Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies
Abstract
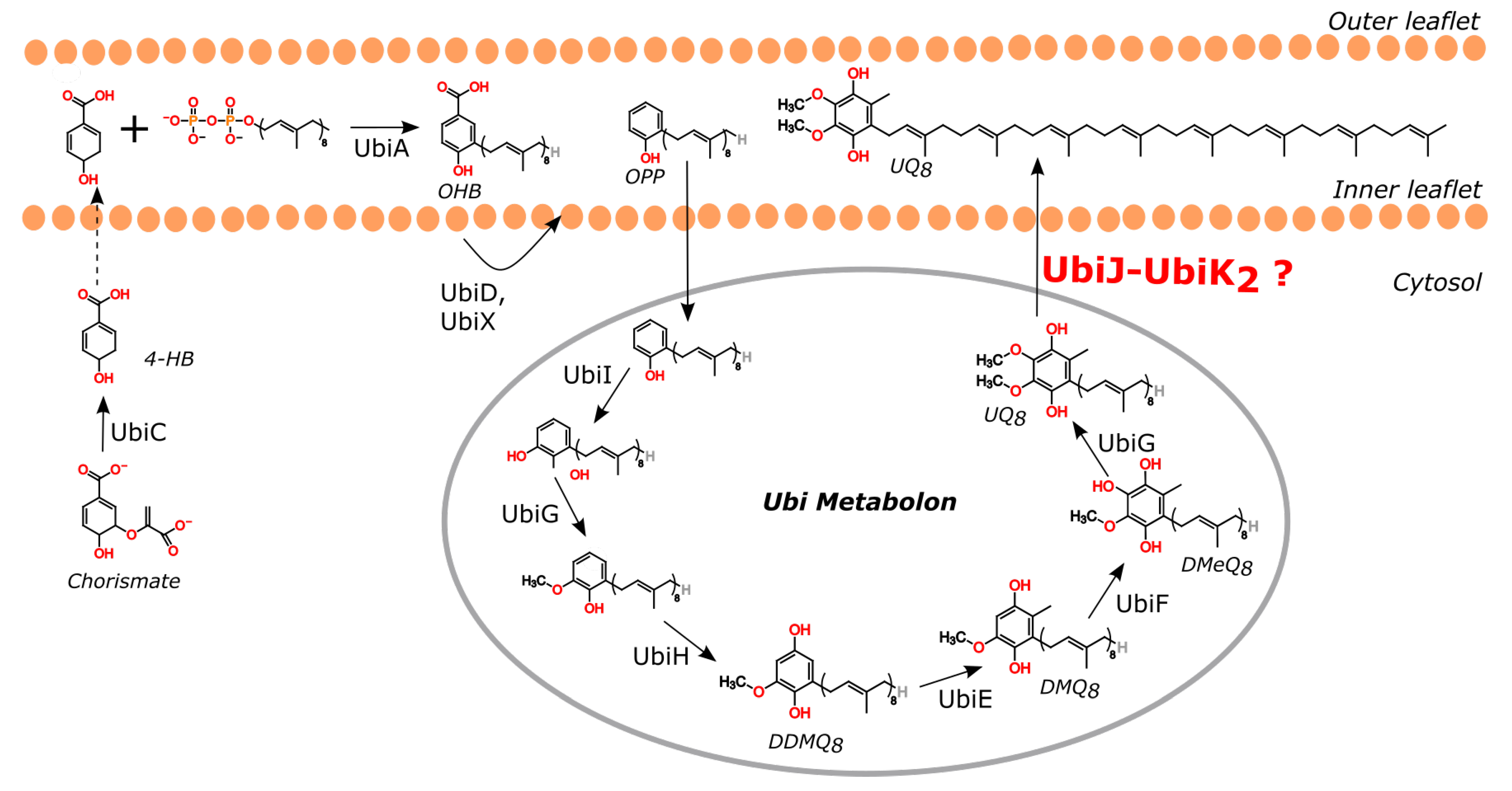
1. Introduction
2. Results and Discussion
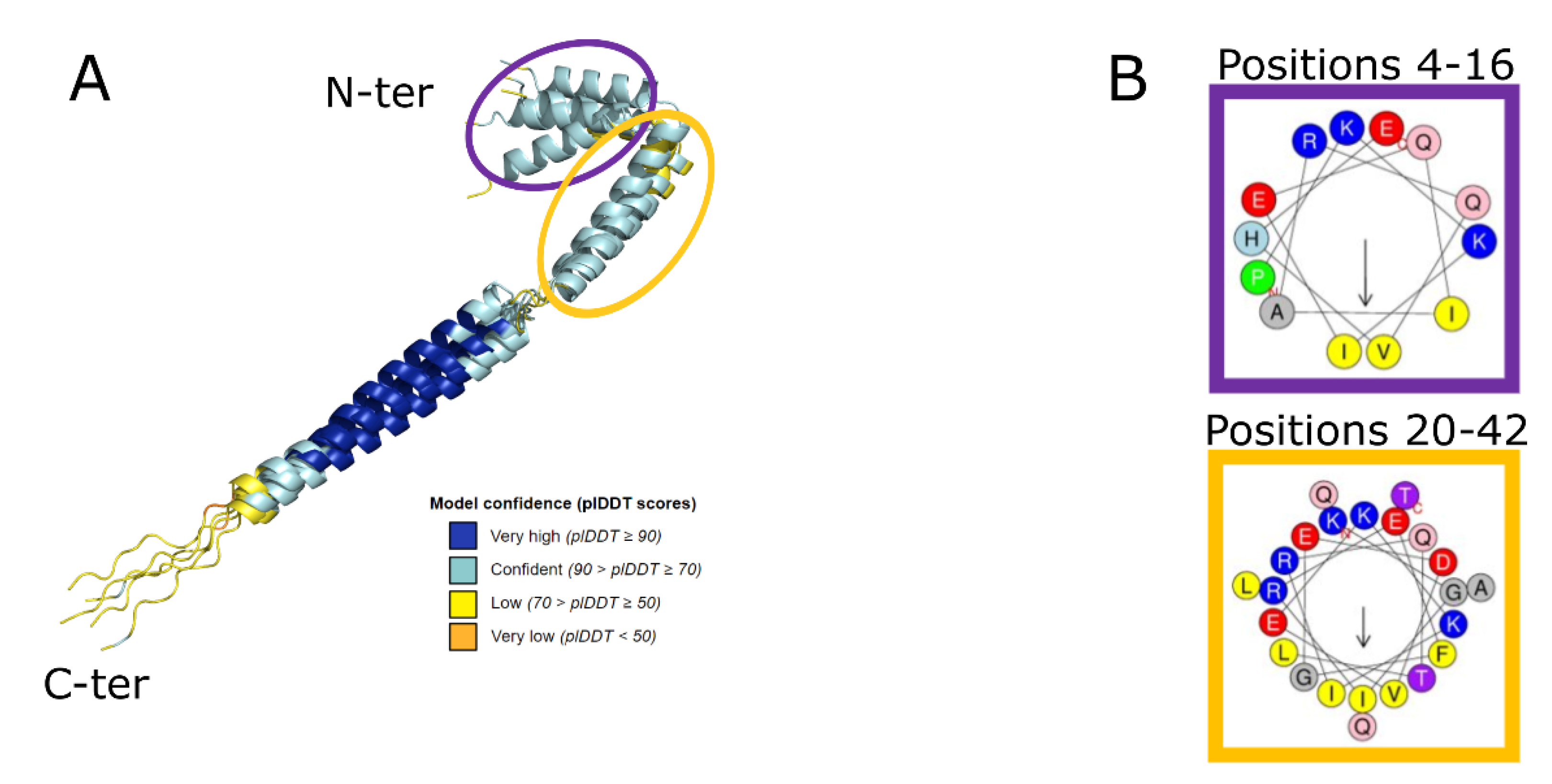
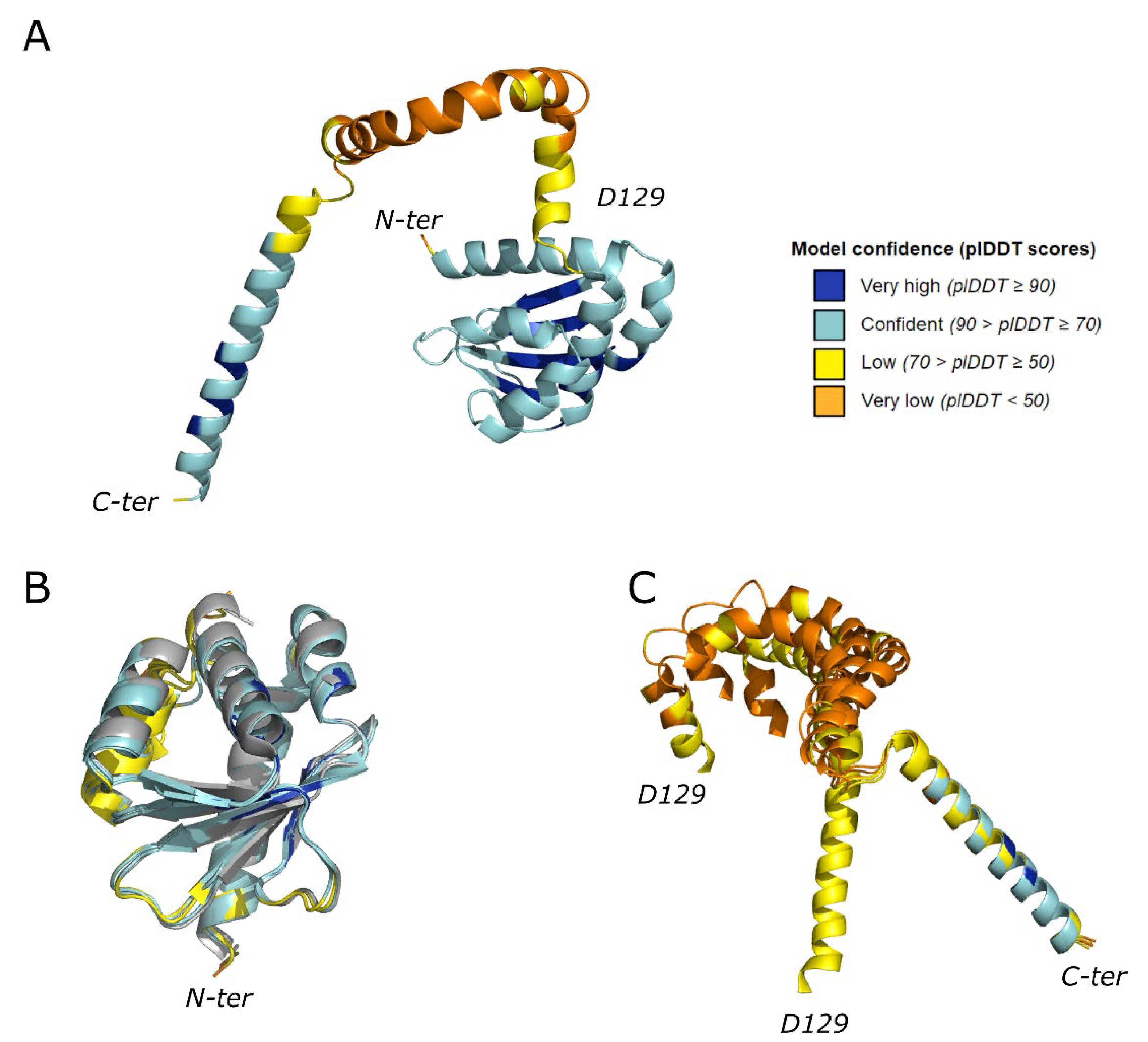
2.1. Protein Monomer Modelling
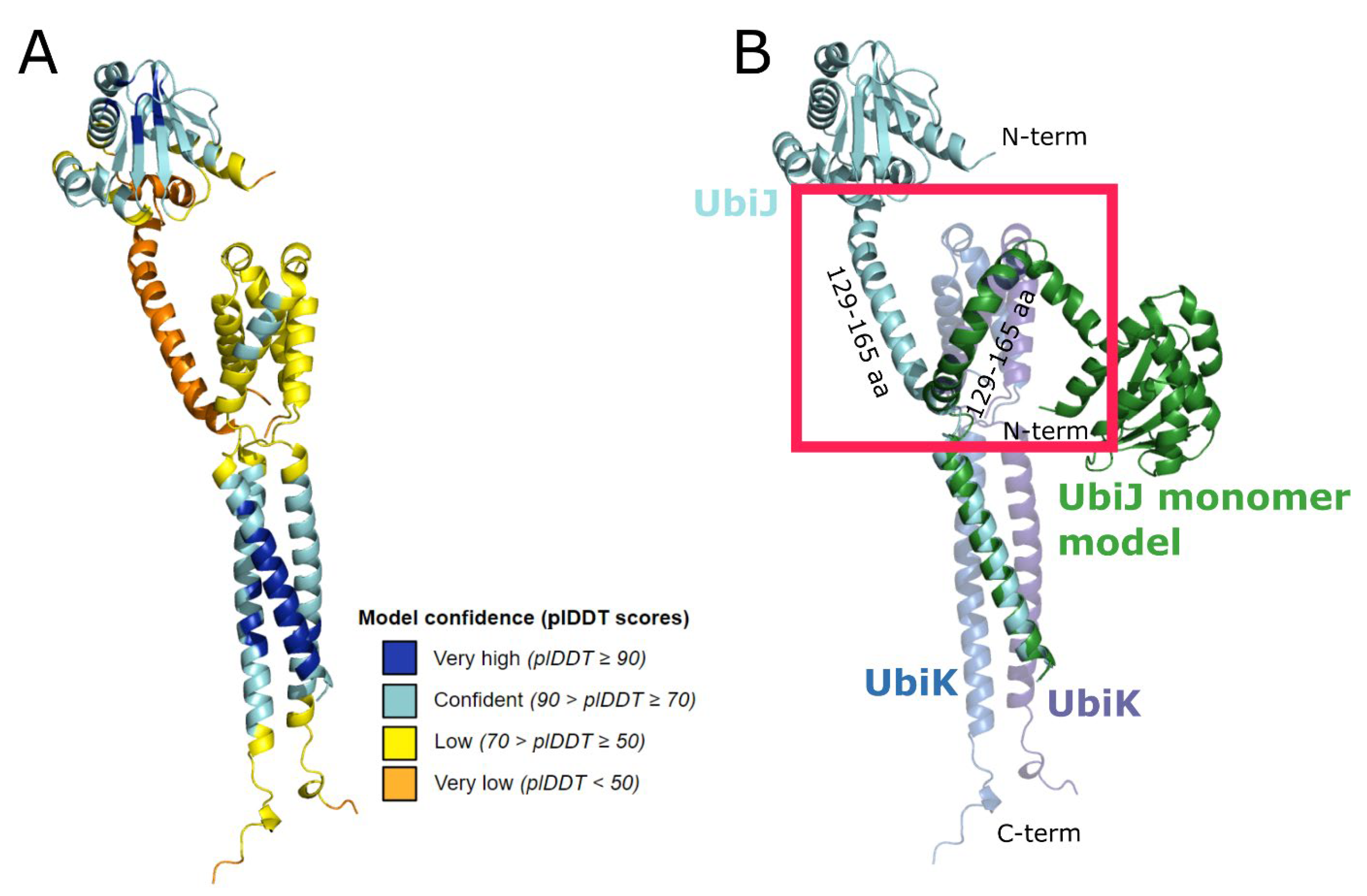
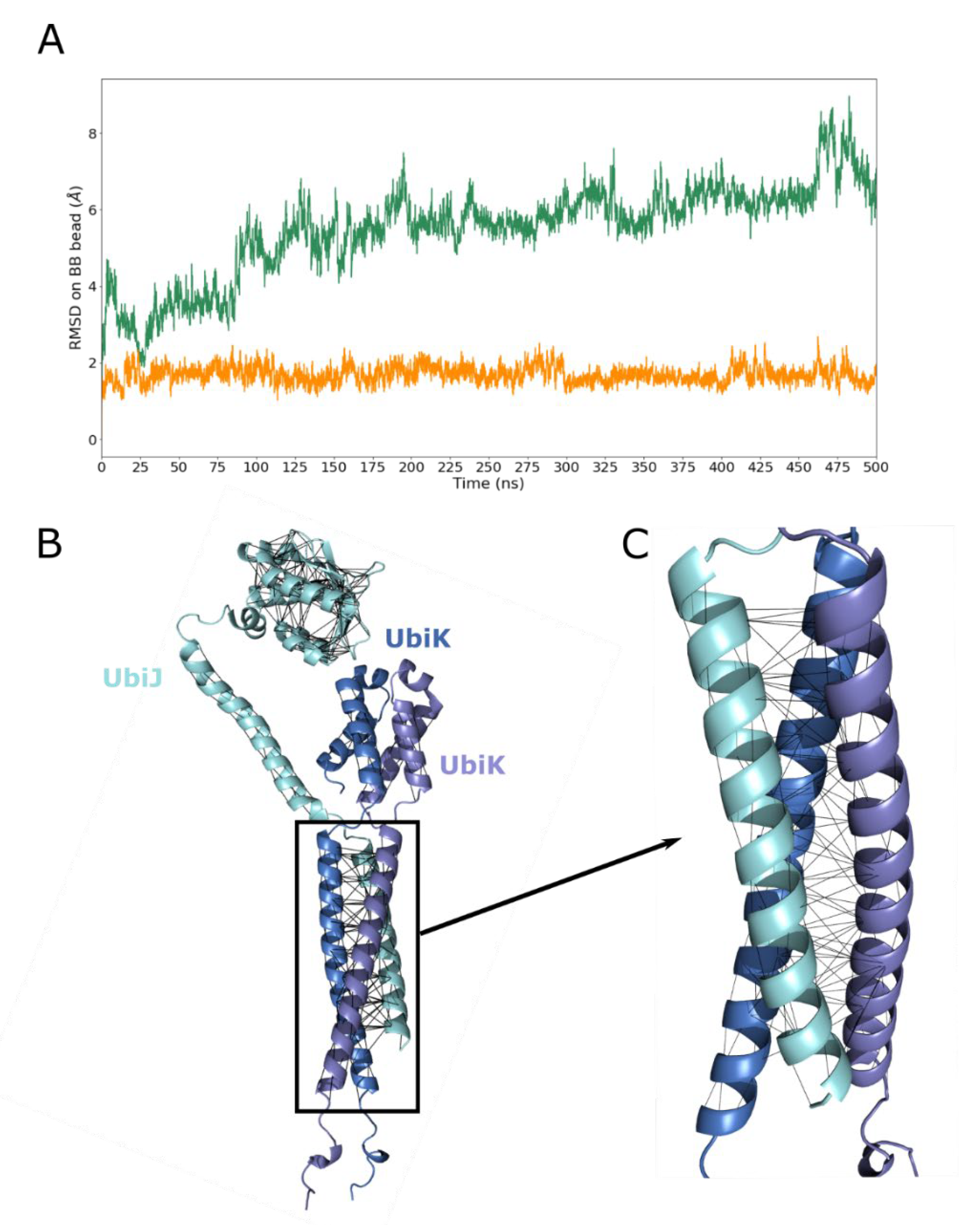
2.2. UbiJ-UbiK2 Heterotrimer Modelling
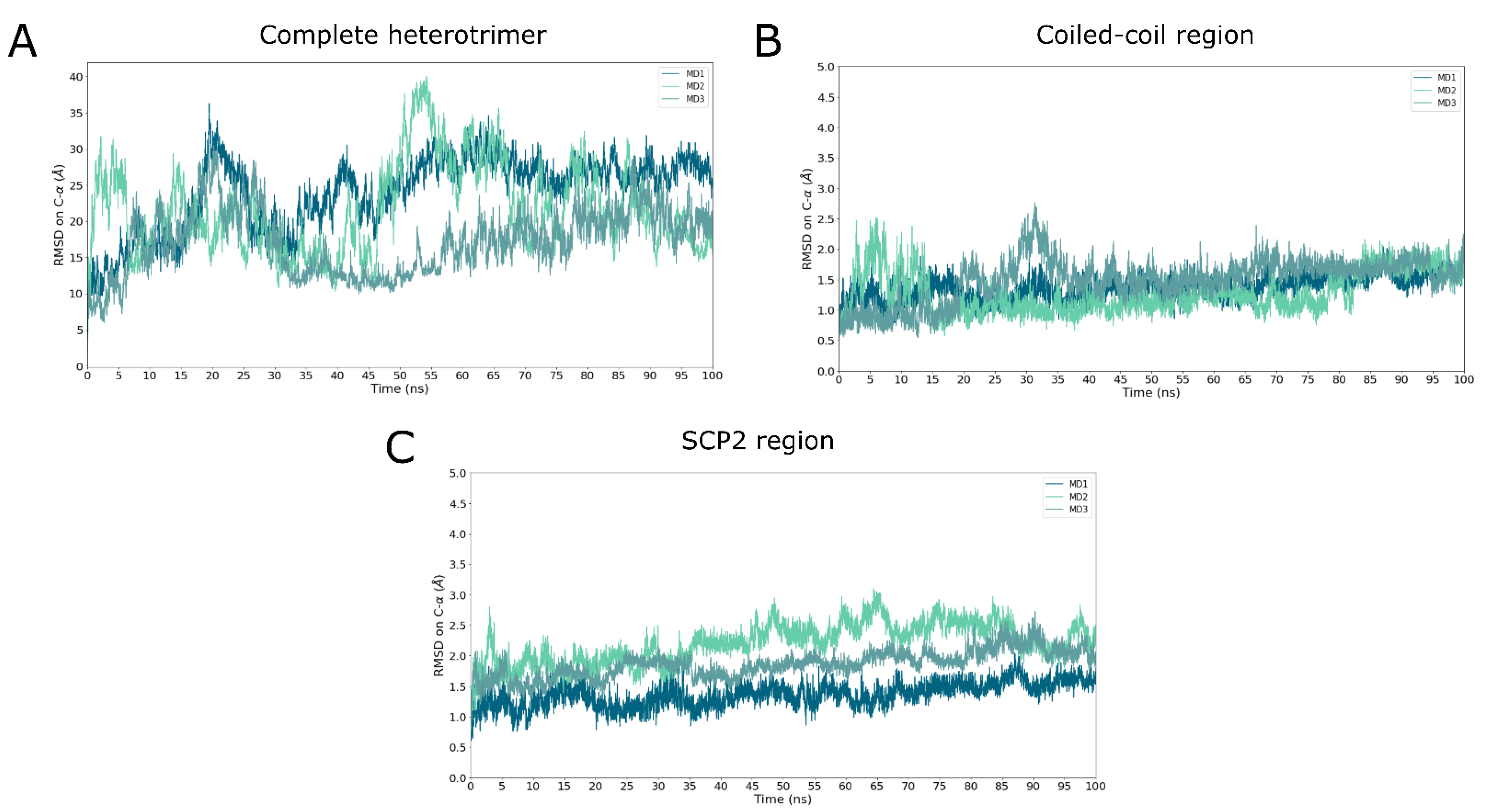
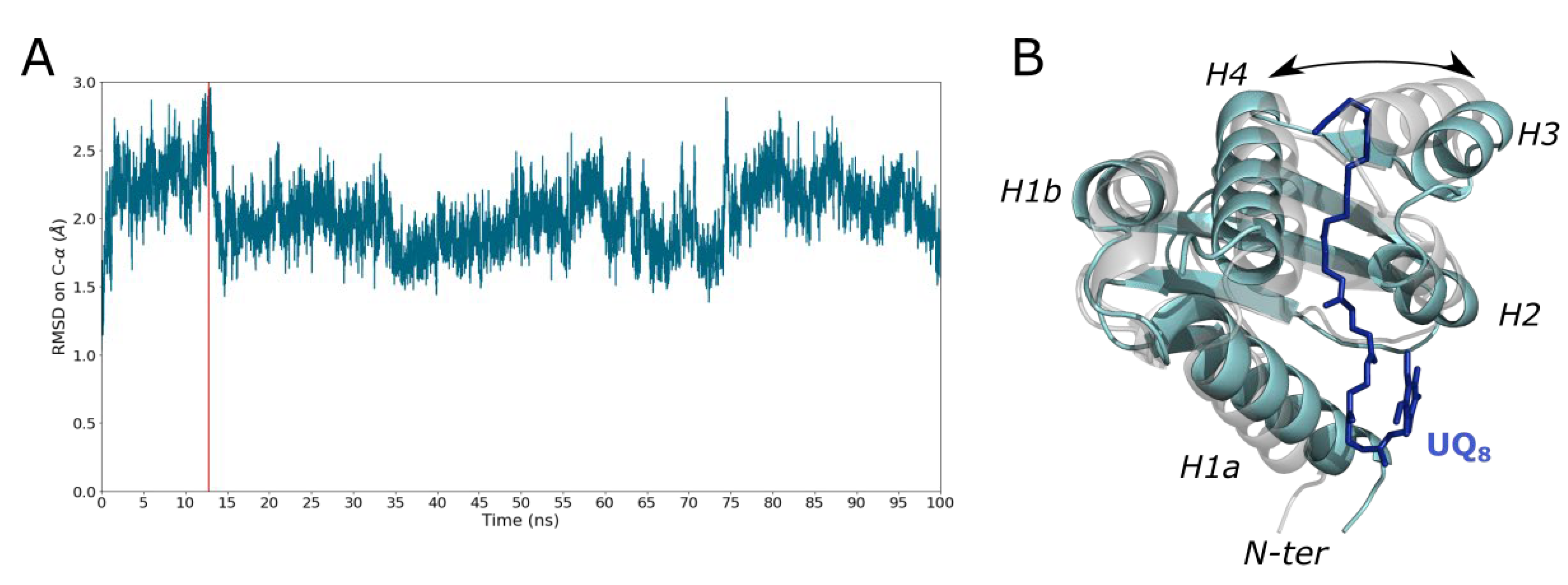
2.3. Heterotrimer Stability Inferred by MD Simulations
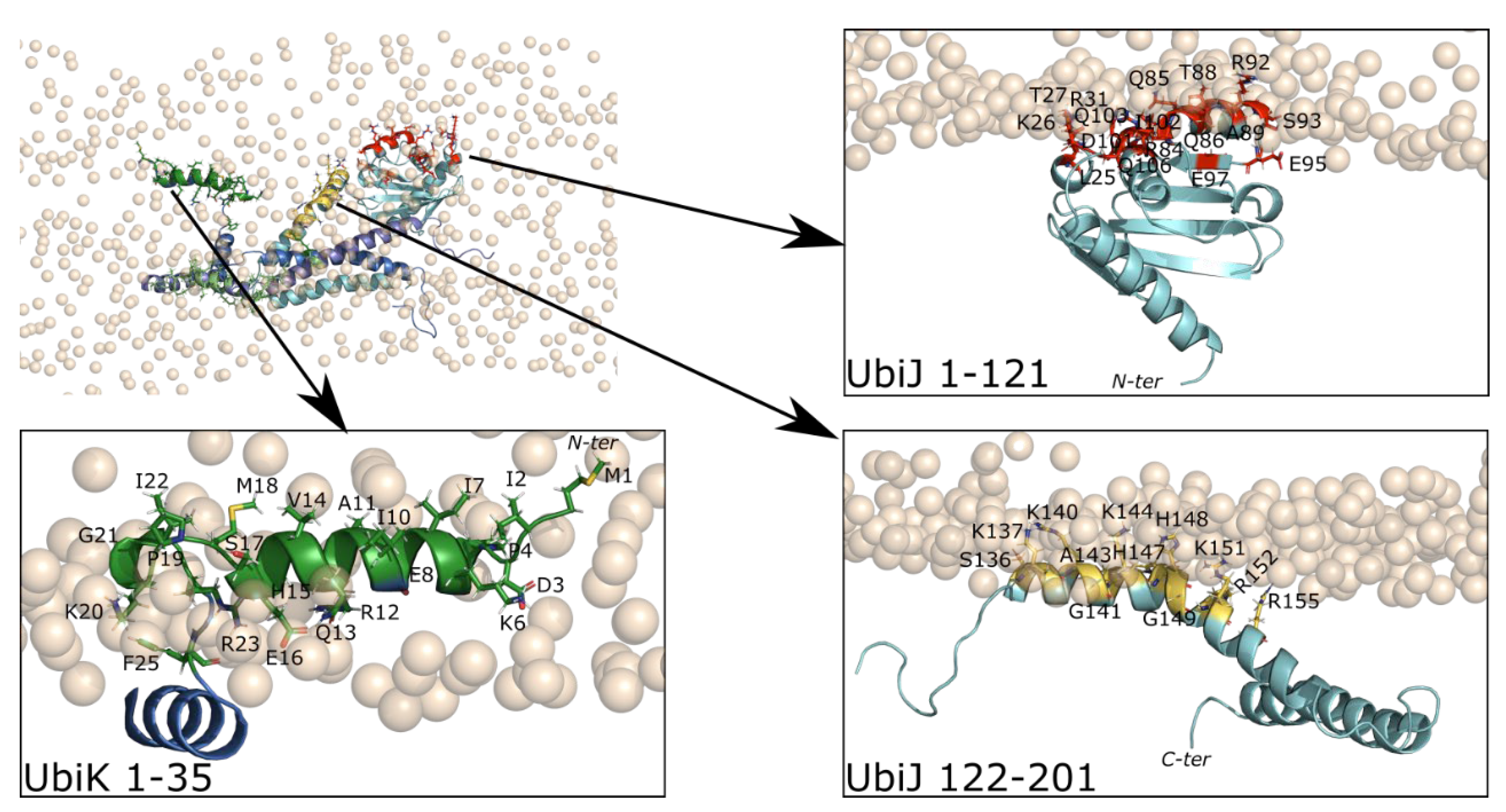
2.4. Heterotrimer-Membrane Interactions
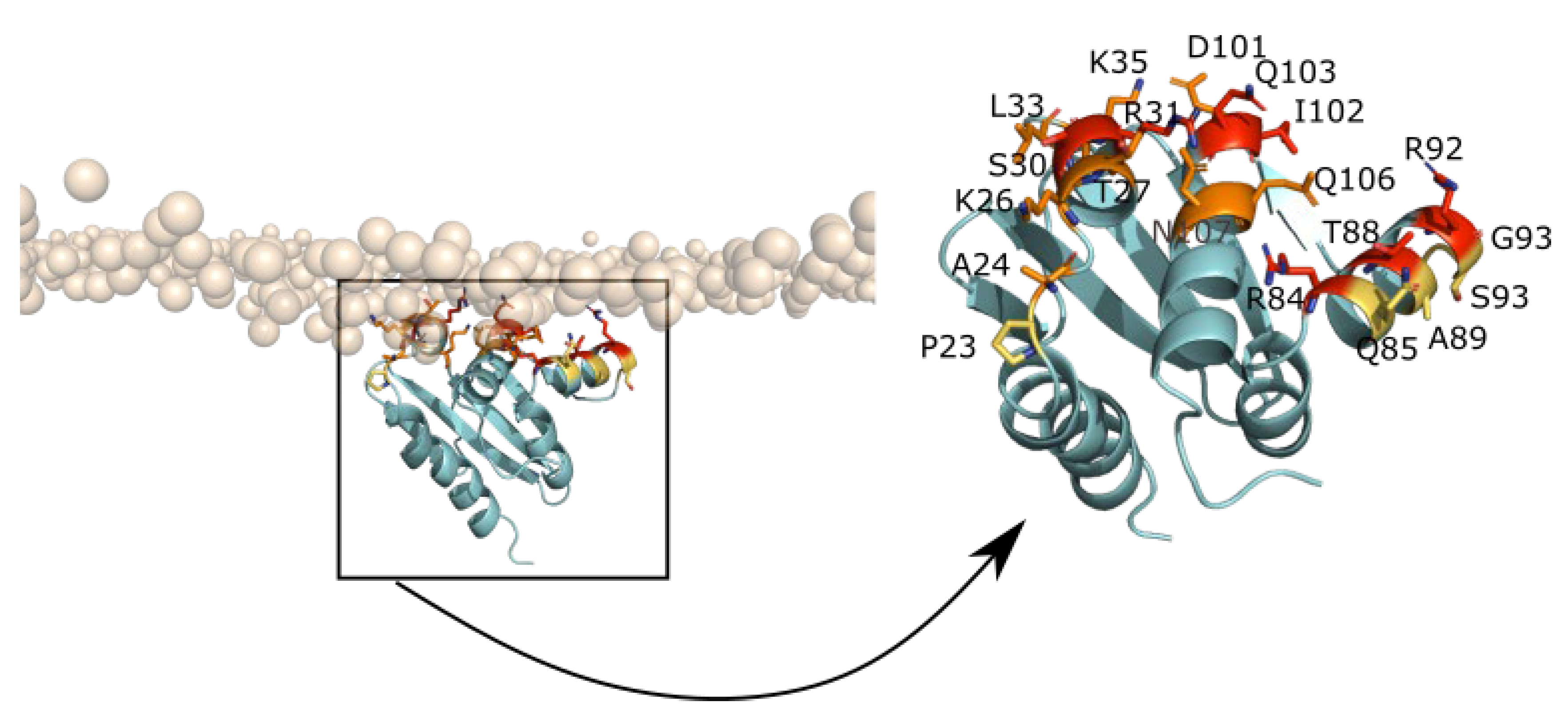
2.4.1. Adsorption of UbiJ SCP2 on the Membrane
2.4.2. Adsorption of Full Heterotrimer on the Membrane
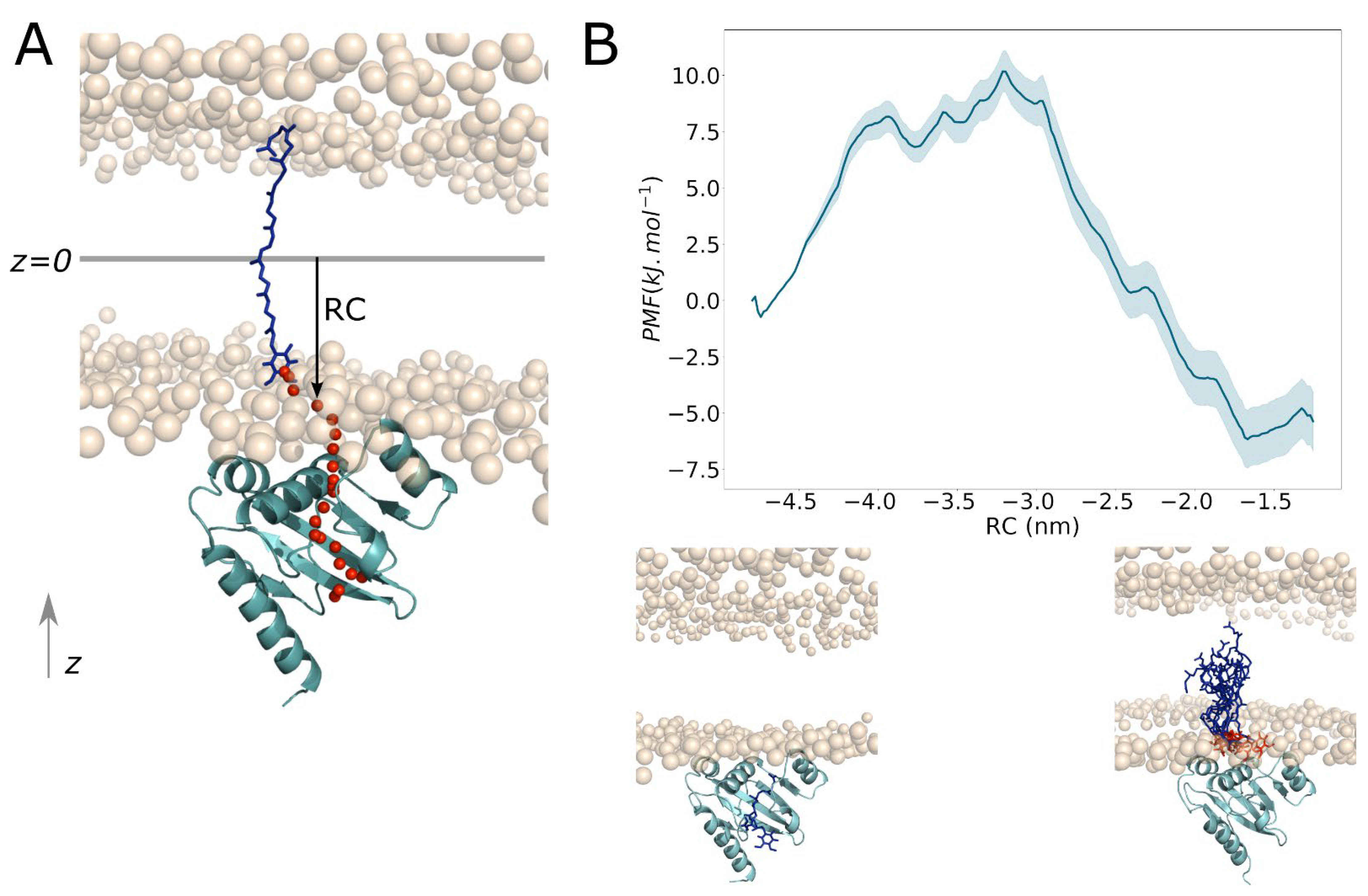
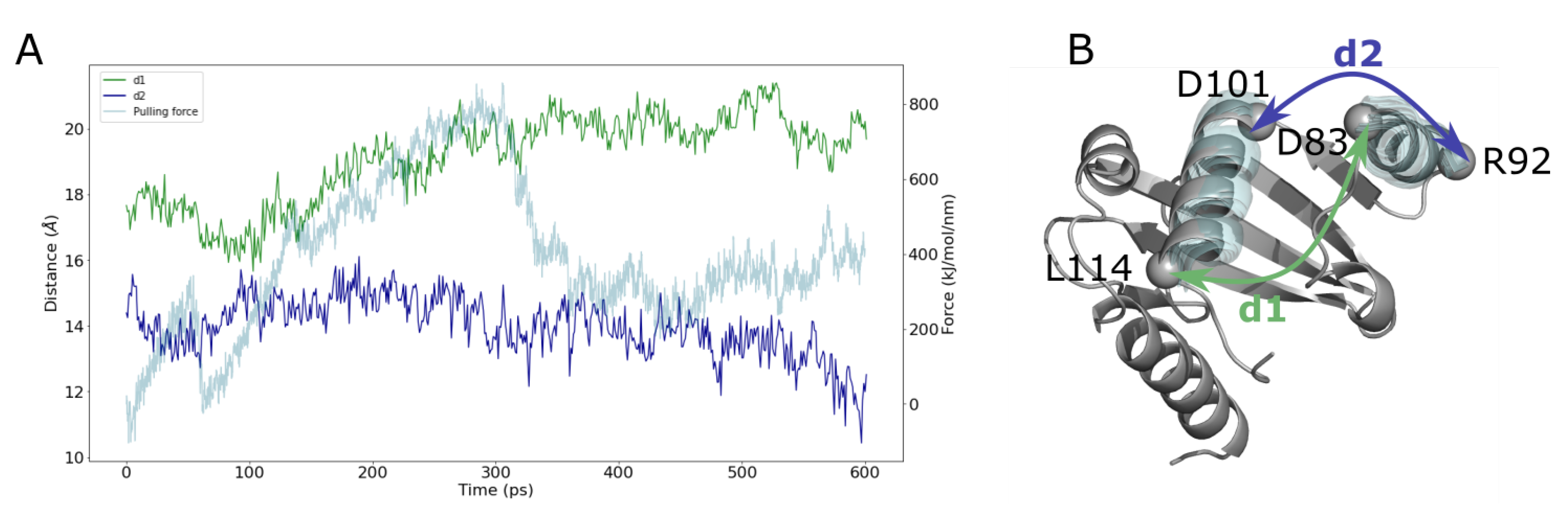
2.5. Ubiquinone Release
3. Material and Methods
3.1. Co-Evolution Sequence Dataset
3.2. Co-Evolution Prediction
3.3. Molecular Modelling Setup
3.4. Molecular Dynamics Simulation Setup
3.4.1. All-Atom MD Simulations
3.4.2. Coarse-Grained MD Simulations
3.5. Binding and Release of UQ8
3.5.1. Ubiquinone Docking Setup
3.5.2. Umbrella Sampling
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stefely, J.A.; Pagliarini, D.J. Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem. Sci. 2017, 42, 824–843. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.S.; Kazemzadeh, K.; Vragniau, C.; Pelosi, L.; Pierrel, F. Advances in Bacterial Pathways for the Biosynthesis of Ubiquinone. Biochim. Et Biophys. Acta BBA Bioenerg. 2020, 1861, 148259. [Google Scholar] [CrossRef] [PubMed]
