Abstract
Genetically encoded caged amino acids can be used to control the dynamics of protein activities and cellular localization in response to external cues. In the present study, we revealed the structural basis for the recognition of O-(2-nitrobenzyl)-L-tyrosine (oNBTyr) by its specific variant of Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (oNBTyrRS), and then demonstrated its potential availability for time-resolved X-ray crystallography. The substrate-bound crystal structure of oNBTyrRS at a 2.79 Å resolution indicated that the replacement of tyrosine and leucine at positions 32 and 65 by glycine (Tyr32Gly and Leu65Gly, respectively) and Asp158Ser created sufficient space for entry of the bulky substitute into the amino acid binding pocket, while Glu in place of Leu162 formed a hydrogen bond with the nitro moiety of oNBTyr. We also produced an oNBTyr-containing lysozyme through a cell-free protein synthesis system derived from the Escherichia coli B95. ΔA strain with the UAG codon reassigned to the nonnatural amino acid. Another crystallographic study of the caged protein showed that the site-specifically incorporated oNBTyr was degraded to tyrosine by light irradiation of the crystals. Thus, cell-free protein synthesis of caged proteins with oNBTyr could facilitate time-resolved structural analysis of proteins, including medically important membrane proteins.
1. Introduction
Photoresponsive caged compounds are useful tools for investigating cellular pathways, biochemical reactions, and enzymatic processes in time-resolved manners [1,2]. The release of caged groups and their subsequent changes in target molecules trigger downstream events that can be analyzed in detailed timeframes [3,4]. The substrates/inhibitors of enzymes and the agonists/antagonists of signaling molecules (receptors and channels) are thus photomanipulated to trigger targeted events in cell signaling and neuronal activities to precisely elucidate downstream responses [5,6]. “Caged proteins” with such photoreactive groups are prepared using complicated methods which are considered a hurdle for applications [7]. In contrast, the genetic encoding of caged amino acids offers a more facile and robust approach for biosynthesizing the photoresponsive proteins. These synthetic amino acids are recognized by specific variants of aminoacyl-tRNA synthetase and attach to tRNA, which in turn translates codons (mostly the UAG stop codon) assigned to the amino acids. The molecular basis for the specific binding of a caged amino acid (o-nitrobenzyl-3,4-dihydroxyphenylalanine, m-oNB-DOPA) has been studied using X-ray crystallography on a variant of Methanocaldococcus jannaschii tyrosyl-tRNA synthetase (MjTyrRS)—designated m-oNBDOPARS—complexed with the substrate [8]. The crystal structure reveals how mutations remodeled the amino acid binding pocket of m-oNBDOPARS to accommodate the tyrosine derivative within a bulky caged group.
In the present study, we determined the crystal structure of another archaeal tyrosyl-tRNA synthetase variant (oNBTyrRS) complexed with O-(2-nitrobenzyl)-L-tyrosine. This amino acid, oNBTyr, has the same caged group as m-oNB-DOPA at a different position on the tyrosine ring [8]. The determined structure not only provides a precise picture of the oNBTyr recognition in terms of its specific variant, but also an interesting comparison between two different synthetases accommodating the same caged group. We also conducted a large-scale preparation of caged proteins and demonstrated the potential applicability of oNBTyr in time-resolved X-ray crystallography. We showed that this amino acid can be efficiently converted into a tyrosine residue within crystals, although the same amino acid is difficult to decage from other enzymes in the solution [9].
2. Results and Discussion
2.1. X-ray Crystallographic Study on oNBTyrRS Complexed with oNBTyr
M. jannaschii TyrRS (MjTyrRS) is one of the aminoacyl-tRNA synthetases commonly used to introduce nonnatural amino acids into proteins by mutating in the substrate recognition sites [10]. oNBTyrRS has five amino acid replacements (Tyr32Gly, Leu65Gly, Phe108Glu, Asp158Ser, and Leu162Glu) at the tyrosine binding pocket of the wild-type MjTyrRS [6,9,11]. The remaining replacement, Asp286Arg, was introduced into the synthetase variant to increase the charging efficiency toward suppressor tRNA with the CUA anticodon [12]. Firstly, 17 mg of oNBTyrRS was prepared from a 1 L Escherichia coli cell culture, and then co-crystallization of oNBTyrRS (10 mg/mL) with oNBTyr (1.4 mg/mL) was carried out using the vapor diffusion method. The diffraction data collected from a single crystal were processed, and the structure of oNBTyrRS with oNBTyr was determined at 2.79 Å resolution (Figure 1A and Table S1). All 306 amino acid residues could be modeled thus, and oNBTyrRS formed a dimer in a similar manner to the wild-type MjTyrRS (PDB:1J1U); the dimer form is reportedly biologically active, as a tRNA substrate binds to two molecules of MjTyrRS [12]. The oNBTyr-bound oNBTyrRS is similar to MjTyrRS complexed with tRNA and L-tyrosine in overall structure, subunit arrangement, and domain architecture; the superposition of the Cα atoms of oNBTyrRS with those of MjTyrRS showed a low average RMSD (0.7 Å) [12,13,14,15]. The electron density in the |2Fo − Fc| map at the substrate binding site of oNBTyrRS unambiguously fits into the chemical structure of oNBTyr (Figure 1B).
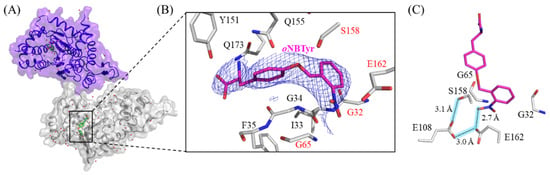
Figure 1.
