Two Short Repeats in the 5′ Untranslated Region of Insulin-like Androgenic Gland Factor in Procambarus clarkii (PcIAG) That Regulate PcIAG Expression
Abstract
:1. Introduction
2. Results
2.1. Isolation of the Insulin-like Androgenic Gland Hormone Gene (PcIAG) from Procambarus Clarkii
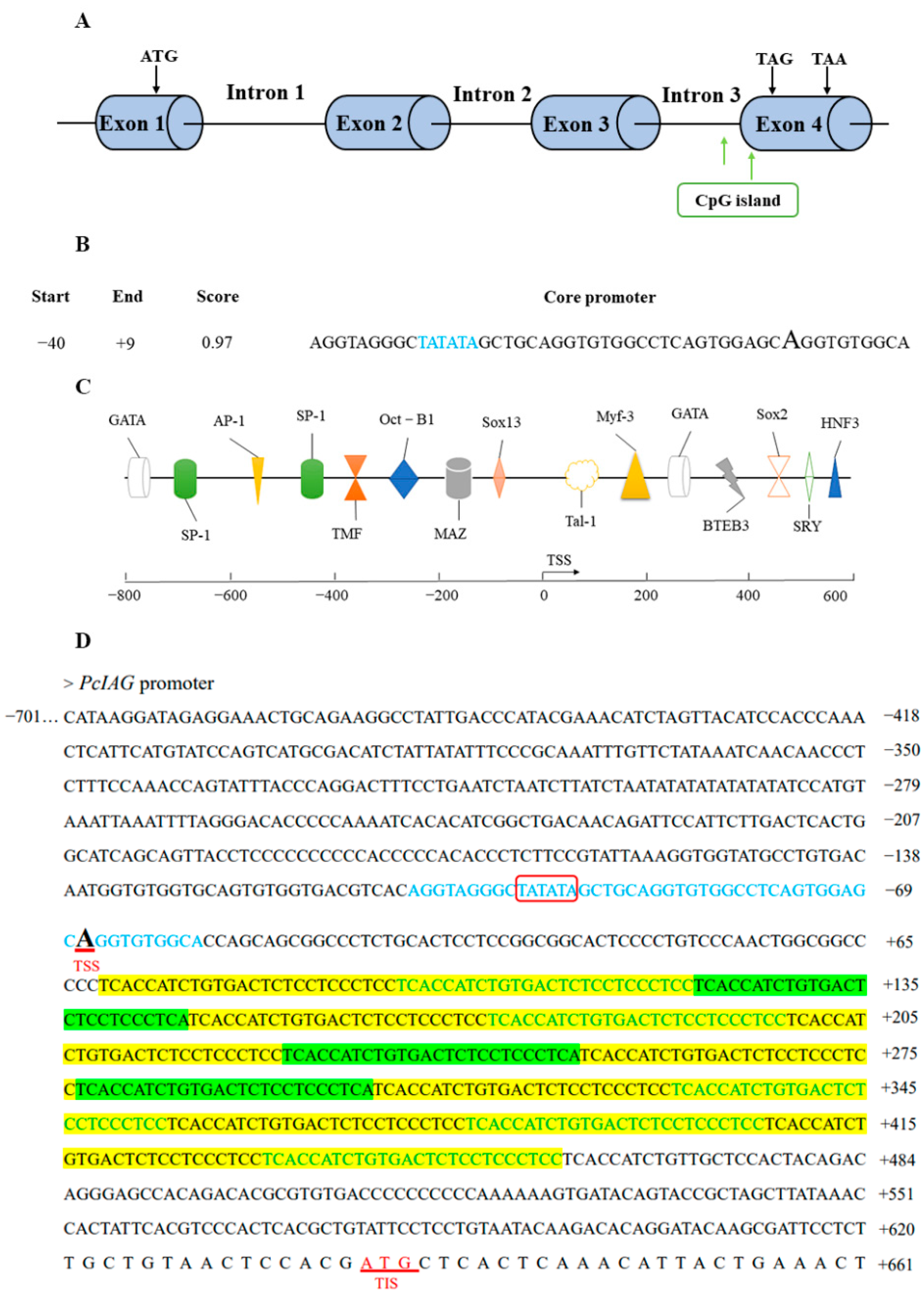
2.2. Bioinformatics Analysis of PcIAG Promoter Sequence
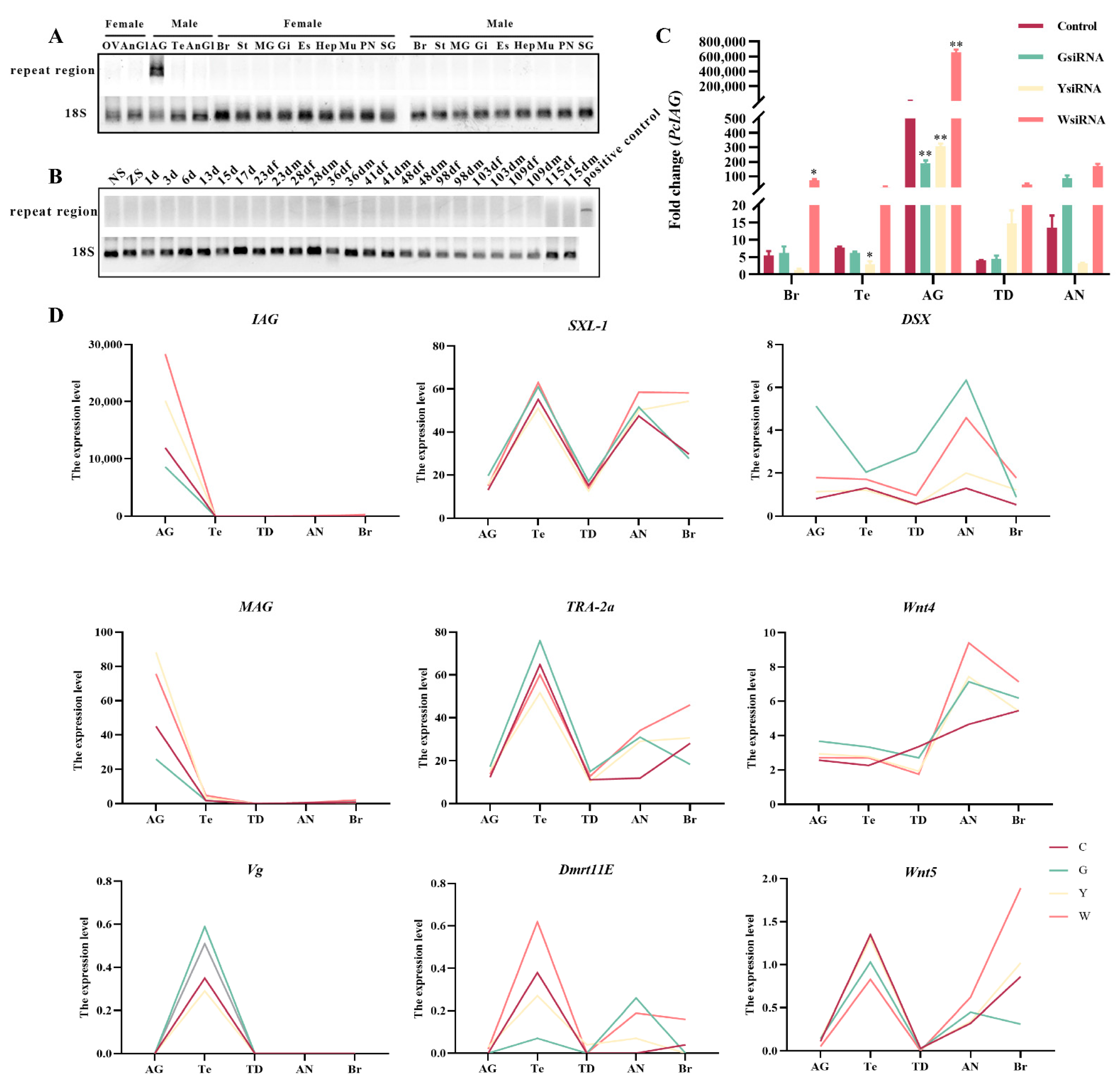
2.3. The Existence Pattern of Repeats in Different Tissues and Developmental Stages of Female and Male Procambarus Clarkii
2.4. Analysis of mRNA Expression Profiles in Gonad and Nerve after siRNA Interference
2.4.1. De Novo Assembly and Annotation
2.4.2. GO and KEGG Enrichment Analyses of Differentially Expressed Genes
2.4.3. Sex-Related Differentially Expressed Genes
2.5. Analysis of miRNA Expression Profiles in Male Gonads after siRNA Interference
2.5.1. Overview of miRNAs in Gonads of Procambarus Clarkii
2.5.2. Differentially Expressed (DE) miRNAs
2.5.3. Target Gene Prediction, GO Classification, and KEGG Pathway Analyses
2.6. Analyses of Exosome miRNA
2.6.1. Comparison of the Top 10 Highly Expressed miRNAs after siRNA Interference with miRNAs in Exosomes
2.6.2. Differentially Expressed (DE) miRNAs
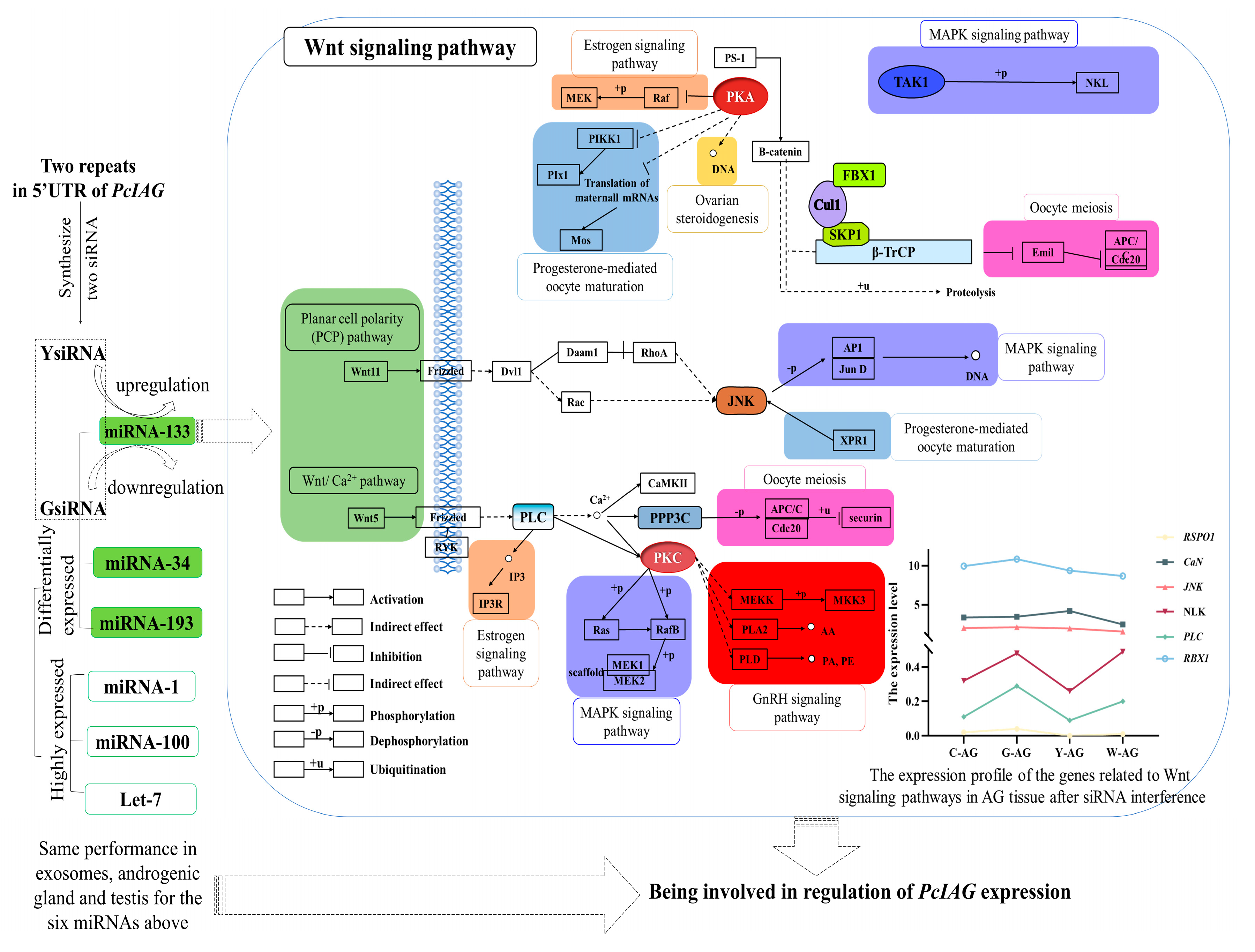
3. Discussion
4. Materials and Methods
4.1. Experimental Crayfish
4.2. Cloning and Analysis of PcIAG Gene
4.3. Cloning and Bioinformatics Analyses of PcIAG Promoter
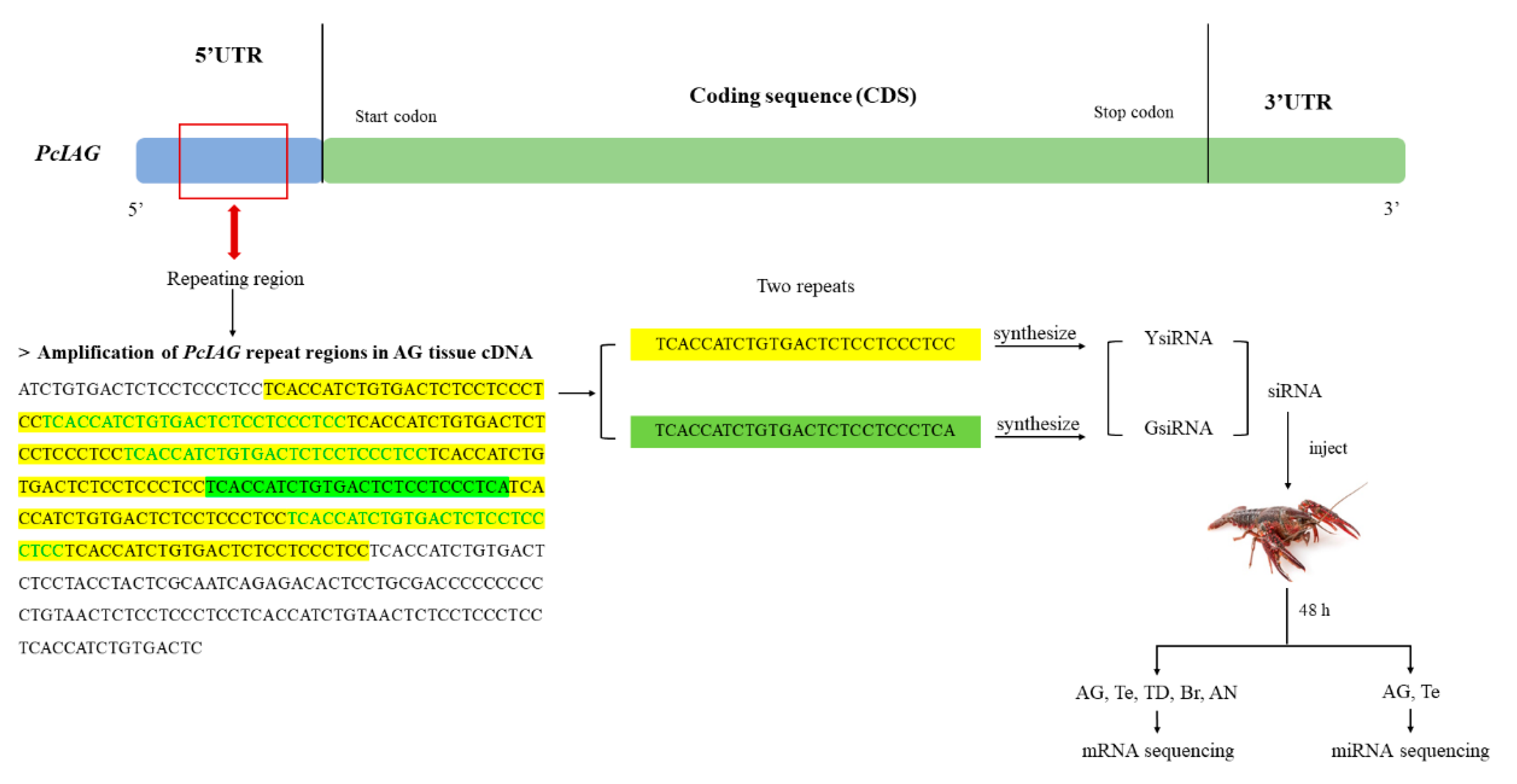
4.4. Amplification of PcIAG Repeat Regions in Male and Female Procambarus Clarkii DNA, and Different Tissues and Different Developmental Stages of Procambarus Clarkii
4.5. RNAi with siRNAs Designed Based on the Wwo Short Repeats in the 5′UTR of PcIAG
4.6. mRNA and miRNA Sequencing
4.7. Exosome miRNA Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jin, S.Y.; Jacquin, L.; Xiong, M.T.; Li, R.J.; Lek, S.; Li, W.; Zhang, T.L. Reproductive pattern and population dynamics of commercial red swamp crayfish (Procambarus clarkii) from China: Implications for sustainable aquaculture management. PeerJ 2019, 7, e6214. [Google Scholar] [CrossRef]