- Bentinger, M.; Tekle, M.; Dallner, G. Coenzyme Q—Biosynthesis and Functions. Biochem. Biophys. Res. Commun. 2010, 396, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Kawamukai, M. Biosynthesis of Coenzyme Q in Eukaryotes. Biosci. Biotechnol. Biochem. 2016, 80, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Meganathan, R. Biosynthesis of Menaquinone (Vitamin K2) and Ubiquinone (Coenzyme Q): A Perspective on Enzymatic Mechanisms. In Vitamins & Hormones; Elsevier: Amsterdam, The Netherlands, 2001; Volume 61, pp. 173–218. ISBN 978-0-12-709861-6. [Google Scholar]
- Tzin, V.; Galili, G.; Aharoni, A. Shikimate Pathway and Aromatic Amino Acid Biosynthesis. In eLS; John Wiley & Sons, Ltd.: Chichester, UK, 2012; ISBN 978-0-470-01617-6. [Google Scholar]
- Aussel, L.; Pierrel, F.; Loiseau, L.; Lombard, M.; Fontecave, M.; Barras, F. Biosynthesis and Physiology of Coenzyme Q in Bacteria. Biochim. Biophys. Acta BBA Bioenerg. 2014, 1837, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Wang, J.-Y.; Wang, J.; Poplawsky, A.; Lin, S.; Zhu, B.; Chang, C.; Zhou, T.; Zhang, L.-H.; He, Y.-W. The Diffusible Factor Synthase XanB2 Is a Bifunctional Chorismatase That Links the Shikimate Pathway to Ubiquinone and Xanthomonadins Biosynthetic Pathways: XanB2 Is a Bifunctional Chorismatase. Mol. Microbiol. 2013, 87, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.C.; Salomon, C.; Mijts, B.; Schmidt-Dannert, C. Biosynthesis of Ubiquinone Compounds with Conjugated Prenyl Side Chains. Appl. Environ. Microbiol. 2008, 74, 6908–6917. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hajj Chehade, M.; Pelosi, L.; Fyfe, C.D.; Loiseau, L.; Rascalou, B.; Brugière, S.; Kazemzadeh, K.; Vo, C.-D.-T.; Ciccone, L.; Aussel, L.; et al. A Soluble Metabolon Synthesizes the Isoprenoid Lipid Ubiquinone. Cell Chem. Biol. 2019, 26, 482–492.e7. [Google Scholar] [CrossRef]
- Burgardt, N.I.; Gianotti, A.R.; Ferreyra, R.G.; Ermácora, M.R. A Structural Appraisal of Sterol Carrier Protein 2. Biochim. Biophys. Acta BBA Proteins Proteom. 2017, 1865, 565–577. [Google Scholar] [CrossRef]