The crystal structure of M. jannaschii oNBTyrRS (Y32G, L65G, F108E, D158S, L162E, and D286R) complexed with oNBTyr. (A) Dimeric structure of M. jannaschii oNBTyrRS bound to oNBTyr in the asymmetric unit. The substrate oNBTyr and water molecules are shown by green sticks and red spheres. (B) Close-up view of oNBTyr. The |2Fo − Fc| electron density map (contoured at 1.0 σ, blue mesh) corresponds to oNBTyr in the amino-acid binding site of oNBTyrRS. Mutated residues are indicated by the red letters. (C) Hydrogen-bonding interactions to recognize the o-nitrobenzyl group by oNBTyrRS. Numbers indicate the distance (Å) between two atoms connected by dashed lines.
As shown in Figure 1C, the five mutated residues play essential roles in the recognition of the structural features of oNBTyr. The substitutions of Tyr32 and Leu65 for Gly and Asp158 for Ser created enough space to accommodate the bulky nitrobenzyl group. This observation corresponds to previous reports that substitutions involving Tyr32 and Asp158 are preferable for MjTyrRS variants to recognize new substrates larger than L-tyrosine (The PDB IDs and references are listed in Table S2). In addition, glutamates replaced with Phe108 and Leu162 form a so-called carboxyl (Glu108)-carboxylate (Glu162) dimer motif [16] (Figure 1C). This is consistent with the pKa values estimated by the PROPKA program, which are 7.81 and 7.55 for Glu108 and 4.76 and 3.50 for Glu162 in the respective monomer of the dimer [17,18]. Furthermore, these residues, together with the oxygen of the nitro group, formed a hydrogen-bonding network, which was stabilized by the participation of Ser158. The nitro group was slightly bent with respect to the planarity of the benzyl group, thus the crucial recognition of the nitro group, which is important for substrate discrimination, was exhibited.
2.2. Structural Comparison with the Wild Type and the Other Mutant of MjTyrRS
The hydroxyl group of the tyrosine ring is recognized by the wild-type MjTyrRS through hydrogen bonding interactions with Tyr32 and Asp158 (Figure 2B, PDB ID: 1J1U) [12]. The hydrophobic environment caused by Phe108 and Leu162 stabilizes these interactions. The replacements of these positions abolished this important mode of discrimination through direct interaction with the side chain of the substrate amino acid, and changed the hydrophobic environment into a hydrophilic one. Thus, these mutations properly switched the substrate specificity between the wild-type and variant synthetases. As this oNBTyrRS is reported to also recognize o-nitropiperonyl-O-tyrosine (oNPTyr), we modeled it into the binding pocket of oNBTyrRS by aligning the main chain of the substrate tyrosine [9,19]. Although the chemical structure of oNPTyr was larger than that of oNBTyr (Figure 2D), the fitting model showed no structural distortion (Supplementary Figure S1). This is because the substrate binding pocket of oNBTyrRS was open toward the surface of the enzyme.
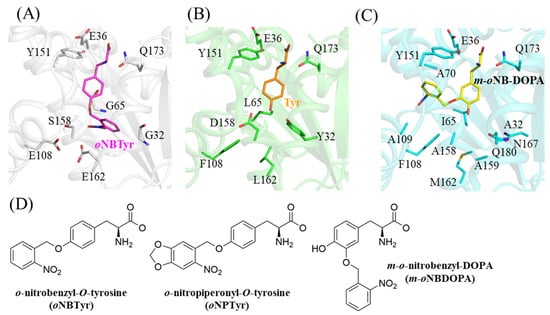
Figure 2.
Structural comparison of the substrate binding modes of various MjTyrRS. Structures of (A) oNBTyr in oNBTyrRS (this study); (B) tyrosine in the wild-type MjTyrRS (PDB: 1J1U [12]); (C) m-oNBDOPA in the corresponding variant of MjTyrRS (PDB: 5L7P [8]); (D) chemical structures of oNBTyr, oNPTyr, and m-oNB-DOPA.
As the other variant TyrRS for caged tyrosine, the crystal structure of m-oNBDOPARS bound to m-oNB-DOPA has been previously reported (PDB ID: 5L7P) [8]. The enzyme has 10 amino acid substitutions of the wild-type MjTyrRS (Supplementary Table S2). The nitrobenzyl moiety of m-oNB-DOPA was attached to position 3 of the tyrosine ring and the hydroxyl group remains as tyrosine; however, the recognition mode varied from those of the wild-type or oNBTyrRS (Figure 2C). Three substituted residues (Tyr32Ala, Leu65Ile, and Asp158Ala) in m-oNBDOPARS form hydrophobic interactions together with Phe108, and the hydroxyl group of tyrosine is stabilized by van der Waals interactions with Ile65 and Ala158. This results in the position of tyrosine becoming similar to the wild-type MjTyrRS. The nitrobenzyl group moiety is packed into a narrow hydrophobic space allowed by replacements with smaller amino acids at two positions (His70Ala and Gln109Ala) and is oriented to the interior of the enzyme, as opposed to oNBTyr. The nitro group of the leaving group forms a polar interaction only with the main chain of Ala70 in the hydrophobic environment. These comparisons indicate that two different mechanisms are achieved among the synthetase variants for the recognition of the same leaving group. Concerning MjTyrRS, previous crystallographic studies (listed in Table S2) have shown that the substrate recognition mode can be altered by amino acid mutations at 19 sites. In combination with the results of this structural analysis, the finding in this present study is expected to provide a basis for the design of photoresponsive components other than the nitrobenzyl group.