- Peng, B.; Tan, Y.F.; Peng, G.H.; Xiong, L.J.; Bai, X.F. Path analysis of effects of phenotypic traits attributes on abdomen meat weight of red swamp crayfish Procambarus clarkii. Fish Sci. 2021, 40, 718–725. (In Chinese) [Google Scholar] [CrossRef]
- Jiang, H.C.; Xing, Z.J.; Lu, W.; Qian, Z.J.; Yu, H.W.; Li, J.L. Transcriptome analysis of red swamp crawfish Procambarus clarkii reveals genes involved in gonadal development. PLoS ONE 2014, 9, e105122. [Google Scholar] [CrossRef] [PubMed]
- Barki, A.; Karplus, I.; Manor, R.; Sagi, A. Intersexuality and behavior in crayfish: The de-masculinization effects of androgenic gland ablation. Horm. Behav. 2006, 50, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Ventura, T.; Sagi, A. The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol. Adv. 2012, 6, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.T.; Wang, W.; Wang, C.Q.; Sun, C.B.; Shi, L.L.; Chan, S.F. Insulin-like androgenic gland hormone from the shrimp Fenneropenaeus merguiensis: Expression, gene organization and transcript variants. Gene 2021, 782, 145529. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Li, Y.H. A sex-reversing factor: Insulin-like androgenic gland hormone in decapods. Rev. Aquac. 2021, 3, 1352–1366. [Google Scholar] [CrossRef]
- Rosen, O.; Manor, R.; Weil, S.; Gafni, O.; Linial, A.; Aflalo, E.D.; Ventura, T.; Sagi, A. A sexual shift induced by silencing of a single insulin-like gene in crayfish: Ovarian upregulation and testicular degeneration. PLoS ONE 2010, 5, e15281. [Google Scholar] [CrossRef]
- Guo, Q.; Li, S.H.; Lv, X.J.; Xiang, J.H.; Sagi, A.; Manor, R.; Li, F.H. A Putative Insulin-like Androgenic Gland Hormone Receptor Gene Specifically Expressed in Male Chinese Shrimp. Endocrinology 2018, 159, 2173–2185. [Google Scholar] [CrossRef]
- Fu, C.Q.; Li, F.J.; Wang, L.F.; Wu, F.R.; Wang, J.M.; Fan, X.L.; Liu, T. Molecular characteristics and abundance of insulin-like androgenic gland hormone and effects of RNA interference in Eriocheir sinensis. Anim. Reprod. Sci. 2020, 215, 106332. [Google Scholar] [CrossRef]
