Electrostatics in Computational Biophysics and Its Implications for Disease Effects
Abstract
:1. Introduction
2. Electrostatics of Wild-Type Biological Macromolecules
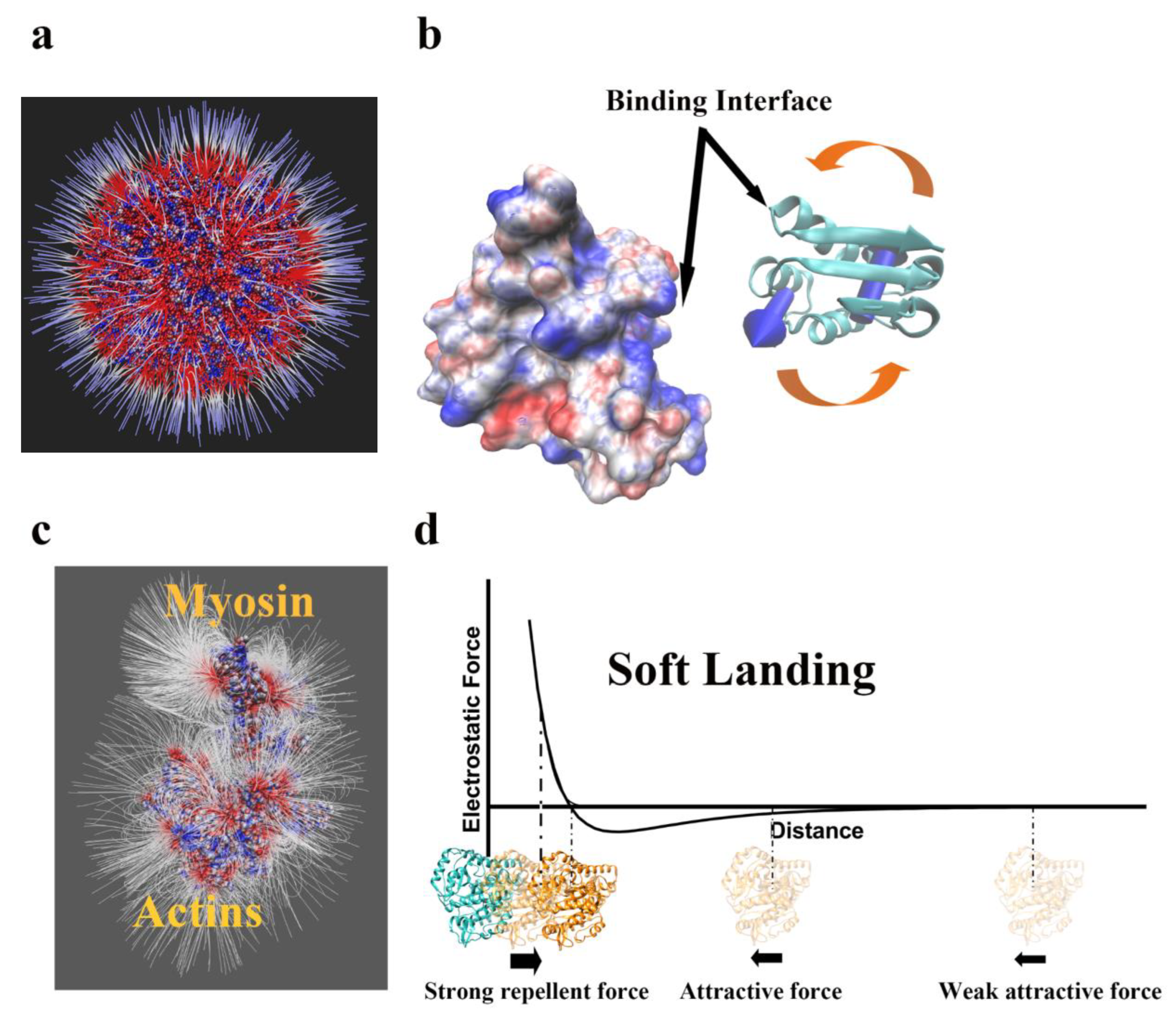
2.1. Long-Range Electrostatic Effects
2.2. Short-Range Electrostatic Effects
2.3. pKa Calculations and pH-Dependent Phenomena
3. Electrostatics and Disease Mechanisms
3.1. Salt-Bridge Disruption
3.2. Hydrogen Bond Disruption
3.3. pH-Dependence Alteration
4. In Silico Drug Discovery
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sagui, C.; Darden, T.A. Molecular dynamics simulations of biomolecules: Long-range electrostatic effects. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 155–179. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, Y.; Li, L.; Jia, Z.; Cao, W.; Alexov, E. DFMD: Fast and effective DelPhiForce steered molecular dynamics approach to model ligand approach toward a receptor: Application to spermine synthase enzyme. Front. Mol. Biosci. 2019, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nussinov, R. Salt bridge stability in monomeric proteins. J. Mol. Biol. 1999, 293, 1241–1255. [Google Scholar] [CrossRef]
- Sinha, N.; Smith-Gill, S.J. Electrostatics in protein binding and function. Curr. Protein Pept. Sci. 2002, 3, 601–614. [Google Scholar] [CrossRef]
- Sun, S.; Karki, C.; Gao, B.Z.; Li, L. Molecular mechanisms of cardiac actomyosin transforming from rigor state to post-rigor state. J. Chem. Phys. 2022, 156, 035101. [Google Scholar] [CrossRef]
- Xie, Y.; Guo, W.; Lopez-Hernadez, A.; Teng, S.; Li, L. The pH Effects on SARS-CoV and SARS-CoV-2 Spike Proteins in the Process of Binding to hACE2. Pathogens 2022, 11, 238. [Google Scholar] [CrossRef] [PubMed]
- Talley, K.; Alexov, E. On the pH-optimum of activity and stability of proteins. Proteins 2010, 78, 2699–2706. [Google Scholar] [CrossRef]
- Koirala, M.; Shashikala, H.M.; Jeffries, J.; Wu, B.; Loftus, S.K.; Zippin, J.H.; Alexov, E. Computational Investigation of the pH Dependence of Stability of Melanosome Proteins: Implication for Melanosome formation and Disease. Int. J. Mol. Sci. 2021, 22, 8273. [Google Scholar] [CrossRef]
- York, D.M.; Darden, T.A.; Pedersen, L.G. The effect of long-range electrostatic interactions in simulations of macromolecular crystals: A comparison of the Ewald and truncated list methods. J. Chem. Phys. 1993, 99, 8345–8348. [Google Scholar] [CrossRef]
- Koirala, M.; Alexov, E. Ab-initio binding of barnase–barstar with DelPhiForce steered Molecular Dynamics (DFMD) approach. J. Theor. Comput. Chem. 2020, 19, 2050016. [Google Scholar] [CrossRef]
- Shashikala, H.M.; Chakravorty, A.; Alexov, E. Modeling electrostatic force in protein-protein recognition. Front. Mol. Biosci. 2019, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Alper, J.; Alexov, E. Cytoplasmic dynein binding, run length, and velocity are guided by long-range electrostatic interactions. Sci. Rep. 2016, 6, 31523. [Google Scholar] [CrossRef]
- Norberg, J.; Nilsson, L. On the truncation of long-range electrostatic interactions in DNA. Biophys. J. 2000, 79, 1537–1553. [Google Scholar] [CrossRef]
- Kodchakorn, K.; Chokepaichitkool, T.; Kongtawelert, P. Mutational scanning of spike RBD protein for enhanced ACE2 affinity emerging Southeast Asia in the late transmission phase. Sci. Rep. 2022, 12, 5896. [Google Scholar] [CrossRef]