2.3. Synthesis of Caged Proteins in a Cell-Free Protein Synthesis System
We used a cell-free protein synthesis system with cell extracts from RFzero-iy, an E. coli strain deprived of the gene coding for the polypeptide release factor 1 gene (prfA) to facilitate efficient site-specific incorporation of nonnatural amino acids into proteins [20,21]. In this study, we employed cell extracts from the E. coli BL21 (DE3) derivative B95.delta A strain (B95.ΔA), in which the prfA gene was deleted and 95 selected UAG codons were replaced with UAA or UGA codons [22]. The large-scale preparation of cell extracts from B95.ΔA was performed as previously reported, except that the preincubation buffer was not used [23]. Efficient protein production with nonnatural amino acids was examined via p-benzoyl phenylalanine (pBpa) incorporation into N11-GFPS2_Y21amber, in which the tyrosine codon at position 21 was replaced with the UAG codon (Supplementary Figure S2B). Similarly, with supplementation of 0.32 mg/mL oNBTyrRS and 0.375 mg/mL oNBTyr, we observed comparable amounts of products were synthesized from the N11-GFPS2_Y21amber gene (Figure 3). This cell-free protein synthesis is a strong option for the production of caged proteins, since medically relevant membrane proteins have been successfully synthesized using cell-free systems.
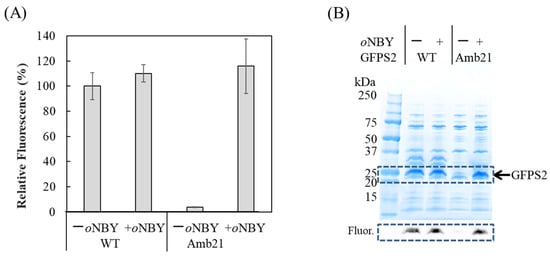
Figure 3.
Cell-free protein synthesis of N11-GFPS2 with (+) or without (−) o-nitrobenzyl Tyr (oNBY) supplementation using the cell extract from the strain B95.deltaA. Wild-type GFPS2 is indicated as “WT” and GFPS2 with Y21amber mutation is indicated as “Amb21”. (A) Fluorescent measurement of GFP in the reaction solutions. The excitation and emission wavelengths are 485 nm and 535 nm, respectively. Data represent the mean ± standard deviation (SD) of four independent experiments; (B) SDS-PAGE analysis of fractions purified by Ni-NTA resin. The 10–20% polyacrylamide gel was stained with CBB after electrophoresis. A portion of a fluorescent image of the gel framed by the dashed line is shown in the lower panel. The fluorescent imaging was performed by Vilber Bio Imaging FUSION FX with 470 nm and 535 nm filters. Molecular weights of protein standards (BIO-RAD Cat. No. 1610363) are indicated.
2.4. Crystallization of Caged Lysozyme and Decaging in the Crystal
To investigate the application of caged proteins to X-ray crystallographic study, we synthesized hen egg-white lysozyme (HEWL) with an oNBTyr substitution for Tyr20 (HEWL_Y20oNBTyr) by using the aforementioned E. coli cell-free protein synthesis system [24,25]. HEWL_Y20oNBTyr was purified from the cell-free reaction solution and crystallized using the sitting drop vapor diffusion method. The HEWL_Y20oNBTyr crystals yielded datasets at 1.44 Å resolution with completeness of 99.6% and a CC1/2 of 0.980. Model building and structure refinement were performed using Phenix (Table S1, PDB ID: 7YNU) [26]. The crystal structure of HEWL_Y20oNBTyr contained one protomer in the asymmetric unit (Figure 4A). Comparison of the HEWL_Y20oNBTyr structure with that of the wild-type lysozyme (PDB ID: 3WUL) shows that these structures are almost the same (RMSD, 0.202 Å) [27]. We observed a continuous density from the tyrosine residue at position 20, in which the nitrobenzyl group could be unambiguously modeled (Figure 4B). The density corresponding to the distal part of oNBTyr appeared somewhat weak, and the occupancy for atoms of the nitrobenzyl group is relatively low (0.5–1.0). This could be caused by the structure flexibility, as with oNPTyr (PDB ID: 6rtz) [9]. The tyrosyl moiety of oNBTyr20 is stabilized by the stacking interactions between Arg21 and Lys96 and the hydrogen bonding with Ser100 (Figure S3). On the other hand, the nitrobenzyl group forms an electrostatic interaction with Arg21, but is exposed on the surface (Figure S3A). Furthermore, the molecular geometry at this position does not affect the crystal packing environment. Alternatively, light contamination during protein synthesis or purification may have caused slight damage to the caged structure. These results confirm that efficient cell-free production of lysozyme with oNBTyr does not occur with any nonspecific suppression of the UAG codon with natural amino acids in the cell-free synthesis reaction. Finally, the crystals were irradiated with 365 nm light for 5 min for decaging. The obtained crystal structure at a 1.4 Å resolution showed that oNBTyr efficiently degraded into Tyr (Figure 4C,D). The overall structure of decaged HEWL was almost identical to that of caged HEWL. The RMSD per residue between structures before and after UV irradiation was 0.13. The release of the nitrobenzyl group resulted in the entry of water molecules into space, whereas a single water molecule was held in the vicinity of the oxygen atom in tyrosine (Figure S4). These results demonstrate that enzymatic reactions and protein structural changes can be induced by light emission on protein crystals, opening doors to time-resolved serial femtosecond crystallography of proteins using X-ray Free Electron Laser (XFEL). Shortening the irradiation time can be achieved through the use of high-intensity lasers and the development of new caged structures that are more efficiently photoreactive than the nitrobenzyl group.