- Liu, F.; Shi, W.Y.; Ye, H.H.; Zeng, C.S.; Zhu, Z.H. Insulin-like androgenic gland hormone 1 (IAG1) regulates sexual differentiation in a hermaphrodite shrimp through feedback to neuroendocrine factors. Gen. Comp. Endocrinol. 2021, 303, 113706. [Google Scholar] [CrossRef]
- Zhang, G.S.; Yin, S.W.; Mao, J.Q.; Liang, F.F.; Zhao, C.; Li, P.; Zhou, G.Q.; Chen, S.Q.; Tang, Z.L. Integrated analysis of mRNA-seq and miRNA-seq in the liver of Pelteobagrus vachelli in response to hypoxia. Sci. Rep. 2016, 6, 22907. [Google Scholar] [CrossRef]
- Hu, Y.; Lan, W.J.; Miller, D. Next-Generation Sequencing for MicroRNA Expression Profile. Methods Mol. Biol. 2017, 1617, 169–177. [Google Scholar] [CrossRef]
- Jin, S.B.; Fu, H.T.; Sun, S.M.; Jiang, S.F.; Xiong, Y.W.; Gong, Y.S.; Qiao, H.; Zhang, W.Y.; Wu, Y. Integrated analysis of microRNA and mRNA expression profiles during the sex-differentiation sensitive period in oriental river prawn, Macrobrachium nipponense. Sci. Rep. 2017, 7, 12011. [Google Scholar] [CrossRef]
- Oliva, G.; Sahr, T.; Buchrieser, C. Small RNAs, 5′ UTR elements and RNA-binding proteins in intracellular bacteria: Impact on metabolism and virulence. FEMS Microbiol. Rev. 2015, 39, 331–349. [Google Scholar] [CrossRef]
- Orom, U.A.; Nielsen, F.C.; Lund, A.H. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell 2008, 30, 460–471. [Google Scholar] [CrossRef]
- Tuller, T.; Ruppin, E.; Kupiec, M. Properties of untranslated regions of the S. cerevisiae genome. BMC Genom. 2009, 10, 391. [Google Scholar] [CrossRef]
- Yang, S.S.; Wang, J.; Li, Z.; Cui, S.; Liu, W. The 5′ Untranslated Region of the Capsid Protein 2 Gene of Mink Enteritis Virus Is Essential for Its Expression. J. Virol. 2018, 92, e00787-18. [Google Scholar] [CrossRef]
- Yu, X.J.; Odenthal, M.; Fries, J.W. Exosomes as miRNA Carriers: Formation-Function-Future. Int. J. Mol. Sci. 2016, 17, 2028. [Google Scholar] [CrossRef]