- Dushanan, R.; Weerasinghe, S.; Dissanayake, D.; Senthilnithy, R. An In-Silico Approach to Evaluate the Inhibitory Potency of Selected Hydroxamic Acid Derivatives on Zinc-Dependent Histone Deacetylase Enzyme. J. Comput. Biophys. Chem. 2021, 20, 603–618. [Google Scholar] [CrossRef]
- King, K.M.; Sharp, A.K.; Davidson, D.S.; Brown, A.M.; Lemkul, J.A. Impact of Electronic Polarization on Preformed, β-Strand Rich Homogenous and Heterogeneous Amyloid Oligomers; World Scientific: Singapore, 2021. [Google Scholar]
- Li, T.; Stevens, A.O.; Gil Pineda, L.I.; Song, S.; Ameyaw Baah, C.A.; He, Y. Changes in structure and flexibility of p53 TAD2 upon binding to p300 Taz2. J. Theor. Comput. Chem. 2020, 19, 2040007. [Google Scholar] [CrossRef]
- Peng, Y.; Alexov, E. Computational investigation of proton transfer, pKa shifts and pH-optimum of protein-DNA and protein-RNA complexes. Proteins 2017, 85, 282–295. [Google Scholar] [CrossRef]
- Arthur, E.J.; Brooks III, C.L. Efficient implementation of constant pH molecular dynamics on modern graphics processors. J. Comput. Chem. 2016, 37, 2171–2180. [Google Scholar] [CrossRef]
- Harris, R.C.; Shen, J. GPU-accelerated implementation of continuous constant pH molecular dynamics in amber: pKa predictions with single-pH simulations. J. Chem. Inf. Model. 2019, 59, 4821–4832. [Google Scholar] [CrossRef]
- Vila-Viçosa, D.; Reis, P.B.; Baptista, A.M.; Oostenbrink, C.; Machuqueiro, M. A pH replica exchange scheme in the stochastic titration constant-pH MD method. J. Chem. Theory Comput. 2019, 15, 3108–3116. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Roitberg, A.E. Constant pH replica exchange molecular dynamics in biomolecules using a discrete protonation model. J. Chem. Theory Comput. 2010, 6, 1401–1412. [Google Scholar] [CrossRef]
- Oliveira, N.F.; Machuqueiro, M. Novel US-CpHMD Protocol to Study the Protonation-Dependent Mechanism of the ATP/ADP Carrier. J. Chem. Inf. Model. 2022, 62, 2550–2560. [Google Scholar] [CrossRef] [PubMed]
- Reilley, D.J.; Wang, J.; Dokholyan, N.V.; Alexandrova, A.N. Titr-DMD—A Rapid, Coarse-Grained Quasi-All-Atom Constant pH Molecular Dynamics Framework. J. Chem. Theory Comput. 2021, 17, 4538–4549. [Google Scholar] [CrossRef] [PubMed]
- Baruah, I.; Borgohain, G. Binding interaction of a potential statin with β-lactoglobulin: An in silico approach. J. Mol. Graph. Model. 2022, 111, 108077. [Google Scholar] [CrossRef]
- Teng, S.; Madej, T.; Panchenko, A.; Alexov, E. Modeling effects of human single nucleotide polymorphisms on protein-protein interactions. Biophys. J. 2009, 96, 2178–2188. [Google Scholar] [CrossRef] [PubMed]
- Stefl, S.; Nishi, H.; Petukh, M.; Panchenko, A.R.; Alexov, E. Molecular mechanisms of disease-causing missense mutations. J. Mol. Biol. 2013, 425, 3919–3936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Martiny, V.; Lagorce, D.; Ikeguchi, Y.; Alexov, E.; Miteva, M.A. Rational design of small-molecule stabilizers of spermine synthase dimer by virtual screening and free energy-based approach. PLoS ONE 2014, 9, e110884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Witham, S.; Petukh, M.; Moroy, G.; Miteva, M.; Ikeguchi, Y.; Alexov, E. A rational free energy-based approach to understanding and targeting disease-causing missense mutations. J. Am. Med. Inform. Assoc. 2013, 20, 643–651. [Google Scholar] [CrossRef]