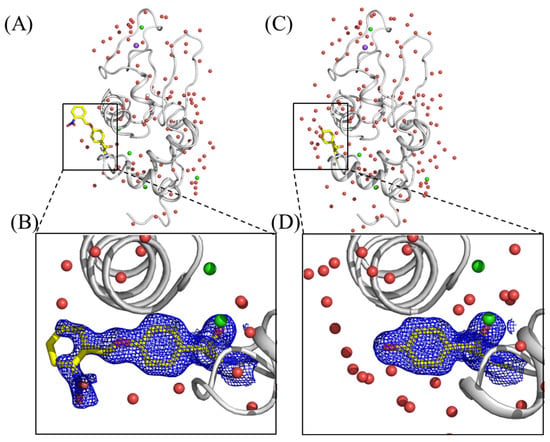
Figure 4.
Crystal Structures of hen egg-white lysozyme (HEWL) with the incorporation of oNBTyr before (A) and after (C) photoactivation. Close-up views of the oNBTyr; (B) and decaged oNBTyr, Tyr; (D) at position 20 of HEWL are shown by stick yellow models. The |2Fo − Fc| electron density maps (0.6 σ) around Tyr20 are depicted by blue mesh. Water molecules, sodium, and chloride atoms are shown by red, purple, and light green spheres, respectively.
3. Materials and Methods
3.1. Strain, Chemicals, and Enzymes
oNBTyr was purchased from Accela ChemBio Inc (San Diego, CA, USA). Almost all chemicals, including isopropyl-ß-D-thiogalactoside (IPTG), dithiothreitol (DTT), Tryptone (Cat. No. 35640-95), Yeast Extract Dried (Cat. No. 15838-45), glutathione oxidized form (GSSG), and glutathione reduced form (GSH), were purchased from Nacalai Tesque Inc. (Kyoto, Japan). E. coli disulfide-bond isomerase C (DsbC) was purified basically according to the previous report [28]. His6-tagged enzymes, small ubiquitin-like modifier (SUMO) protease, and TEV protease were purified according to previous studies [29,30]. The p-benzoyl-L-phenylalanyl-tRNA synthetase (pBpaRS), a mutant MjTyrRS (Y32G, E107S, D158T, I159S, and D286R) was cloned to the pET21b vector with a TEV protease site and His6 tag fused to the C terminus.
3.2. Construction of Plasmids Vectors for Protein Production
Codon-optimized gene encoding residues 19-147 of chicken egg white lysozyme (HEWL, UniProt ID: P00698) were chemically synthesized (Thermo Fisher Scientific, Waltham, MA, USA). The gene was N-terminally fused with a modified natural poly-histidine affinity tag (N11) and a SUMO protease recognition site and cloned into the plasmid vector pCR2.1-TOPO (Thermo Fisher Scientific, Waltham, MA, USA) [31]. The amber codon TAG was substituted for Tyr20 of HEWL by PCR mutagenesis method to construct pCR-HEWL-Y20amber.
Methanocalodococcus jannaschii tyrosyl-tRNA synthetase (MjTyrRS, UniProt ID: Q57834) which evolved to recognize oNBTyr had mutations Y32G, L65G, F108E, D158S, L162E, and D286R (oNBTyrRS) [11]. For the construction of pET21a-oNBTyrRS-TEV-His6, the gene encoding oNBTyrRS was chemically synthesized (Thermo Fisher Scientific, Waltham, MA, USA) and inserted into the pET-21a vector using the In-Fusion HD Cloning kit (Clontech, Shiga, Japan).
DNA sequences of synthetic genes and primers used for PCR are listed in Supplemental Table S3.
3.3. Expression, Purification, and Crystallization of the oNBTyr-tRNA Synthetase
E. coli Rosetta2 (DE3) cells transformed with the plasmid pET21a-oNBTyrRS-TEV-His6 were grown at 37 °C in a 2 L Luria–Bertani (LB) medium. When the optical density at 600 nm (OD600) reached 0.7–0.8, IPTG was added at the final concentration of 0.3 mM and the cells were cultivated at 20 °C for an additional 24 h. Cells were harvested at 8000× g for 10 min at 4 °C.
The cell pellet was suspended in lysis buffer A [20 mM Tris-HCl (pH 8.0), 1 M NaCl, 20 mM imidazole, and 20% glycerol] at 10 mL per gram and lysed by sonication on ice. The lysate was centrifuged at 16,000× g at 4 °C for 30 min. The supernatant was applied to HisTrap HP column (Cytiva, Tokyo, Japan), washed with a 10-column volume of buffer A, and eluted with a linear gradient of imidazole (20–500 mM imidazole). The eluate was dialyzed against sterile buffer A. After cleavage of the His-tag by TEV protease (0.08 mg/mL) for 16 h at 4 °C, the cleaved tag and His-tagged TEV protease were removed via passage through the HisTrap column. The flow-through fractions were pooled and applied onto a HiPrep 26/10 Desalting (Sephadex G25) column (Cytiva, Tokyo, Japan), equilibrated in 20 mM Tris-HCl (pH 8.0) buffer containing 0.1 M NaCl, 10% glycerol, and 1 mM DTT. Further purification was performed by a HiTrap Q column (Cytiva, Tokyo, Japan) with a linear gradient of 0–0.3 M NaCl. Finally, the pooled oNBTyrRS was dialyzed against a storage buffer [20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 1 mM DTT, 200 mM KCl, and 25% glycerol] for 16 h at 4 °C. The purified oNBTyrRS was concentrated to approximately 15 mg/mL using an Amicon Ultra-centrifuge device 10-kDa MWCO (Millipore, Burlington, MA, USA).
Co-crystallization of oNBTyrRS with the oNBTyr was performed by vapor diffusion using sitting drops of the crystallization solution at 20 °C. A 2 µL droplet of 10 mg/mL protein solution containing 1.4 mg/mL of oNBTyr mixed with the same amount of reservoir solution was equilibrated against 0.5 mL reservoir solution [20% PEG300, 3% PEG8000, 10% glycerol, and 100 mM Tris-HCl (pH 8.0)].
