck, M.; Auphan-Anezin, N.; et al. Membrane Cholesterol Efflux Drives Tumor-Associated Macrophage Reprogramming and Tumor Progression. Cell Metab. 2019, 29, 1376–1389.e4. [Google Scholar] [CrossRef]
- Kuo, C.-Y.; Ann, D.K. When fats commit crimes: Fatty acid metabolism, cancer stemness and therapeutic resistance. Cancer Commun. 2018, 38, 47. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Zheng, L.; Zhao, H.; Yan, G.; Zhang, Q.; Zhou, Y.; Lei, J.; Zhang, J.; Wang, J.; et al. RIPK3 Orchestrates Fatty Acid Metabolism in Tumor-Associated Macrophages and Hepatocarcinogenesis. Cancer Immunol. Res. 2020, 8, 710–721. [Google Scholar] [CrossRef]
- Luo, J.; Luo, J.; Yao, J.F.; Deng, X.F.; Zheng, X.D.; Jia, M.; Wang, Y.Q.; Zhu, J.H. 14, 15-EET induces breast cancer cell EMT and cisplatin resistance by up-regulating integrin alphavbeta3 and activating FAK/PI3K/AKT signaling. J. Exp. Clin. Cancer Res. 2018, 37, 23. [Google Scholar] [CrossRef]
- Sun, S.C. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017, 17, 545–558. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.M.; Xu, Y.L.; Yuan, L.W.; Zhang, L.L.; Huang, M.Y.; Ye, Z.H.; Lu, J.J. TGFbeta2-mediated epithelial-mesenchymal transition and NF-kappaB pathway activation contribute to osimertinib resistance. Acta Pharmacol. Sin. 2021, 42, 451–459. [Google Scholar] [CrossRef]
- Chen, J.-S.; Su, I.-J.; Leu, Y.-W.; Young, K.-C.; Sun, H.S. Expression of T-cell lymphoma invasion and metastasis 2 (TIAM2) promotes proliferation and invasion of liver cancer. Int. J. Cancer 2012, 130, 1302–1313. [Google Scholar] [CrossRef]
- He, H.; Huang, J.; Wu, S.; Jiang, S.; Liang, L.; Liu, Y.; Liu, W.; Xie, L.; Tao, Y.; Jiang, Y.; et al. The roles of GTPase-activating proteins in regulated cell death and tumor immunity. J. Hematol. Oncol. 2021, 14, 1–15. [Google Scholar] [CrossRef]
- Honda, A.; Ito, Y.; Takahashi-Niki, K.; Matsushita, N.; Nozumi, M.; Tabata, H.; Takeuchi, K.; Igarashi, M. Extracellular Signals Induce Glycoprotein M6a Clustering of Lipid Rafts and Associated Signaling Molecules. J. Neurosci. 2017, 37, 4046–4064. [Google Scholar] [CrossRef]
- Rooney, C.; White, G.; Nazgiewicz, A.; Woodcock, A.S.; Anderson, K.; Ballestrem, C.; Malliri, A. The Rac activator STEF (Tiam2) regulates cell migration by microtubule-mediated focal adhesion disassembly. EMBO Rep. 2010, 11, 292–298. [Google Scholar] [CrossRef]
- Cooke, M.; Kreider-Letterman, G.; Baker, M.J.; Zhang, S.; Sullivan, N.T.; Eruslanov, E.; Abba, M.C.; Goicoechea, S.M.; García-Mata, R.; Kazanietz, M.G. FARP1, ARHGEF39, and TIAM2 are essential receptor tyrosine kinase effectors for Rac1-dependent cell motility in human lung adenocarcinoma. Cell Rep. 2021, 37, 109905. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Ou, Y.; Liu, S.; Yin, J.; Zhuo, W.; Huang, M.; Zhu, T.; Zhang, W.; Zhou, H.; Liu, Z. The Fibroblast TIAM2 Promotes Lung Cancer Cell Invasion and Metastasis. J. Cancer 2019, 10, 1879–1889. [Google Scholar] [CrossRef] [PubMed]