- Zhou, Y.J.; Zhang, Y.; Gong, H.; Luo, S.Q.; Cui, Y. The Role of Exosomes and Their Applications in Cancer. Int. J. Mol. Sci. 2021, 22, 12204. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.J.; Aswath, K.; Schroeder, S.G.; Lippolis, J.D.; Reinhardt, T.A.; Sonstegard, T.S. MicroRNA expression profiles of bovine milk exosomes in response to Staphylococcus aureus infection. BMC Genom. 2015, 16, 806. [Google Scholar] [CrossRef]
- Zheng, J.B.; Cheng, S.; Jia, Y.Y.; Gu, Z.M.; Li, F.; Chi, M.L.; Liu, S.L.; Jiang, W.P. Molecular identification and expression profiles of four splice variants of Sex-lethal gene in Cherax quadricarinatus. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2019, 234, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Fu, H.T.; Zhou, Q.; Sun, S.M.; Jiang, S.F.; Xiong, Y.W.; Gong, Y.; Qiao, H.S.; Zhang, W.Y. Transcriptome analysis of androgenic gland for discovery of novel genes from the oriental river prawn, Macrobrachium nipponense, using Illumina Hiseq 2000. PLoS ONE 2013, 8, e76840. [Google Scholar] [CrossRef]
- Peng, J.X.; Wei, P.Y.; Zhang, B.; Zhao, Y.Z.; Zeng, D.G.; Chen, X.L.; Li, M.; Chen, X.H. Gonadal transcriptomic analysis and differentially expressed genes in the testis and ovary of the Pacific white shrimp (Litopenaeus vannamei). BMC Genom. 2015, 16, 1006. [Google Scholar] [CrossRef]
- Jin, S.B.; Zhang, W.Y.; Xiong, Y.W.; Jiang, S.F.; Qiao, H.; Gong, Y.S.; Wu, Y.; Fu, H.T. Identification of Important Genes Involved in the Sex-Differentiation Mechanism of Oriental River Prawn, Macrobrachium nipponense, During the Gonad Differentiation and Development Period. Front. Genet. 2022, 13, 797796. [Google Scholar] [CrossRef]
- Jin, S.B.; Fu, H.T.; Jiang, S.F.; Xiong, Y.W.; Qiao, H.; Zhang, W.Y.; Gong, Y.S.; Wu, Y. Identification of androgenic gland microRNA and their target genes to discover sex-related microRNA in the oriental river prawn, Macrobrachium nipponense. Genet. Mol. Res. 2015, 14, 18396–18406. [Google Scholar] [CrossRef]
- Luo, B.Y.; Xiong, X.Y.; Liu, X.; He, X.Y.; Qiu, G.F. Identification and characterization of sex-biased and differentially expressed miRNAs in gonadal developments of the Chinese mitten crab, Eriocheir sinensis. Mol. Reprod. Dev. 2021, 88, 217–227. [Google Scholar] [CrossRef]