3.4. Data Collection, Structure Determination, and Refinement of the oNBTyr-tRNA Synthetase
The datasets of oNBTyrRS with oNBTyr were collected on SPring-8 BL32XU using a wavelength of 1.0 Å with the EIGER X 9M [32]. The crystal size was evaluated using the automated data collection system ZOO, developed at SPring-8 BL32XU [33]. After dose estimation using the crystal size, a helical data collection scheme was used with a beam size of 15 × 10 μm and an oscillation step of 0.1° (total 360°, 3600 images). The collected images were processed every 300 images (corresponding to 30°) using KAMO [34] with the XDS program [35]. The structure was solved by molecular replacement using the Phaser program [36] in the Phenix suite [26], with wild-type M. jannaschii tyrosyl-tRNA synthetase without RNA (PDB ID: 1J1U) as the search model [12]. The refinement was conducted using the Phenix programs [26], and the structure was manually rebuilt with the Coot program [37]. Data collection and refinement statistics are presented in Supplemental Table S1.
3.5. Preparation of E. coli S30 Extract and Construction of Cell-Free Protein Synthesis System for Site-Specific Incorporation of Nonnatural Amino Acids
The E. coli BL21 (DE3) derivative B95. delta A strain (B95.ΔA) was transformed by the plasmid vector piodoTyrRS-MJR1-gent(pACYC), which harbors MjTyrRS recognizes tRNATyr possessing the anticodon CUA, and minor tRNAs [22]. The transformed cells grew efficiently at 30 °C in 20 L of “3xYT” medium (24 g Tryptone, 15 g yeast extract, and 7.5 g NaCl per liter) supplemented with 0.15 mg/mL iodo-Tyr and reached 3.5–4.0 OD600 within 6 h (Supplementary Figure S2A). Cultivation was performed in a fermenter (TS-MW-30, TAKASUGI MFG. Co., Ltd., Tokyo, Japan) under the control of temperature (30 °C), aeration (20 L/min), pressure (0.02 MPa—near atmospheric pressure), and agitation (220 rpm). Cells were harvested using a tangential flow filtration (TFF) system and centrifuged at 7500× g for 23 min at 4 °C [23]. Two hundred grams of the cells were harvested from the 20 L culture. Large-scale preparation of cell extracts from B95.ΔA was performed as previously reported, except that the preincubation buffer was not used [23].
Efficient protein production with nonnatural amino acids was examined by p-benzoyl phenylalanine (pBpa) incorporation into N11-GFPS2_Y21amber, in which the tyrosine codon at position 21 was replaced with the UAG codon of N11-GFPS2 (GenBank: LC185343.1) (Supplemental Figure S2B). The pBpaRS-TEV-His6 was purified in a similar manner to oNBTyrRS but using only HisTrap columns (Cytiva, Tokyo, Japan). After cleavage of the His-tag by TEV protease (0.08 mg/mL) for 16 h at 4 °C, the cleaved tag and His-tagged TEV protease were removed via passage through the HisTrap column once more. The flow-through fractions were pooled and dialyzed against a storage buffer consisting of 20 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 6 mM 2-ME, 40 mM KCl, and 50% glycerol. Approximately 11.4 g of cells yielded 115 mg of pBpaRS (23 mg/mL, 5 mL).
The small-scale dialysis-mode cell-free protein synthesis with a 30 μL reaction mixture was performed at 25 °C and shaken at 240 rpm for 4 h, as previously described [23,38]. The concentration of pBpa was 1 mM both inside (reaction solution) and outside (feed solution). pBpaRS was added only to the reaction solution at a concentration of 0.1–0.3 mg/mL. The GFPS2 fluorescence at 535 nm was measured using a multi-label counter, ARVO SX (PerkinElmer, Waltham, MA, USA) with excitation at 485 nm.
3.6. A Site-Specific Introduction of oNBTyr to HEWL by a Cell-Free System and Its Purification
To incorporate oNBTyr at position 20 of HEWL, cell-free protein synthesis was performed in the same way as in the production of N11-GFPS2_Y21amber, except that pCR-HEWL-Y20amber (2 μg/mL) was used as a template and 0.32 mg/mL oNBTyrRS and 0.375 mg/mL oNBTyr were added. No reducing reagent (DTT) was used, but instead, 0.4 mg/mL E. coli DsbC, 1 mM GSSG, and 4 mM GSH were added to correctly form cysteine disulfide bonds under oxidative conditions. After 6 h of incubation at 25 °C, the reaction solution was centrifuged at 15,000× g for 10 min. The supernatant was applied to the HisTrap column (Cytiva, Tokyo, Japan) equilibrated with 20 mM Tris-HCl (pH 8.0) and 1 M NaCl, which was washed extensively with 20 mM imidazole in the buffer and eluted with 20 mM Tris-HCl buffer (pH 8.0) containing 0.5 M NaCl and 500 mM imidazole. The His-tag was cleaved with a final concentration of 0.4 mg/mL C-terminal His-tagged SUMO protease and dialyzed against 20 mM Tris-HCl buffer (pH 8.0), which contained 0.5 M NaCl and 20 mM imidazole, for 16 h at 4 °C. The cleaved tag and the His-tagged SUMO protease were removed via passage through the HisTrap column. The flow-through fractions were concentrated with a 3 kDa MWCO Amicon Ultra filter unit and applied to Superdex 200 10/300 column (Cytiva, Tokyo, Japan), equilibrated in Milli-Q-water. The resulting peak fractions were collected and concentrated again. We obtained over 1.0 mg of HEWL_Y20oNBTyr from the 54 mL reaction solution.