- Loiseau, L.; Fyfe, C.; Aussel, L.; Hajj Chehade, M.; Hernández, S.B.; Faivre, B.; Hamdane, D.; Mellot-Draznieks, C.; Rascalou, B.; Pelosi, L.; et al. The UbiK Protein Is an Accessory Factor Necessary for Bacterial Ubiquinone (UQ) Biosynthesis and Forms a Complex with the UQ Biogenesis Factor UbiJ. J. Biol. Chem. 2017, 292, 11937–11950. [Google Scholar] [CrossRef]
- del Carmen Carrica, M.; Craig, P.O.; del Valle Alonso, S.; Goldbaum, F.A.; Cravero, S.L. Brucella Abortus MFP: A Trimeric Coiled-Coil Protein with Membrane Fusogenic Activity. Biochemistry 2008, 47, 8165–8175. [Google Scholar] [CrossRef]
- Carrica, M.C.; Craig, P.O.; García-Angulo, V.A.; Aguirre, A.; García-Véscovi, E.; Goldbaum, F.A.; Cravero, S.L. YqiC of Salmonella Enterica Serovar Typhimurium Is a Membrane Fusogenic Protein Required for Mice Colonization. BMC Microbiol. 2011, 11, 95. [Google Scholar] [CrossRef]
- Truebestein, L.; Leonard, T.A. Coiled-coils: The Long and Short of It. BioEssays 2016, 38, 903–916. [Google Scholar] [CrossRef]
- Grigoryan, G.; Keating, A. Structural Specificity in Coiled-Coil Interactions. Curr. Opin. Struct. Biol. 2008, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pereira, J.; Simpkin, A.J.; Hartmann, M.D.; Rigden, D.J.; Keegan, R.M.; Lupas, A.N. High-accuracy Protein Structure Prediction in CASP14. Proteins 2021, 89, 1687–1699. [Google Scholar] [CrossRef]
- Schaarschmidt, J.; Monastyrskyy, B.; Kryshtafovych, A.; Bonvin, A.M.J.J. Assessment of Contact Predictions in CASP12: Co-Evolution and Deep Learning Coming of Age. Proteins 2018, 86, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bei, Z.; Xi, W.; Hao, M.; Ju, Z.; Saravanan, K.M.; Zhang, H.; Guo, N.; Wei, Y. Evaluation of Residue-Residue Contact Prediction Methods: From Retrospective to Prospective. PLoS Comput. Biol. 2021, 17, e1009027. [Google Scholar] [CrossRef]