- Peng, Y.; Norris, J.; Schwartz, C.; Alexov, E. Revealing the effects of missense mutations causing Snyder-Robinson syndrome on the stability and dimerization of spermine synthase. Int. J. Mol. Sci. 2016, 17, 77. [Google Scholar] [CrossRef]
- Zhang, Z.; Norris, J.; Kalscheuer, V.; Wood, T.; Wang, L.; Schwartz, C.; Alexov, E.; Van Esch, H. A Y328C missense mutation in spermine synthase causes a mild form of Snyder–Robinson syndrome. Hum. Mol. Genet. 2013, 22, 3789–3797. [Google Scholar] [CrossRef]
- Zhang, Z.; Norris, J.; Schwartz, C.; Alexov, E. In silico and in vitro investigations of the mutability of disease-causing missense mutation sites in spermine synthase. PLoS ONE 2011, 6, e20373. [Google Scholar] [CrossRef]
- Zhang, Z.; Teng, S.; Wang, L.; Schwartz, C.E.; Alexov, E. Computational analysis of missense mutations causing Snyder-Robinson syndrome. Hum. Mutat. 2010, 31, 1043–1049. [Google Scholar] [CrossRef]
- Andac, C.A.; Cakmak, O.; Okten, S.; Caglar-Andac, S.; IŞILDAK, İ. In-silico pharmacokinetic and affinity studies of piperazine/morpholine substituted quinolines in complex with GAK as promising anti-HCV agent. J. Comput. Biophys. Chem. 2021, 20, 869–879. [Google Scholar] [CrossRef]
- Xue, W.; Pan, D.; Yang, Y.; Liu, H.; Yao, X. Molecular modeling study on the resistance mechanism of HCV NS3/4A serine protease mutants R155K, A156V and D168A to TMC435. Antivir. Res. 2012, 93, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Sharafdini, R.; Mosaddeghi, H. Inhibition of Insulin Amyloid Fibrillation by Salvianolic Acids and Calix [n] arenes: Molecular Docking Insight. J. Comput. Biophys. Chem. 2021, 20, 539–555. [Google Scholar] [CrossRef]
- da Fonseca, A.M.; de Araújo, F.A.M.; Carvalho, R.M.M.; Silva de Menezes, J.F.; Sa Pires Silva, A.M. Molecular Docking Study of Antibiotics, Anti-Inflammatory Drugs and [Eu (TTA)3⋅AMX] Complex as COVID-19 Biomarker through Interaction of Its Main Protease (Mpro). J. Comput. Biophys. Chem. 2021, 20, 405–415. [Google Scholar] [CrossRef]
- Alibakhshi, A.; Ahangarzadeh, S.; Beikmohammadi, L.; Soltanmohammadi, B.; Bahrami, A.A.; Ranjbar, M.M.; Mohammadi, E. Computational design of a potential therapeutic peptide against spike protein of SARS-CoV-2. J. Comput. Biophys. Chem. 2021, 20, 337–346. [Google Scholar] [CrossRef]
- Llamas-García, M.L.; Páez-Pérez, E.D.; Benitez-Cardoza, C.G.; Montero-Morán, G.M.; Lara-González, S. Improved Stability of Human CGI-58 Induced by Phosphomimetic S237E Mutation. ACS Omega 2022, 7, 12643–12653. [Google Scholar] [CrossRef]
- Xian, Y.; Xie, Y.; Silva, S.M.; Karki, C.B.; Qiu, W.; Li, L. StructureMan: A structure manipulation tool to study large scale biomolecular interactions. Front. Mol. Biosci. 2021, 7, 627087. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Buckle, A.M.; Schreiber, G.; Fersht, A.R. Protein-protein recognition: Crystal structural analysis of a barnase-barstar complex at 2.0-. ANG. resolution. Biochemistry 1994, 33, 8878–8889. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Garcia, R.N.; Adachi, M.; Maruyama, Y.; Tecson-Mendoza, E.M.; Mikami, B.; Utsumi, S. Structure of 8Sα globulin, the major seed storage protein of mung bean. Acta Crystallogr. D 2006, 62, 824–832. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Wu, L.; Niu, S.; Song, C.; Zhang, Z.; Lu, G.; Qiao, C.; Hu, Y.; Yuen, K.-Y. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 2020, 181, 894–904.e9. [Google Scholar] [CrossRef]