3.7. Crystallization, Data Collection, Structure Determination, and Refinement of HEWL_Y20oNBTyr
HEWL_Y20oNBTyr was crystallized using the sitting drop vapor diffusion method. Next, 2 μL of the protein solution (13.5 mg/mL) was mixed with an equal volume of the precipitant solution, composed of 100 mM Na acetate (pH 3.6), 8% PEG6000, and 28% NaCl. The crystallization plates were incubated at 20 °C in the dark.
Data collection, processing, and refinement are the same as in Section 3.4. The structure of wild-type lysozyme (PDB ID: 4YM8) was used as the search model. Before data collection of “decaged” HEWL_Y20oNBTyr, the crystal was illuminated by a LED lamp (OptoCode LED-EXTA, 365 nm) for five minutes at 20 °C. Data collection and refinement statistics are presented in Supplemental Table S1.
4. Conclusions
In this study, the crystal structure of oNBTyrRS-oNBTyr was determined. It provides a structural basis for the recognition of caged tyrosine, oNBTyr by mutant TyrRS. The o-nitrobenzyl group is commonly used as a photoremovable protecting group. However, the o-nitrobenzyl group has lower efficiency and slower kinetics in photoreaction than other caging groups [p-Hydroxyphenacy, 2-(2-nitrophenyl)ethyl, and (Coumarin-4-yl)methyl, 7-nitroindoline] [39]. To apply time-resolved serial femtosecond crystallography at the XFEL facility, the introduction of nonnatural amino acids using a more reactive photoremovable protecting group is required. The structural information obtained in this study and the structural comparison with similar caged-tyrosine structures will provide the basis for the development of a new caged amino acid introduction system.
In addition, we confirmed for the first time that the photoremovable protecting group was removed via light irradiation of protein crystals, in which oNBTyr was introduced by the E. coli cell-free synthesis system. The E. coli cell-free synthesis system is capable of synthesizing proteins that are difficult to produce in cellular systems, such as membrane proteins and highly toxic proteins. The synthesis reaction volume is smaller than that of the E. coli culture system, which has the advantage of reducing the number of expensive nonnatural amino acids, making it useful for a variety of applied research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810399/s1. References [40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55] are cited in the supplementary materials.
Author Contributions
T.H., K.K. and C.T. designed the research; T.H., K.K., Y.I.-K., K.H., K.I., Y.T. and M.I. performed the research; T.H., K.K. and C.T. analyzed the data; R.A. maintained the computing environments and database for the research; T.H., K.K., C.T. and M.S. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by MEXT/JSPS KAKENHI Grant Numbers 20H05450 and 22H04759 (T.H.) and the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED, under Grant Number JP21am0101070 (support number 1536 and 1667).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Protein Data Bank (PDB) entry codes were 7YNW for oNBTyrRS, 7YNU for HEWL_Y20oNBTyr, and 7YNV for “decaged” HEWL.
Acknowledgments
We thank Kunio Hirata, Naoki Sakai, and Yoshiaki Kawano at the BL32XU of SPring-8 with for their technical assistance with data collection. We thank Kensaku Sakamoto for the critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Engels, J.; Schlaeger, E.J. Synthesis, structure, and reactivity of adenosine cyclic 3′,5′-phosphate benzyl triesters. J. Med. Chem. 1977, 20, 907–911. [Google Scholar] [CrossRef]
- Kaplan, J.H.; Forbush, B., 3rd; Hoffman, J.F. Rapid photolytic release of adenosine 5′-triphosphate from a protected analogue: Utilization by the Na: K pump of human red blood cell ghosts. Biochemistry 1978, 17, 1929–1935. [Google Scholar] [CrossRef]
- Mayer, G.; Heckel, A. Biologically active molecules with a “light switch”. Angew. Chem. Int. Ed. Engl. 2006, 45, 4900–4921. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.M.; Larson, D.R.; Lawrence, D.S. Illuminating the chemistry of life: Design, synthesis, and applications of “caged” and related photoresponsive compounds. ACS Chem. Biol. 2009, 4, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A.; Gauron, C.; Volovitch, M.; Bensimon, D.; Jullien, L.; Vriz, S. How to control proteins with light in living systems. Nat. Chem. Biol. 2014, 10, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Y.; Liu, Y.; Zheng, S.; Wang, X.; Zhao, J.; Yang, F.; Zhang, G.; Wang, C.; Chen, P.R. Time-resolved protein activation by proximal decaging in living systems. Nature 2019, 569, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Ellis-Davies, G.C. Caged compounds: Photorelease technology for control of cellular chemistry and physiology. Nat. Met. 2007, 4, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Hauf, M.; Richter, F.; Schneider, T.; Faidt, T.; Martins, B.M.; Baumann, T.; Durkin, P.; Dobbek, H.; Jacobs, K.; Möglich, A.; et al. Photoactivatable Mussel-Based Underwater Adhesive Proteins by an Expanded Genetic Code. ChemBioChem 2017, 18, 1819–1823. [Google Scholar] [CrossRef] [PubMed]