- Lensink, M.F.; Nadzirin, N.; Velankar, S.; Wodak, S.J. Modeling Protein-protein, Protein-peptide, and Protein-oligosaccharide Complexes: CAPRI 7th Edition. Proteins 2020, 88, 916–938. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein Complex Prediction with AlphaFold-Multimer. BioRxiv 2021. [Google Scholar] [CrossRef]
- Leach, A.R. Molecular Modelling: Principles and Applications; Pearson Education: London, UK, 2001; ISBN 978-0-582-38210-7. [Google Scholar]
- Mariani, V.; Biasini, M.; Barbato, A.; Schwede, T. LDDT: A Local Superposition-Free Score for Comparing Protein Structures and Models Using Distance Difference Tests. Bioinformatics 2013, 29, 2722–2728. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Thornton, J.M. The Impact of AlphaFold2 One Year On. Nat. Methods 2022, 19, 15–20. [Google Scholar] [CrossRef]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A Web Server to Screen Sequences with Specific—Helical Properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef]
- Zhang, Y. TM-Align: A Protein Structure Alignment Algorithm Based on the TM-Score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]
- Gabler, F.; Nam, S.; Till, S.; Mirdita, M.; Steinegger, M.; Söding, J.; Lupas, A.N.; Alva, V. Protein Sequence Analysis Using the MPI Bioinformatics Toolkit. Curr. Protoc. Bioinform. 2020, 72, e108. [Google Scholar] [CrossRef]
- Ludwiczak, J.; Winski, A.; Szczepaniak, K.; Alva, V.; Dunin-Horkawicz, S. DeepCoil—A Fast and Accurate Prediction of Coiled-Coil Domains in Protein Sequences. Bioinformatics 2019, 35, 2790–2795. [Google Scholar] [CrossRef]