- Khan, S.; Ipsen, R.; Almdal, K.; Svensson, B.; Harris, P. Revealing the Dimeric Crystal and Solution Structure of β-Lactoglobulin at pH 4 and Its pH and Salt Dependent Monomer–Dimer Equilibrium. Biomacromolecules 2018, 19, 2905–2912. [Google Scholar] [CrossRef]
- Piana, S.; Lindorff-Larsen, K.; Dirks, R.M.; Salmon, J.K.; Dror, R.O.; Shaw, D.E. Evaluating the effects of cutoffs and treatment of long-range electrostatics in protein folding simulations. PLoS ONE 2012, 7, e39918. [Google Scholar]
- Li, L.; Li, C.; Sarkar, S.; Zhang, J.; Witham, S.; Zhang, Z.; Wang, L.; Smith, N.; Petukh, M.; Alexov, E. DelPhi: A comprehensive suite for DelPhi software and associated resources. BMC Biophys. 2012, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Lopez, J.A.; Xie, Y.; Guo, W.; Liu, D.; Li, L. HIT web server: A hybrid method to improve electrostatic calculations for biomolecules. Comput. Struct. Biotechnol. J. 2022, 20, 1580–1583. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Karki, C.; Xie, Y.; Xian, Y.; Guo, W.; Gao, B.Z.; Li, L. Hybrid method for representing ions in implicit solvation calculations. Comput. Struct. Biotechnol. J. 2021, 19, 801–811. [Google Scholar] [CrossRef]
- Sarkar, S.; Witham, S.; Zhang, J.; Zhenirovskyy, M.; Rocchia, W.; Alexov, E. DelPhi Web Server: A comprehensive online suite for electrostatic calculations of biological macromolecules and their complexes. Commun. Comput. Phys. 2013, 13, 269–284. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chakravorty, A.; Alexov, E. DelPhiForce, a tool for electrostatic force calculations: Applications to macromolecular binding. J. Comput. Chem. 2017, 38, 584–593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Alper, J.; Alexov, E. Multiscale method for modeling binding phenomena involving large objects: Application to kinesin motor domains motion along microtubules. Sci. Rep. 2016, 6, 23249. [Google Scholar] [CrossRef] [PubMed]
- Tajielyato, N.; Li, L.; Peng, Y.; Alper, J.; Alexov, E. E-hooks provide guidance and a soft landing for the microtubule binding domain of dynein. Sci. Rep. 2018, 8, 13266. [Google Scholar] [CrossRef] [PubMed]
- Pabbathi, A.; Coleman, L.; Godar, S.; Paul, A.; Garlapati, A.; Spencer, M.; Eller, J.; Alper, J.D. Long-range electrostatic interactions significantly modulate the affinity of dynein for microtubules. Biophys. J. 2022, 121, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Jia, Z.; Peng, Y.; Chakravorty, A.; Sun, L.; Alexov, E. DelPhiForce web server: Electrostatic forces and energy calculations and visualization. Bioinformatics 2017, 33, 3661–3663. [Google Scholar] [CrossRef] [PubMed]
- Salas, G.G.S.; Hernandez, A.E.L.; He, J.; Karki, C.; Xie, Y.; Sun, S.; Xian, Y.; Li, L. Using computational approaches to study dengue virus capsid assembly. Comput. Math. Math. Phys. 2019, 7, 64–72. [Google Scholar] [CrossRef]
- Xian, Y.; Karki, C.B.; Silva, S.M.; Li, L.; Xiao, C. The roles of electrostatic interactions in capsid assembly mechanisms of giant viruses. Int. J. Mol. Sci. 2019, 20, 1876. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Terrón, M.C.; Khandokar, Y.; Aragao, D.; Hardy, J.M.; Radjainia, M.; Jiménez-Zaragoza, M.; De Pablo, P.J.; Coulibaly, F.; Luque, D. Structural insights into the assembly and regulation of distinct viral capsid complexes. Nat. Commun. 2016, 7, 13014. [Google Scholar] [CrossRef]