- Meng, X.L.; Zhang, X.H.; Li, J.; Liu, P. Identification and comparative profiling of ovarian and testicular microRNAs in the swimming crab Portunus trituberculatus. Gene 2018, 640, 6–13. [Google Scholar] [CrossRef]
- Song, Y.N.; Shi, L.L.; Liu, Z.Q.; Qiu, G.F. Global analysis of the ovarian microRNA transcriptome: Implication for miR-2 and miR-133 regulation of oocyte meiosis in the Chinese mitten crab, Eriocheir sinensis (Crustacea: Decapoda). BMC Genom. 2014, 15, 547. [Google Scholar] [CrossRef] [Green Version]
- Li, S.H.; Li, F.H.; Sun, Z.; Xiang, J.H. Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis. Gen. Comp. Endocrinol. 2012, 177, 246–255. [Google Scholar] [CrossRef]
- Huang, X.S.; Ye, H.H.; Huang, H.Y.; Yang, Y.N.; Gong, J. An insulinlike androgenic gland hormone gene in the mud crab, Scylla paramamosain, extensively expressed and involved in the processes of growth and female reproduction. Gen. Comp. Endocrinol. 2014, 204, 229–238. [Google Scholar] [CrossRef]
- Li, F.J.; Jiang, F.W.; Bai, H.K.; Fu, H.T.; Jin, S.B.; Sun, S.M.; Qiao, H.; Zhang, W.Y. Genomic cloning, expression, and single nucleotide polymorphism association analysis of the insulin-like androgenic gland hormone gene in the oriental river prawn (Macrobrachium nipponense). Genet. Mol. Res. 2015, 14, 5910–5921. [Google Scholar] [CrossRef]
- Levy, T.; Rosen, O.; Simons, O.; Savaya Alkalay, A.; Sagi, A. The gene encoding the insulin-like androgenic gland hormone in an all-female parthenogenetic crayfish. PLoS ONE 2017, 12, e0189982. [Google Scholar] [CrossRef]
- Ma, K.Y.; Li, J.L.; Qiu, G.F. Identification of putative regulatory region of insulin-like androgenic gland hormone gene (IAG) in the prawn Macrobrachium nipponense and proteins that interact with IAG by using yeast two-hybrid system. Gen. Comp. Endocrinol. 2016, 229, 112–118. [Google Scholar] [CrossRef]
- Modi, D.; Shah, C.; Sachdeva, G.; Gadkar, S.; Bhartiya, D.; Puri, C. Ontogeny and cellular localization of SRY transcripts in the human testes and its detection in spermatozoa. Reproduction 2005, 130, 603–613. [Google Scholar] [CrossRef]