- Seemayer, S.; Gruber, M.; Söding, J. CCMpred—Fast and Precise Prediction of Protein Residue–Residue Contacts from Correlated Mutations. Bioinformatics 2014, 30, 3128–3130. [Google Scholar] [CrossRef]
- Esque, J.; Oguey, C.; de Brevern, A.G. Comparative Analysis of Threshold and Tessellation Methods for Determining Protein Contacts. J. Chem. Inf. Model. 2011, 51, 493–507. [Google Scholar] [CrossRef]
- Gruber, J.; Zawaira, A.; Saunders, R.; Barrett, C.P.; Noble, M.E.M. Computational Analyses of the Surface Properties of Protein–Protein Interfaces. Acta Crystallogr. D Biol. Crystallogr. 2007, 63, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.D. Functional and Structural Roles of Coiled Coils. Subcell Biochem. 2017, 82, 63–93. [Google Scholar] [CrossRef]
- Poma, A.B.; Cieplak, M.; Theodorakis, P.E. Combining the MARTINI and Structure-Based Coarse-Grained Approaches for the Molecular Dynamics Studies of Conformational Transitions in Proteins. J. Chem. Theory Comput. 2017, 13, 1366–1374. [Google Scholar] [CrossRef]
- Larsen, A.H.; John, L.H.; Sansom, M.S.P.; Corey, R.A. Specific Interactions of Peripheral Membrane Proteins with Lipids: What Can Molecular Simulations Show Us? Biosci. Rep. 2022, 42, BSR20211406. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-H.; Guan, Z.; Zhao, J.; Raetz, C.R.H. Three Phosphatidylglycerol-Phosphate Phosphatases in the Inner Membrane of Escherichia Coli. J. Biol. Chem. 2011, 286, 5506–5518. [Google Scholar] [CrossRef]
- Galassi, V.V.; Arantes, G.M. Partition, Orientation and Mobility of Ubiquinones in a Lipid Bilayer. Biochim. Biophys. Acta BBA Bioenerg. 2015, 1847, 1560–1573. [Google Scholar] [CrossRef]
- Warnau, J.; Sharma, V.; Gamiz-Hernandez, A.P.; Di Luca, A.; Haapanen, O.; Vattulainen, I.; Wikström, M.; Hummer, G.; Kaila, V.R.I. Redox-Coupled Quinone Dynamics in the Respiratory Complex I. Proc. Natl. Acad. Sci. USA 2018, 115, E8413–E8420. [Google Scholar] [CrossRef]