- Ganguly, D.; Otieno, S.; Waddell, B.; Iconaru, L.; Kriwacki, R.W.; Chen, J. Electrostatically accelerated coupled binding and folding of intrinsically disordered proteins. J. Mol. Biol. 2012, 422, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yuwen, T.; Zhu, F.; Skrynnikov, N.R. Role of electrostatic interactions in binding of peptides and intrinsically disordered proteins to their folded targets. 1. NMR and MD characterization of the complex between the c-Crk N-SH3 domain and the peptide Sos. Biochemistry 2014, 53, 6473–6495. [Google Scholar] [CrossRef] [PubMed]
- Mendis, J.; Kaya, E.; Kucukkal, T.G. Identification of Hotspot Residues in Binding of SARS-CoV-2 Spike and Human ACE2 Proteins. J. Comput. Biophys. Chem. 2021, 20, 729–739. [Google Scholar] [CrossRef]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.Q.; Folstein, M.F. Insulin, insulin-degrading enzyme and amyloid-β peptide in Alzheimer’s disease: Review and hypothesis. Neurobiol. Aging 2006, 27, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Nandi, T.; Ainavarapu, S.R.K. Native Salt Bridges Are a Key Regulator of Ubiquitin’s Mechanical Stability. J. Phys. Chem. B 2022, 126, 3505–3511. [Google Scholar] [CrossRef]
- Ji, S.; Yang, W.; Yu, W. Understanding the role of the CB1 toggle switch in interaction networks using molecular dynamics simulation. Sci. Rep. 2021, 11, 22369. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, N.; Danger, R.; Chesneau, M.; Brouard, S.; Laurent, A.D. Structural insights into the catalytic mechanism of granzyme B upon substrate and inhibitor binding. J. Mol. Graph. Model. 2022, 114, 108167. [Google Scholar] [CrossRef] [PubMed]
- Rauh, O.; Opper, J.; Sturm, M.; Drexler, N.; Scheub, D.D.; Hansen, U.-P.; Thiel, G.; Schroeder, I. Role of Ion Distribution and Energy Barriers for Concerted Motion of Subunits in Selectivity Filter Gating of a K+ Channel. J. Mol. Biol. 2022, 434, 167522. [Google Scholar] [CrossRef]
- Yao, S.; Ma, B.; Yi, Q.; Guan, M.-X.; Cang, X. Investigating the Broad Matrix-Gate Network in the Mitochondrial ADP/ATP Carrier through Molecular Dynamics Simulations. Molecules 2022, 27, 1071. [Google Scholar] [CrossRef] [PubMed]
- Shahoei, R.; Pangeni, S.; Sanders, M.A.; Zhang, H.; Mladenovic-Lucas, L.; Roush, W.R.; Halvorsen, G.; Kelly, C.V.; Granneman, J.G.; Huang, Y.-M.M. Molecular Modeling of ABHD5 Structure and Ligand Recognition. Front. Mol. Biosci. 2022, 9, 935375. [Google Scholar] [CrossRef]
- Rollins, Z.A.; Faller, R.; George, S.C. Using molecular dynamics simulations to interrogate T cell receptor non-equilibrium kinetics. Comput. Struct. Biotechnol. J. 2022, 20, 2124–2133. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Q.; Pang, Y.; Yuan, C.; Tan, H.; Li, X.; Chen, G. Theoretical investigation on the reaction mechanism of UTP cyclohydrolase. Phys. Chem. Chem. Phys. 2022, 24, 17641–17653. [Google Scholar] [CrossRef] [PubMed]
- Méndez, E.; Videla, P.E.; Laria, D. Equilibrium and Dynamical Characteristics of Hydrogen Bond Bifurcations in Water–Water and Water–Ammonia Dimers: A Path Integral Molecular Dynamics Study. J. Phys. Chem. A 2022, 126, 4721–4733. [Google Scholar] [CrossRef] [PubMed]
- Yun, Y.; Khaliullin, R.Z.; Jung, Y. Correlated Local Fluctuations in the Hydrogen Bond Network of Liquid Water. J. Am. Chem. Soc. 2022, 144, 13127–13136. [Google Scholar] [CrossRef]