- Kneuttinger, A.C.; Straub, K.; Bittner, P.; Simeth, N.A.; Bruckmann, A.; Busch, F.; Rajendran, C.; Hupfeld, E.; Wysocki, V.H.; Horinek, D.; et al. Light Regulation of Enzyme Allostery through Photo-responsive Unnatural Amino Acids. Cell Chem. Biol. 2019, 26, 1501–1514.e9. [Google Scholar] [CrossRef]
- Liu, C.C.; Schultz, P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [Green Version]
- Deiters, A.; Groff, D.; Ryu, Y.; Xie, J.; Schultz, P.G. A genetically encoded photocaged tyrosine. Angew. Chem. Int. Ed. Engl. 2006, 45, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nureki, O.; Ishitani, R.; Yaremchuk, A.; Tukalo, M.; Cusack, S.; Sakamoto, K.; Yokoyama, S. Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion. Nat. Struct. Biol. 2003, 10, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Calendar, R.; Berg, P. Purification and physical characterization of tyrosyl ribonucleic acid synthetases from Escherichia coli and Bacillus subtilis. Biochemistry 1966, 5, 1681–1690. [Google Scholar] [CrossRef]
- Koch, G.L. Tyrosyl transfer ribonucleic acid synthetase from Bacillus stearothermophilus. Preparation and properties of the crystallizable enzyme. Biochemistry 1974, 13, 2307–2312. [Google Scholar] [CrossRef] [PubMed]
- Brick, P.; Bhat, T.N.; Blow, D.M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J. Mol. Biol. 1989, 208, 83–98. [Google Scholar] [CrossRef]
- D’Ascenzo, L.; Auffinger, P. A comprehensive classification and nomenclature of carboxyl-carboxyl(ate) supramolecular motifs and related catemers: Implications for biomolecular systems. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 2015, 71, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.; Rostkowski, M.; Jensen, J.H. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Olsson, M.H.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Luo, J.; Torres-Kolbus, J.; Liu, J.; Deiters, A. Genetic Encoding of Photocaged Tyrosines with Improved Light-Activation Properties for the Optical Control of Protease Function. ChemBioChem 2017, 18, 1442–1447. [Google Scholar] [CrossRef]
- Mukai, T.; Yanagisawa, T.; Ohtake, K.; Wakamori, M.; Adachi, J.; Hino, N.; Sato, A.; Kobayashi, T.; Hayashi, A.; Shirouzu, M.; et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem. Biophys. Res. Commun. 2011, 411, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Adachi, J.; Katsura, K.; Seki, E.; Takemoto, C.; Shirouzu, M.; Terada, T.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Cell-Free Protein Synthesis Using S30 Extracts from Escherichia coli RFzero Strains for Efficient Incorporation of Non-Natural Amino Acids into Proteins. Int. J. Mol. Sci. 2019, 20, 492. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Hoshi, H.; Ohtake, K.; Takahashi, M.; Yamaguchi, A.; Hayashi, A.; Yokoyama, S.; Sakamoto, K. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 2015, 5, 9699. [Google Scholar] [CrossRef] [PubMed]
- Katsura, K.; Matsuda, T.; Tomabechi, Y.; Yonemochi, M.; Hanada, K.; Ohsawa, N.; Sakamoto, K.; Takemoto, C.; Shirouzu, M. A reproducible and scalable procedure for preparing bacterial extracts for cell-free protein synthesis. J. Biochem. 2017, 162, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Kigawa, T.; Yabuki, T.; Matsuda, N.; Matsuda, T.; Nakajima, R.; Tanaka, A.; Yokoyama, S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genom. 2004, 5, 63–68. [Google Scholar] [CrossRef]
- Matsuda, T.; Ito, T.; Takemoto, C.; Katsura, K.; Ikeda, M.; Wakiyama, M.; Kukimoto-Niino, M.; Yokoyama, S.; Kurosawa, Y.; Shirouzu, M. Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction. PLoS ONE 2018, 13, e0193158. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.-W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Biol. Crystallogr. 2019, 75, 861–877. [Google Scholar] [CrossRef]
- Sugahara, M.; Mizohata, E.; Nango, E.; Suzuki, M.; Tanaka, T.; Masuda, T.; Tanaka, R.; Shimamura, T.; Tanaka, Y.; Suno, C.; et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods 2015, 12, 61–63. [Google Scholar] [CrossRef]
- Kim, D.-M.; Swartz, J.R. Efficient production of a bioactive, multiple disulfide-bonded protein using modified extracts of Escherichia coli. Biotechnol. Bioeng. 2004, 85, 122–129. [Google Scholar] [CrossRef]
- Malakhov, M.P.; Mattern, M.R.; Malakhova, O.A.; Drinker, M.; Weeks, S.D.; Butt, T.R. SUMO fusions and SUMO-specific protease for efficient expression and purification of proteins. J. Struct. Funct. Genom. 2004, 5, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Katsura, K.; Tomabechi, Y.; Matsuda, T.; Yonemochi, M.; Mikuni, J.; Ohsawa, N.; Terada, T.; Yokoyama, S.; Kukimoto-Niino, M.; Takemoto, C.; et al. Phosphorylated and non-phosphorylated HCK kinase domains produced by cell-free protein expression. Protein Expr. Purif. 2018, 150, 92–99. [Google Scholar] [CrossRef]
- Shinoda, T.; Shinya, N.; Ito, K.; Ishizuka-Katsura, Y.; Ohsawa, N.; Terada, T.; Hirata, K.; Kawano, Y.; Yamamoto, M.; Tomita, T.; et al. Cell-free methods to produce structurally intact mammalian membrane proteins. Sci. Rep. 2016, 6, 30442. [Google Scholar] [CrossRef] [PubMed]