- Pelosi, L.; Vo, C.-D.-T.; Abby, S.S.; Loiseau, L.; Rascalou, B.; Chehade, M.H.; Faivre, B.; Goussé, M.; Chenal, C.; Touati, N.; et al. Ubiquinone Biosynthesis over the Entire O2 Range: Characterization of a Conserved O2-Independent Pathway. Mbio 2019, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino Acid Substitution Matrices from Protein Blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Iserte, J.; Simonetti, F.L.; Zea, D.J.; Teppa, E.; Marino-Buslje, C. I-COMS: Interprotein-COrrelated Mutations Server. Nucleic Acids Res. 2015, 43, W320–W325. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov Model Speed Heuristic and Iterative HMM Search Procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Remmert, M.; Biegert, A.; Hauser, A.; Söding, J. HHblits: Lightning-Fast Iterative Protein Sequence Searching by HMM-HMM Alignment. Nat. Methods 2012, 9, 173–175. [Google Scholar] [CrossRef] [PubMed]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H.; The UniProt Consortium. UniRef Clusters: A Comprehensive and Scalable Alternative for Improving Sequence Similarity Searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Almeida, A.; Beracochea, M.; Boland, M.; Burgin, J.; Cochrane, G.; Crusoe, M.R.; Kale, V.; Potter, S.C.; Richardson, L.J.; et al. MGnify: The Microbiome Analysis Resource in 2020. Nucleic Acids Res. 2019, 48, D570–D578. [Google Scholar] [CrossRef]
- Mirdita, M.; von den Driesch, L.; Galiez, C.; Martin, M.J.; Söding, J.; Steinegger, M. Uniclust Databases of Clustered and Deeply Annotated Protein Sequences and Alignments. Nucleic Acids Res. 2017, 45, D170–D176. [Google Scholar] [CrossRef] [PubMed]