- Koirala, M.; Alexov, E. Computational chemistry methods to investigate the effects caused by DNA variants linked with disease. J. Theor. Comput. Chem. 2020, 19, 1930001. [Google Scholar] [CrossRef]
- Liu, W.; Yao, C.; Shang, Q.; Liu, Y.; Liu, C.; Meng, F. Insights into the binding of dorzagliatin with glucokinase: A molecular dynamics simulation. J. Theor. Comput. Chem. 2020, 19, 2050027. [Google Scholar] [CrossRef]
- Bai, H.; Li, J.; Zhang, H.; Liu, S. Simulative Analysis of a Family of DNA Tetrahedrons Produced by Changing the Twisting Number of Each Double Helix. J. Comput. Biophys. Chem. 2021, 20, 529–537. [Google Scholar] [CrossRef]
- Zhou, T.; Wang, H.; Song, L.; Zhao, Y. Computational study of switching mechanism in add A-riboswitch. J. Theor. Comput. Chem. 2020, 19, 2040001. [Google Scholar] [CrossRef]
- Garcia-Moreno, B. Adaptations of proteins to cellular and subcellular pH. J. Biol. 2009, 8, 98. [Google Scholar] [CrossRef] [PubMed]
- Alexov, E. Special Issue on Computational Chemistry Methods to Predict pKa’s of Ionizable Groups in Proteins, RNAs, DNAs and Small Molecules. J. Comput. Biophys. Chem. 2021, 20, 109–110. [Google Scholar] [CrossRef]
- Mitra, R.C.; Zhang, Z.; Alexov, E. In silico modeling of pH-optimum of protein-protein binding. Proteins 2011, 79, 925–936. [Google Scholar] [CrossRef]
- Ma, S.; Henderson, J.A.; Shen, J. Exploring the pH-Dependent Structure–Dynamics–Function Relationship of Human Renin. J. Chem. Inf. Model. 2020, 61, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Verma, N.; Henderson, J.A.; Zhan, S.; Shen, J. Profiling MAP kinase cysteines for targeted covalent inhibitor design. RSC Med. Chem. 2022, 13, 54–63. [Google Scholar] [CrossRef]
- Santos, H.A.; Vila-Viçosa, D.; Teixeira, V.H.; Baptista, A.M.; Machuqueiro, M. Constant-pH MD simulations of DMPA/DMPC lipid bilayers. J. Chem. Theory Comput. 2015, 11, 5973–5979. [Google Scholar] [CrossRef]
- Teixeira, V.H.; Vila-Viçosa, D.; Reis, P.B.; Machuqueiro, M. pKa Values of Titrable Amino Acids at the Water/Membrane Interface. J. Chem. Theory Comput. 2016, 12, 930–934. [Google Scholar] [CrossRef]
- Khaniya, U.; Mao, J.; Wei, R.J.; Gunner, M. Characterizing protein protonation microstates using Monte Carlo sampling. J. Phys. Chem. B 2022, 126, 2476–2485. [Google Scholar] [CrossRef] [PubMed]
- Pahari, S.; Sun, L.; Basu, S.; Alexov, E. DelPhiPKa: Including salt in the calculations and enabling polar residues to titrate. Proteins 2018, 86, 1277–1283. [Google Scholar] [CrossRef]
- Suhail, M. The Target Determination and the Mechanism of Action of Chiral-Antimalarial Drugs: A Docking Approach. J. Comput. Biophys. Chem. 2021, 20, 501–516. [Google Scholar] [CrossRef]
- Chen, J.; Hu, J.; Xu, Y.; Krasny, R.; Geng, W. Computing Protein pKas Using the TABI Poisson–Boltzmann Solver. J. Comput. Biophys. Chem. 2021, 20, 175–187. [Google Scholar] [CrossRef]
- Peng, Y.; Kelle, R.; Little, C.; Michonova, E.; Kornev, K.G.; Alexov, E. pH-dependent interactions of Apolipophorin-III with a lipid disk. J. Comput. Biophys. Chem. 2021, 20, 153–164. [Google Scholar] [CrossRef]
- Sargolzaei, M.; Nikoofard, H.; Jorabchi, M.N. Quantum-chemical ab initio study of side chain pKa of linear and cyclic lysine dipeptides. J. Comput. Biophys. Chem. 2021, 20, 131–139. [Google Scholar] [CrossRef]