- Hirata, K.; Kawano, Y.; Ueno, G.; Hashimoto, K.; Murakami, H.; Hasegawa, K.; Hikima, T.; Kumasaka, T.; Yamamoto, M. Achievement of protein micro-crystallography at SPring-8 beamline BL32XU. J. Phys. Conf. Ser. 2013, 425, 012002. [Google Scholar] [CrossRef]
- Hirata, K.; Yamashita, K.; Ueno, G.; Kawano, Y.; Hasegawa, K.; Kumasaka, T.; Yamamoto, M. ZOO: An automatic data-collection system for high-throughput structure analysis in protein microcrystallography. Acta Crystallogr. D Biol. Crystallogr. 2019, 75, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, K.; Hirata, K.; Yamamoto, M. KAMO: Towards automated data processing for microcrystals. Acta Crystallogr. D Biol. Crystallogr. 2018, 74, 441–449. [Google Scholar] [CrossRef]
- Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Seki, E.; Yanagisawa, T.; Yokoyama, S. Cell-Free Protein Synthesis for Multiple Site-Specific Incorporation of Noncanonical Amino Acids Using Cell Extracts from RF-1 Deletion E. coli Strains. Methods Mol. Biol. 2018, 1728, 49–65. [Google Scholar]
- San Miguel, V.; Bochet, C.G.; del Campo, A. Wavelength-selective caged surfaces: How many functional levels are possible? J. Am. Chem. Soc. 2011, 133, 5380–5388. [Google Scholar] [CrossRef]
- Liu, Q.; He, Q.T.; Lyu, X.; Yang, F.; Zhu, Z.L.; Xiao, P.; Yang, Z.; Zhang, F.; Yang, Z.Y.; Wang, X.Y.; et al. DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl (1)H-NMR probe. Nat. Commun. 2020, 11, 4857. [Google Scholar] [CrossRef] [PubMed]
- Beyer, J.N.; Hosseinzadeh, P.; Gottfried-Lee, I.; Van Fossen, E.M.; Zhu, P.; Bednar, R.M.; Karplus, P.A.; Mehl, R.A.; Cooley, R.B. Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase. J. Mol. Biol. 2020, 432, 4690–4704. [Google Scholar] [CrossRef]
- Schiefner, A.; Nastle, L.; Landgraf, M.; Reichert, A.J.; Skerra, A. Structural Basis for the Specific Cotranslational Incorporation of p-Boronophenylalanine into Biosynthetic Proteins. Biochemistry 2018, 57, 2597–2600. [Google Scholar] [CrossRef]
- Baumann, T.; Hauf, M.; Schildhauer, F.; Eberl, K.B.; Durkin, P.M.; Deniz, E.; Löffler, J.G.; Acevedo-Rocha, C.G.; Jaric, J.; Martins, B.M.; et al. Site-Resolved Observation of Vibrational Energy Transfer Using a Genetically Encoded Ultrafast Heater. Angew. Chem. Int. Ed. Engl. 2019, 58, 2899–2903. [Google Scholar] [CrossRef]
- Luo, X.; Fu, G.; Wang, R.E.; Zhu, X.; Zambaldo, C.; Liu, R.; Liu, T.; Lyu, X.; Du, J.; Xuan, W.; et al. Genetically encoding phosphotyrosine and its nonhydrolyzable analog in bacteria. Nat. Chem. Biol. 2017, 13, 845–849. [Google Scholar] [CrossRef]
- Liu, X.; Jiang, L.; Li, J.; Wang, L.; Yu, Y.; Zhou, Q.; Lv, X.; Gong, W.; Lu, Y.; Wang, J. Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers. J. Am. Chem. Soc. 2014, 136, 13094–13097. [Google Scholar] [CrossRef]
- Cooley, R.B.; Karplus, P.A.; Mehl, R.A. Gleaning unexpected fruits from hard-won synthetases: Probing principles of permissivity in non-canonical amino acid-tRNA synthetases. ChemBioChem 2014, 15, 1810–1819. [Google Scholar] [CrossRef] [PubMed]
- Cooley, R.B.; Feldman, J.L.; Driggers, C.M.; Bundy, T.A.; Stokes, A.L.; Karplus, P.A.; Mehl, R.A. Structural basis of improved second-generation 3-nitro-tyrosine tRNA synthetases. Biochemistry 2014, 53, 1916–1924. [Google Scholar] [CrossRef]
- Li, F.; Shi, P.; Li, J.; Yang, F.; Wang, T.; Zhang, W.; Gao, F.; Ding, W.; Li, D.; Li, J.; et al. A genetically encoded 19F NMR probe for tyrosine phosphorylation. Angew. Chem. Int. Ed. Engl. 2013, 52, 3958–3962. [Google Scholar] [CrossRef]
- Young, D.D.; Young, T.S.; Jahnz, M.; Ahmad, I.; Spraggon, G.; Schultz, P.G. An evolved aminoacyl-tRNA synthetase with atypical polysubstrate specificity. Biochemistry 2011, 50, 1894–1900. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, W.; Song, W.; Wang, Y.; Yu, Z.; Li, J.; Wu, M.; Wang, L.; Zang, J.; Lin, Q. A biosynthetic route to photoclick chemistry on proteins. J. Am. Chem. Soc. 2010, 132, 14812–14818. [Google Scholar] [CrossRef]
- Tippmann, E.M.; Liu, W.; Summerer, D.; Mack, A.V.; Schultz, P.G. A genetically encoded diazirine photocrosslinker in Escherichia coli. ChemBioChem 2007, 8, 2210–2214. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Liu, W.; Schultz, P.G. A genetically encoded bidentate, metal-binding amino acid. Angew. Chem. Int. Ed. Engl. 2007, 46, 9239–9242. [Google Scholar] [CrossRef]
- Liu, W.; Alfonta, L.; Mack, A.V.; Schultz, P.G. Structural basis for the recognition of para-benzoyl-L-phenylalanine by evolved aminoacyl-tRNA synthetases. Angew. Chem. Int. Ed. Engl. 2007, 46, 6073–6075. [Google Scholar] [CrossRef]
- Turner, J.M.; Graziano, J.; Spraggon, G.; Schultz, P.G. Structural plasticity of an aminoacyl-tRNA synthetase active site. Proc. Natl. Acad. Sci. USA 2006, 103, 6483–6488. [Google Scholar] [CrossRef]
- Turner, J.M.; Graziano, J.; Spraggon, G.; Schultz, P.G. Structural characterization of a p-acetylphenylalanyl aminoacyl-tRNA synthetase. J. Am. Chem. Soc. 2005, 127, 14976–14977. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).