- wwPDB Consortium; Burley, S.K.; Berman, H.M.; Bhikadiya, C.; Bi, C.; Chen, L.; Costanzo, L.D.; Christie, C.; Duarte, J.M.; Dutta, S.; et al. Protein Data Bank: The Single Global Archive for 3D Macromolecular Structure Data. Nucleic Acids Res. 2019, 47, D520–D528. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 All-Atom Additive Protein Force Field: Validation Based on Comparison to NMR Data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Souza, P.C.T.; Alessandri, R.; Barnoud, J.; Thallmair, S.; Faustino, I.; Grünewald, F.; Patmanidis, I.; Abdizadeh, H.; Bruininks, B.M.H.; Wassenaar, T.A.; et al. Martini 3: A General Purpose Force Field for Coarse-Grained Molecular Dynamics. Nat. Methods 2021, 18, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, J.; Gumbart, J.C. Preparing Membrane Proteins for Simulation Using CHARMM-GUI. In Structure and Function of Membrane Proteins; Schmidt-Krey, I., Gumbart, J.C., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2302, pp. 237–251. ISBN 978-1-07-161393-1. [Google Scholar]
- Wassenaar, T.A.; Ingólfsson, H.I.; Böckmann, R.A.; Tieleman, D.P.; Marrink, S.J. Computational Lipidomics with Insane: A Versatile Tool for Generating Custom Membranes for Molecular Simulations. J. Chem. Theory Comput. 2015, 11, 2144–2155. [Google Scholar] [CrossRef] [PubMed]
- Lemkul, J.A. From Proteins to Perturbed Hamiltonians: A Suite of Tutorials for the GROMACS-2018 Molecular Simulation Package [Article v1.0]. Living J. Comput. Mol. Sci. 2018, 1, 5068. [Google Scholar] [CrossRef]
- Kroon, P.C. Martinize 2—VerMoUTH. In Aggregate, Automate, Assemble; University of Groningen: Groningen, The Netherlands, 2020; pp. 16–53. ISBN 978-94-034-2581-8. [Google Scholar]