- Graham, J.D.; Hunt, S.M.; Tran, N.; Clarke, C.L. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J. Mol. Endocrinol. 1999, 22, 295–304. [Google Scholar] [CrossRef]
- Manek, R.; Nelson, T.; Tseng, E.; Rodriguez-Lebron, E. 5′ UTR-mediated regulation of Ataxin-1 expression. Neurobiol. Dis. 2020, 134, 104564. [Google Scholar] [CrossRef]
- Créancier, L.; Morello, D.; Mercier, P.; Prats, A.C. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 2000, 150, 275–281. [Google Scholar] [CrossRef]
- Golestaneh, N.; Beauchamp, E.; Fallen, S.; Kokkinaki, M.; Uren, A.; Dym, M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction 2009, 138, 151–162. [Google Scholar] [CrossRef] [Green Version]
- Biason-Lauber, A.; Konrad, D.; Navratil, F.; Schoenle, E.J. A WNT4 mutation associated with Müllerian-duct regression and virilization in a 46, XX woman. N. Engl. J. Med. 2004, 351, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.A.; Rhee, W.J. Exosomes: Biogenesis, Composition, Functions, and Their Role in Pre-metastatic Niche Formation. Biotechnol. Bioprocess Eng. 2019, 24, 689–701. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Yu, S.N.; Handler, A.M.; Tu, Z.J.; Saccone, G.; Xi, Z.; Zhang, H.Y. miRNA-1-3p is an early embryonic male sex-determining factor in the Oriental fruit fly Bactrocera dorsalis. Nat. Commun. 2020, 11, 932. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, Y.L.; Li, Q.; Yang, H.D.; Duan, Z.L.; Wang, Q. Profiling microRNAs in the testis during sexual maturation stages in Eriocheir sinensis. Anim. Reprod. Sci. 2015, 162, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.H.; Wang, W.M.; Liu, X.L.; Luo, W.; Zhang, J.; Gul, Y. DNA extraction from crayfish exoskeleton. Indian J. Exp. Biol. 2011, 49, 953–957. [Google Scholar]
- Shi, L.L.; Han, S.X.; Fei, J.M.; Zhang, L.; Ray, J.W.; Wang, W.M.; Li, Y.H. Molecular Characterization and Functional Study of Insulin-Like Androgenic Gland Hormone Gene in the Red Swamp Crayfish, Procambarus clarkii. Genes 2019, 10, 645. [Google Scholar] [CrossRef] [Green Version]