- Lopez-Hernandez, A.E.; Xie, Y.; Guo, W.; Li, L. The electrostatic features of dengue virus capsid assembly. J. Comput. Biophys. Chem. 2021, 20, 201–207. [Google Scholar] [CrossRef]
- Zhang, Y.; Haider, K.; Kaur, D.; Ngo, V.A.; Cai, X.; Mao, J.; Khaniya, U.; Zhu, X.; Noskov, S.; Lazaridis, T. Characterizing the water wire in the Gramicidin channel found by Monte Carlo sampling using continuum electrostatics and in molecular dynamics trajectories with conventional or polarizable force fields. J. Comput. Biophys. Chem. 2021, 20, 111–130. [Google Scholar] [CrossRef]
- MacKenzie, D.W.; Schaefer, A.; Steckner, J.; Leo, C.A.; Naser, D.; Artikis, E.; Broom, A.; Ko, T.; Shah, P.; Ney, M.Q. A fine balance of hydrophobic-electrostatic communication pathways in a pH-switching protein. Proc. Natl. Acad. Sci. USA 2022, 119, e2119686119. [Google Scholar] [CrossRef] [PubMed]
- Petukh, M.; Kucukkal, T.G.; Alexov, E. On human disease-causing amino acid variants: Statistical study of sequence and structural patterns. Hum. Mutat. 2015, 36, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Alexov, E. Investigating the linkage between disease-causing amino acid variants and their effect on protein stability and binding. Proteins 2016, 84, 232–239. [Google Scholar] [CrossRef]
- Li, L.; Jia, Z.; Peng, Y.; Godar, S.; Getov, I.; Teng, S.; Alper, J.; Alexov, E. Forces and Disease: Electrostatic force differences caused by mutations in kinesin motor domains can distinguish between disease-causing and non-disease-causing mutations. Sci. Rep. 2017, 7, 8237. [Google Scholar] [CrossRef]
- Karki, C.; Xian, Y.; Xie, Y.; Sun, S.; Lopez-Hernandez, A.E.; Juarez, B.; Wang, J.; Sun, J.; Li, L. A computational model of ESAT-6 complex in membrane. J. Theor. Comput. Chem. 2020, 19, 2040002. [Google Scholar] [CrossRef]
- Wang, L.-Q.; Zhao, K.; Yuan, H.-Y.; Li, X.-N.; Dang, H.-B.; Ma, Y.; Wang, Q.; Wang, C.; Sun, Y.; Chen, J. Genetic prion disease–related mutation E196K displays a novel amyloid fibril structure revealed by cryo-EM. Sci. Adv. 2021, 7, eabg9676. [Google Scholar] [CrossRef]
- Zhou, T.; Tsybovsky, Y.; Gorman, J.; Rapp, M.; Cerutti, G.; Chuang, G.-Y.; Katsamba, P.S.; Sampson, J.M.; Schön, A.; Bimela, J. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe 2020, 28, 867–879.e5. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Escobedo, A.; Topal, B.; Kunze, M.; Aranda, J.; Chiesa, G.; Mungianu, D.; Bernardo-Seisdedos, G.; Eftekharzadeh, B.; Gairí, M.; Pierattelli, R. Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor. Nat. Commun. 2019, 10, 2034. [Google Scholar] [CrossRef] [Green Version]
- Mompeán, M.; Baralle, M.; Buratti, E.; Laurents, D.V. An amyloid-like pathological conformation of TDP-43 is stabilized by hypercooperative hydrogen bonds. Front. Mol. Neurosci. 2016, 9, 125. [Google Scholar] [CrossRef]
- Boyer, D.R.; Li, B.; Sun, C.; Fan, W.; Sawaya, M.R.; Jiang, L.; Eisenberg, D.S. Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol 2019, 26, 1044–1052. [Google Scholar] [CrossRef]
- White, K.A.; Ruiz, D.G.; Szpiech, Z.A.; Strauli, N.B.; Hernandez, R.D.; Jacobson, M.P.; Barber, D.L. Cancer-associated arginine-to-histidine mutations confer a gain in pH sensing to mutant proteins. Sci. Signal. 2017, 10, eaam9931. [Google Scholar] [CrossRef]
- Liu, Y.; White, K.A.; Barber, D.L. Intracellular pH regulates cancer and stem cell behaviors: A protein dynamics perspective. Front. Oncol. 2020, 10, 1401. [Google Scholar] [CrossRef]