- Vickery, O.N.; Stansfeld, P.J. CG2AT2: An Enhanced Fragment-Based Approach for Serial Multi-Scale Molecular Dynamics Simulations. J. Chem. Theory Comput. 2021, 17, 6472–6482. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Hub, J.S.; de Groot, B.L.; van der Spoel, D. G_wham—A Free Weighted Histogram Analysis Implementation Including Robust Error and Autocorrelation Estimates. J. Chem. Theory Comput. 2010, 6, 3713–3720. [Google Scholar] [CrossRef]
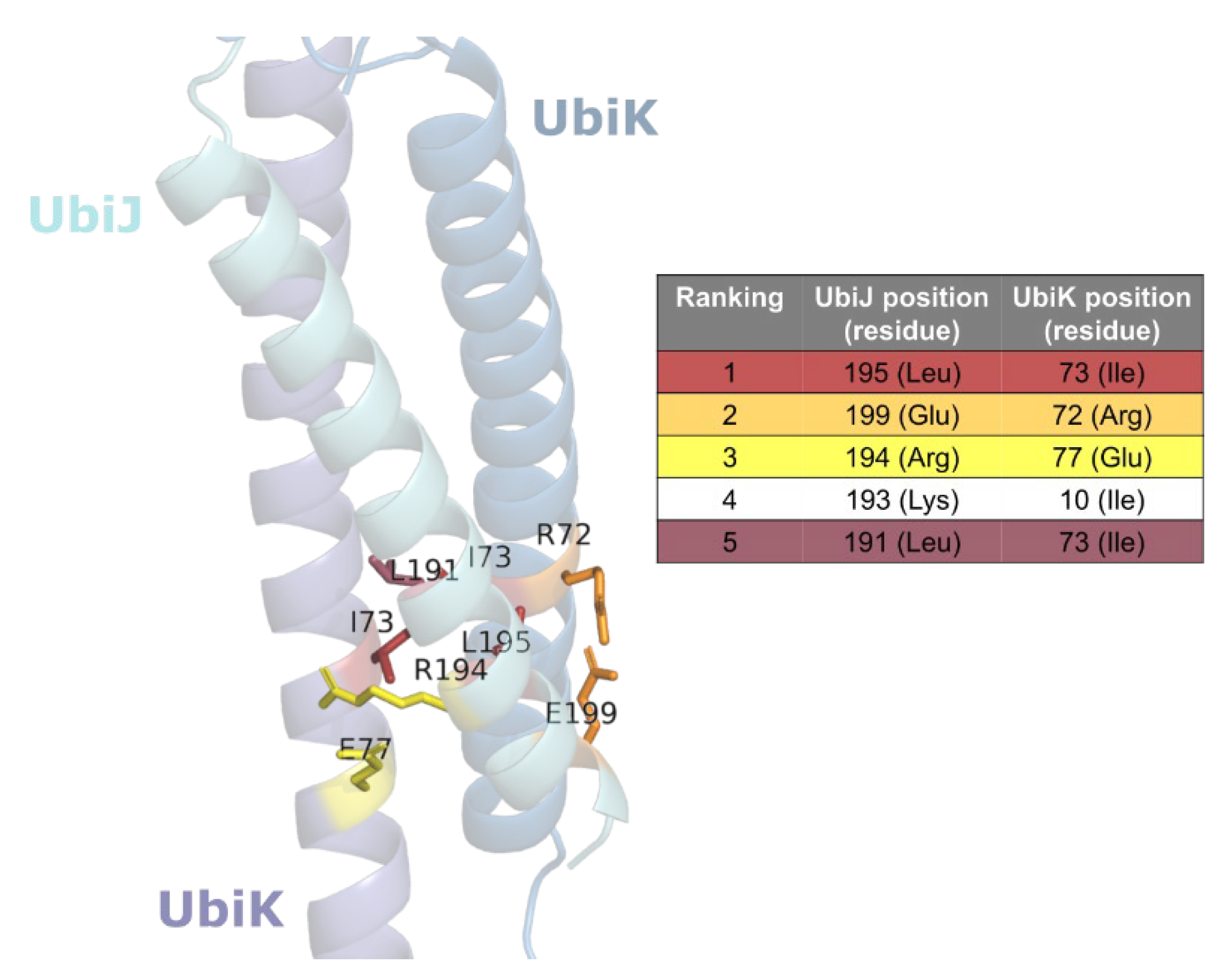

| Ranking | UbiJ Residue Position | UbiK Residue Position | Distance A (Å) | Distance B (Å) |
|---|---|---|---|---|
| 1 | 195 | 73 | 4.25 | 3.39 |
| 2 | 199 | 72 | 11.08 | 2.79 |
| 3 | 194 | 77 | 2.72 | 11.59 |
| 4 | 193 | 10 | 46.13 | 43.07 |
| 5 | 191 | 73 | 3.53 | 5.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Launay, R.; Teppa, E.; Martins, C.; Abby, S.S.; Pierrel, F.; André, I.; Esque, J. Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies. Int. J. Mol. Sci. 2022, 23, 10323. https://doi.org/10.3390/ijms231810323
Launay R, Teppa E, Martins C, Abby SS, Pierrel F, André I, Esque J. Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies. International Journal of Molecular Sciences. 2022; 23(18):10323. https://doi.org/10.3390/ijms231810323
Chicago/Turabian StyleLaunay, Romain, Elin Teppa, Carla Martins, Sophie S. Abby, Fabien Pierrel, Isabelle André, and Jérémy Esque. 2022. "Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies" International Journal of Molecular Sciences 23, no. 18: 10323. https://doi.org/10.3390/ijms231810323
APA StyleLaunay, R., Teppa, E., Martins, C., Abby, S. S., Pierrel, F., André, I., & Esque, J. (2022). Towards Molecular Understanding of the Functional Role of UbiJ-UbiK2 Complex in Ubiquinone Biosynthesis by Multiscale Molecular Modelling Studies. International Journal of Molecular Sciences, 23(18), 10323. https://doi.org/10.3390/ijms231810323