| miRNA | Sequence | Whether Differential Expression | |
|---|---|---|---|
| bantam-3p | aae-bantam-3p | UGAGAUCAUUUUGAAAGCUGAU | yes |
| bmo-bantam-3p | UGAGAUCAUUGUGAAAGCUAAUU | yes | |
| Let-7 | aae-let-7 | UGAGGUAGUUGGUUGUAUAGU | |
| bdo-let-7 | UGAGGUAGUAGGUUGUAUAGU | ||
| miR-7 | dpu-miR-7 | UGGAAGACUAGUGAUUUUGUUGU | |
| tcf-miR-7 | UGGAAGACUAGUGAUUUUGUUGUU | ||
| miR-9a | aae-miR-9a | UCUUUGGUUAUCUAGCUGUAUGA | |
| miR-9a-5p | bmo-miR-9a-5p | UCUUUGGUUAUCUAGCUGUAUGA | |
| dme-miR-9a-5p | UCUUUGGUUAUCUAGCUGUAUGA | ||
| tcf-miR-9a-5p | UCUUUGGUUAUCUAGCUGUAUGA | ||
| miR-10 | dpu-miR-10 | UACCCUGUAGAUCCGAAUUUGU | |
| miR-34 | dpu-miR-34 | UGGCAGUGUGGUUAGCUGGUUGUG | |
| tcf-miR-34 | UGGCAGUGUGGUUAGCUGGUUG | yes | |
| miR-133 | aae-miR-133 | UUGGUCCCCUUCAACCAGCUGU | |
| dpu-miR-133 | UUGGUCCCCUUCAACCAGCUGU | ||
| tcf-miR-133 | UUGGUCCCCUUCAACCAGCUGU | yes | |
| miR-190 | aae-miR-190 | AGAUAUGUUUGAUAUUCUUGGUUG | |
| bdo-miR-190 | AGAUAUGUUUGAUAUUCUUGGUUG | ||
| tcf-miR-190 | AGAUAUGUUUGAUAUUCUUGGUUG | ||
| miR-190-5p | bmo-miR-190-5p | AGAUAUGUUUGAUAUUCUUGGUU | |
| miR-263a | dpu-miR-263a | AAUGGCACUGGAAGAAUUCAC | |
| tcf-miR-263a | AAUGGCACUGGAAGAAUUCACGGG | yes | |
| bdo-miR-263a | AAUGGCACUGGAAGAAUUCACGGG | ||
| miR-263b | bdo-miR-263b | CUUGGCACUGGGAGAAUUCACAG | yes |
| tcf-miR-263b | CUUGGCACUGGAAGAAUUCACAGA | yes | |
| dpu-miR-263b | CUUGGCACUGGAAGAAUUCACA | yes | |
| miR-307 | dpu-miR-307 | UCACAACCUCCUUGAGUGAG | |
| tcf-miR-307 | UCACAACCUCCUUGAGUGAGUG |
| After siRNA Interference with PcIAG, the Top 10 miRNAs Expressed | miRNA Sequence | miRNAs in Exosomes before and after the Androgenic Gland Ablation |
|---|---|---|
| tcf-miR-1 | UGGAAUGUAAAGAAGUAUGGAG | pte-miR-1-3p |
| tcf-miR-100 | AACCCGUAGAUCCGAACUUGUGU | hpo-miR-100-5p |
| tcf-let-7-5p | UGAGGUAGUAGGUUGUAUGGUU | oga-let-7c |
| tcf-miR-9a-5p | UCUUUGGUUAUCUAGCUGUAUGA | |
| tcf-miR-9b-5p | UCUUUGGUGGUCUAGCUGUAUGA | |
| tcf-miR-279a | UGACUAGAUCCACACUCATCCA | |
| SEQ14455_4438 | UUGAGCAAAGCUUCAGGGGGUUU | animal-m0537-3p |
| SEQ23317_6170 | AAAUAUCAGCUGGUAAAUUUGG | |