- Kosti, C.N.; Vaitsi, P.C.; Pappas, A.G.; Iliopoulou, M.P.; Psarra, K.K.; Magkouta, S.F.; Kalomenidis, I.T. CSF1/CSF1R signaling mediates malignant pleural effusion formation. JCI Insight 2022, 7, e155300. [Google Scholar] [CrossRef] [PubMed]
- Stempin, C.C.; Geysels, R.C.; Park, S.; Palacios, L.M.; Volpini, X.; Motran, C.C.; Rodríguez, E.V.A.; Nicola, J.P.; Cheng, S.-Y.; Pellizas, C.G.; et al. Secreted Factors by Anaplastic Thyroid Cancer Cells Induce Tumor-Promoting M2-like Macrophage Polarization through a TIM3-Dependent Mechanism. Cancers 2021, 13, 4821. [Google Scholar] [CrossRef]
- Ruffell, B.; Coussens, L.M. Macrophages and Therapeutic Resistance in Cancer. Cancer Cell 2015, 27, 462–472. [Google Scholar] [CrossRef]
- Sun, Y.; Dong, Y.; Liu, X.; Zhang, Y.; Bai, H.; Duan, J.; Wang, Z. Blockade of STAT3/IL-4 overcomes EGFR T790M-cis-L792F-induced resistance to osimertinib via suppressing M2 macrophages polarization. EBioMedicine 2022, 83, 104200. [Google Scholar] [CrossRef]
- Rocher, C.; Singla, D.K. SMAD-PI3K-Akt-mTOR pathway mediates BMP-7 polarization of monocytes into M2 macrophages. PLoS ONE 2013, 8, e84009. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Scagliotti, G.V.; Mulshine, J.L.; Kwon, R.; Curran, W.J., Jr.; Wu, Y.-L.; Paz-Ares, L. Lung cancer: Current therapies and new targeted treatments. Lancet 2017, 389, 299–311. [Google Scholar] [CrossRef]
- To, K.K.W.; Fong, W.; Cho, W.C.S. Immunotherapy in Treating EGFR-Mutant Lung Cancer: Current Challenges and New Strategies. Front. Oncol. 2021, 11, 635007. [Google Scholar] [CrossRef]
- Zhao, P.; Zhang, J.; Wu, A.; Zhang, M.; Zhao, Y.; Tang, Y.; Wang, B.; Chen, T.; Li, F.; Zhao, Q.; et al. Biomimetic codelivery overcomes osimertinib-resistant NSCLC and brain metastasis via macrophage-mediated innate immunity. J. Control. Release 2021, 329, 1249–1261. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. The lasso method for variable selection in the Cox model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Wilkerson, M.D.; Hayes, D.N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010, 26, 1572–1573. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Hübschmann, D. Make Interactive Complex Heatmaps in R. Bioinformatics 2021, 38, 1460–1462. [Google Scholar] [CrossRef]
- Ito, K.; Murphy, D. Application of ggplot2 to Pharmacometric Graphics. CPT Pharmacomet. Syst. Pharmacol. 2013, 2, e79. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.T.; Neely, J.G.; Paniello, R.C.; Voelker, C.C.J.; Nussenbaum, B.; Wang, E. A practical guide to understanding Kaplan-Meier curves. Otolaryngol. Neck Surg. 2010, 143, 331–336. [Google Scholar] [CrossRef]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Trevino, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Ait Blal, H.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ding, Y.; Song, J.; Kan, J. Hypolipidaemic effect and mechanism of paprika seed oil on Sprague-Dawley rats. J. Sci. Food Agric. 2017, 97, 4242–4249. [Google Scholar] [CrossRef]