- White, K.A.; Kisor, K.; Barber, D.L. Intracellular pH dynamics and charge-changing somatic mutations in cancer. Cancer Metastasis Rev. 2019, 38, 17–24. [Google Scholar] [CrossRef]
- Takano, K.; Liu, D.; Tarpey, P.; Gallant, E.; Lam, A.; Witham, S.; Alexov, E.; Chaubey, A.; Stevenson, R.E.; Schwartz, C.E. An X-linked channelopathy with cardiomegaly due to a CLIC2 mutation enhancing ryanodine receptor channel activity. Hum. Mol. Genet. 2012, 21, 4497–4507. [Google Scholar] [CrossRef]
- Warwicker, J. A model for pH coupling of the SARS-CoV-2 spike protein open/closed equilibrium. Brief. Bioinform. 2021, 22, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.R.; Warwicker, J. Predicted pH-dependent stability of SARS-CoV-2 spike protein trimer from interfacial acidic groups. Comput. Struct. Biotechnol. J. 2021, 19, 5140–5148. [Google Scholar] [CrossRef] [PubMed]
- Kucukkal, T.G.; Yang, Y.; Uvarov, O.; Cao, W.; Alexov, E. Impact of rett syndrome mutations on MeCP2 MBD stability. Biochemistry 2015, 54, 6357–6368. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Kucukkal, T.G.; Li, J.; Alexov, E.; Cao, W. Binding analysis of methyl-CpG binding domain of MeCP2 and Rett syndrome mutations. ACS Chem. Biol. 2016, 11, 2706–2715. [Google Scholar] [CrossRef] [PubMed]
- Wassman, C.D.; Baronio, R.; Demir, Ö.; Wallentine, B.D.; Chen, C.-K.; Hall, L.V.; Salehi, F.; Lin, D.-W.; Chung, B.P.; Wesley Hatfield, G. Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun. 2013, 4, 1407. [Google Scholar] [CrossRef] [PubMed]
- İş, Y.S. Elucidation of Ligand/Protein Interactions between BCR-ABL Tyrosine Kinase and Some Commercial Anticancer Drugs Via DFT Methods. J. Comput. Biophys. Chem. 2021, 20, 433–447. [Google Scholar] [CrossRef]
- Panigrahi, D. Molecular Docking Analysis of the Phytochemicals from Tinospora Cordifolia as Potential Inhibitor Against Multi Targeted SARS-CoV-2 & Cytokine Storm. J. Comput. Biophys. Chem. 2021, 20, 559–580. [Google Scholar]
- Ghamsari, P.A.; Samadizadeh, M.; Mirzaei, M. Halogenated derivatives of cytidine: Structural analysis and binding affinity. J. Theor. Comput. Chem. 2020, 19, 2050033. [Google Scholar] [CrossRef]
- Qader, S.W.; Abdallah, H.H.; Zahid, M.; Chua, L.S. In vitro Acetylcholinesterase inhibitory activity of polyphenolic compounds identified from Matricaria recutita. J. Theor. Comput. Chem. 2020, 19, 2050029. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, S.; Poudel, P.; Alexov, E.; Li, L. Electrostatics in Computational Biophysics and Its Implications for Disease Effects. Int. J. Mol. Sci. 2022, 23, 10347. https://doi.org/10.3390/ijms231810347
Sun S, Poudel P, Alexov E, Li L. Electrostatics in Computational Biophysics and Its Implications for Disease Effects. International Journal of Molecular Sciences. 2022; 23(18):10347. https://doi.org/10.3390/ijms231810347
Chicago/Turabian StyleSun, Shengjie, Pitambar Poudel, Emil Alexov, and Lin Li. 2022. "Electrostatics in Computational Biophysics and Its Implications for Disease Effects" International Journal of Molecular Sciences 23, no. 18: 10347. https://doi.org/10.3390/ijms231810347
APA StyleSun, S., Poudel, P., Alexov, E., & Li, L. (2022). Electrostatics in Computational Biophysics and Its Implications for Disease Effects. International Journal of Molecular Sciences, 23(18), 10347. https://doi.org/10.3390/ijms231810347