| SEQ52799_9422 | UAUUAUGCUAAGAUUCGUGUAU | |
| SEQ52799_9421 | ||
| SEQ39002_8168 | CAUCACAGUGAUAGUACCUUACU | animal-m0495-3p |
| SEQ39002_8166 | ||
| SEQ396075_21005 | CCAAAAGGCCGAGAAGCGAUCACAU | |
| SEQ304206_18791 | CCCUCAGGAUAGCUGGAAC | |
| SEQC9500399_28213 | UGACUAGAGGACUACUCAUCC | animal-m0377-3p |
| Differentially Expressed miRNAs after siRNA Interference | miRNA Sequence | Differentially Expressed miRNA in Exosomes | miRNA Sequence |
|---|---|---|---|
| tcf-miR-133 | UUGGUCCCCUUCAACCAGCUGU | animal-mir-133-3 | UUGGUCCCCUUCAACCAGCUGU |
| aae-miR-193 | UACUGGCCUACUAAGUCCCAAC | animal-mir-193-5 | AACUGGCCCUCAAAGUCCCGCU |
| tcf-miR-193 | UACUGGCCUGCUAAGUCCCAAG | ||
| tcf-miR-34 | UGGCAGUGUGGUUAGCUGGUUG | animal-mir-34-5 | AGGCAGUGUAGUUAGCUGAUUGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Sun, R.; Chen, X.; Wang, Q.; Feng, P.; Zhao, Y.; Li, Y. Two Short Repeats in the 5′ Untranslated Region of Insulin-like Androgenic Gland Factor in Procambarus clarkii (PcIAG) That Regulate PcIAG Expression. Int. J. Mol. Sci. 2022, 23, 10348. https://doi.org/10.3390/ijms231810348
Yang S, Sun R, Chen X, Wang Q, Feng P, Zhao Y, Li Y. Two Short Repeats in the 5′ Untranslated Region of Insulin-like Androgenic Gland Factor in Procambarus clarkii (PcIAG) That Regulate PcIAG Expression. International Journal of Molecular Sciences. 2022; 23(18):10348. https://doi.org/10.3390/ijms231810348
Chicago/Turabian StyleYang, Siqi, Rong Sun, Xiuli Chen, Qishuai Wang, Pengfei Feng, Yongzhen Zhao, and Yanhe Li. 2022. "Two Short Repeats in the 5′ Untranslated Region of Insulin-like Androgenic Gland Factor in Procambarus clarkii (PcIAG) That Regulate PcIAG Expression" International Journal of Molecular Sciences 23, no. 18: 10348. https://doi.org/10.3390/ijms231810348
APA StyleYang, S., Sun, R., Chen, X., Wang, Q., Feng, P., Zhao, Y., & Li, Y. (2022). Two Short Repeats in the 5′ Untranslated Region of Insulin-like Androgenic Gland Factor in Procambarus clarkii (PcIAG) That Regulate PcIAG Expression. International Journal of Molecular Sciences, 23(18), 10348. https://doi.org/10.3390/ijms231810348