- Liang, L.; Liu, Y.; Jiang, S.; Huang, J.; He, H.; Shen, L.; Cong, L.; Jiang, Y. Novel Circulating Tumour Cell-Related Risk Model Indicates Prognosis and Immune Infiltration in Lung Adenocarcinoma. J. Immunol. Res. 2022, 2022, 6521290. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Zhang, Y.; Huang, H.; Cao, J.; Fu, X. DFMG reverses proliferation and migration of vascular smooth muscle cells induced by co-culture with injured vascular endothelial cells via suppression of the TLR4-mediated signaling pathway. Mol. Med. Rep. 2018, 17, 5692–5699. [Google Scholar] [CrossRef]
- Liang, L.; Yan, B.; Liu, Y.; Jiang, S.; He, H.; Huang, J.; Liu, W.; Xie, L. FOXP3 Contributes to TMZ Resistance, Prognosis, and Immune Infiltration in GBM from a Novel Pyroptosis-Associated Risk Signature. Dis. Markers 2022, 2022, 4534080. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Wang, X.; Mao, C.; Jiang, Y.; Shi, Y.; Chen, L.; Liu, S.; Wang, B.; Pan, S.; Tao, Y.; et al. Long non-coding RNA linc01433 promotes migration and invasion in non-small cell lung cancer. Thorac. Cancer 2018, 9, 589–597. [Google Scholar] [CrossRef]
- Sun, W.; Li, S.; Tang, G.; Sun, S.; Luo, Y.; Bai, R.; Han, L.; Jiang, X.; Gao, Y.; Huang, Z.; et al. HHLA2 deficiency inhibits non-small cell lung cancer progression and THP-1 macrophage M2 polarization. Cancer Med. 2021, 10, 5256–5269. [Google Scholar] [CrossRef]


| Primers | Forward | Reverse |
|---|---|---|
| CD86 | 5′-CCATCAGCTTGTCTGTTTCATTCC-3′ | 5′-GCTGTAATCCAAGGAATGTGGTC-3′ |
| IL-12 | 5′-CCTTGCACTTCTGAAGAGATTGA-3′ | 5′-ACAGGGCCATCATAAAAGAGGT-3′ |
| CD206 | 5′-AGCCAACACCAGCTCCTCAAGA-3′ | 5′-CAAAACGCTCGCGCATTGTCCA-3′ |
| IL-10 | 5′-GACTTTAAGGGTTACCTGGGTTG-3′ | 5′-TCACATGCGCCTTGATGTCTG-3′ |
| β-actin | 5′-CACCATTGGCAATGAGCGGTTC-3′ | 5′-AGGTCTTTGCGGATGTCCACGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, L.; He, H.; Jiang, S.; Liu, Y.; Huang, J.; Sun, X.; Li, Y.; Jiang, Y.; Cong, L. TIAM2 Contributes to Osimertinib Resistance, Cell Motility, and Tumor-Associated Macrophage M2-like Polarization in Lung Adenocarcinoma. Int. J. Mol. Sci. 2022, 23, 10415. https://doi.org/10.3390/ijms231810415
Liang L, He H, Jiang S, Liu Y, Huang J, Sun X, Li Y, Jiang Y, Cong L. TIAM2 Contributes to Osimertinib Resistance, Cell Motility, and Tumor-Associated Macrophage M2-like Polarization in Lung Adenocarcinoma. International Journal of Molecular Sciences. 2022; 23(18):10415. https://doi.org/10.3390/ijms231810415
Chicago/Turabian StyleLiang, Lu, Hua He, Shiyao Jiang, Yueying Liu, Jingjing Huang, Xiaoyan Sun, Yi Li, Yiqun Jiang, and Li Cong. 2022. "TIAM2 Contributes to Osimertinib Resistance, Cell Motility, and Tumor-Associated Macrophage M2-like Polarization in Lung Adenocarcinoma" International Journal of Molecular Sciences 23, no. 18: 10415. https://doi.org/10.3390/ijms231810415
APA StyleLiang, L., He, H., Jiang, S., Liu, Y., Huang, J., Sun, X., Li, Y., Jiang, Y., & Cong, L. (2022). TIAM2 Contributes to Osimertinib Resistance, Cell Motility, and Tumor-Associated Macrophage M2-like Polarization in Lung Adenocarcinoma. International Journal of Molecular Sciences, 23(18), 10415. https://doi.org/10.3390/ijms231810415







