Modulation of Specific Sphingosine-1-Phosphate Receptors Augments a Repair Mediating Schwann Cell Phenotype
Abstract
1. Introduction
2. Results
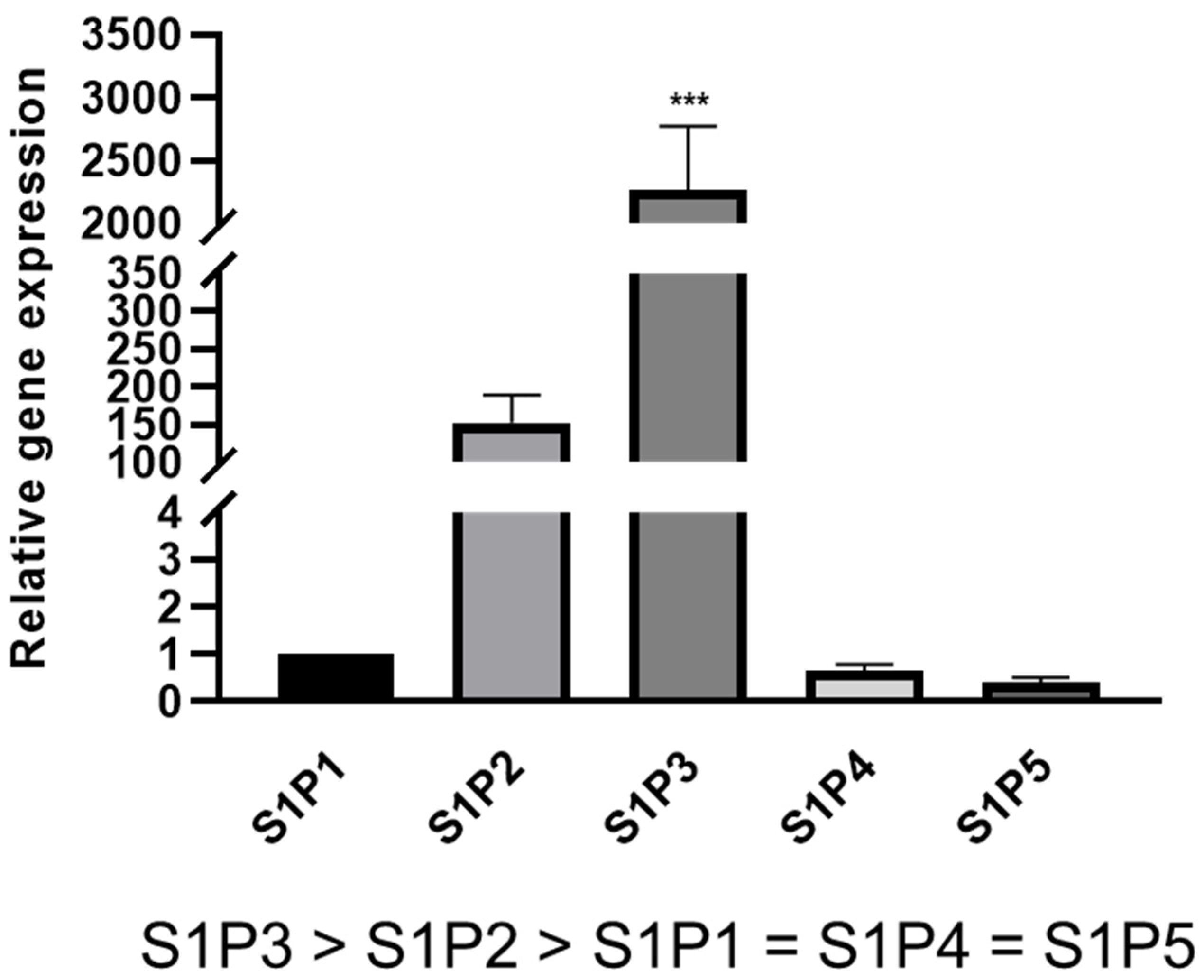
2.1. S1P Receptor Expression in Rat Schwann Cells
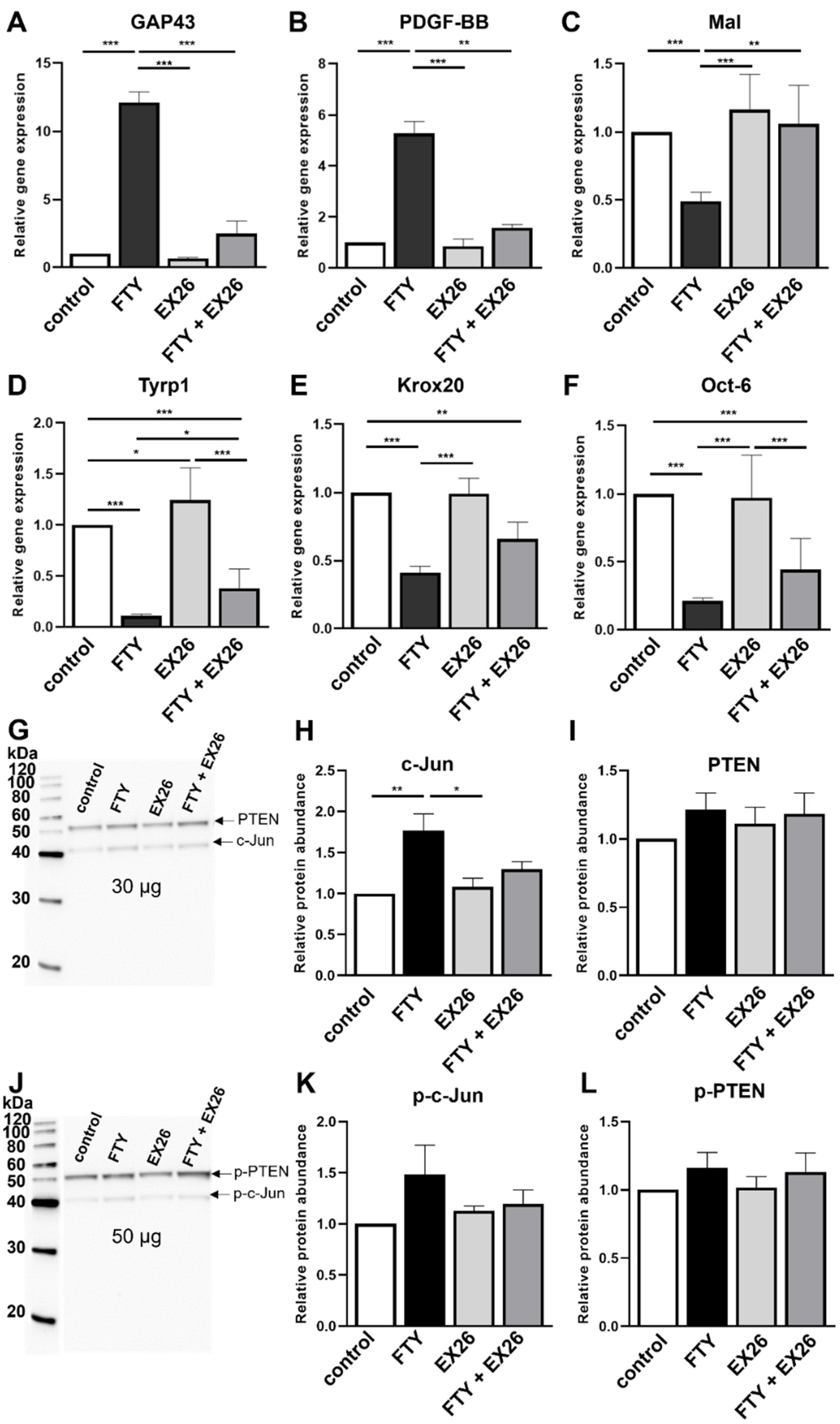
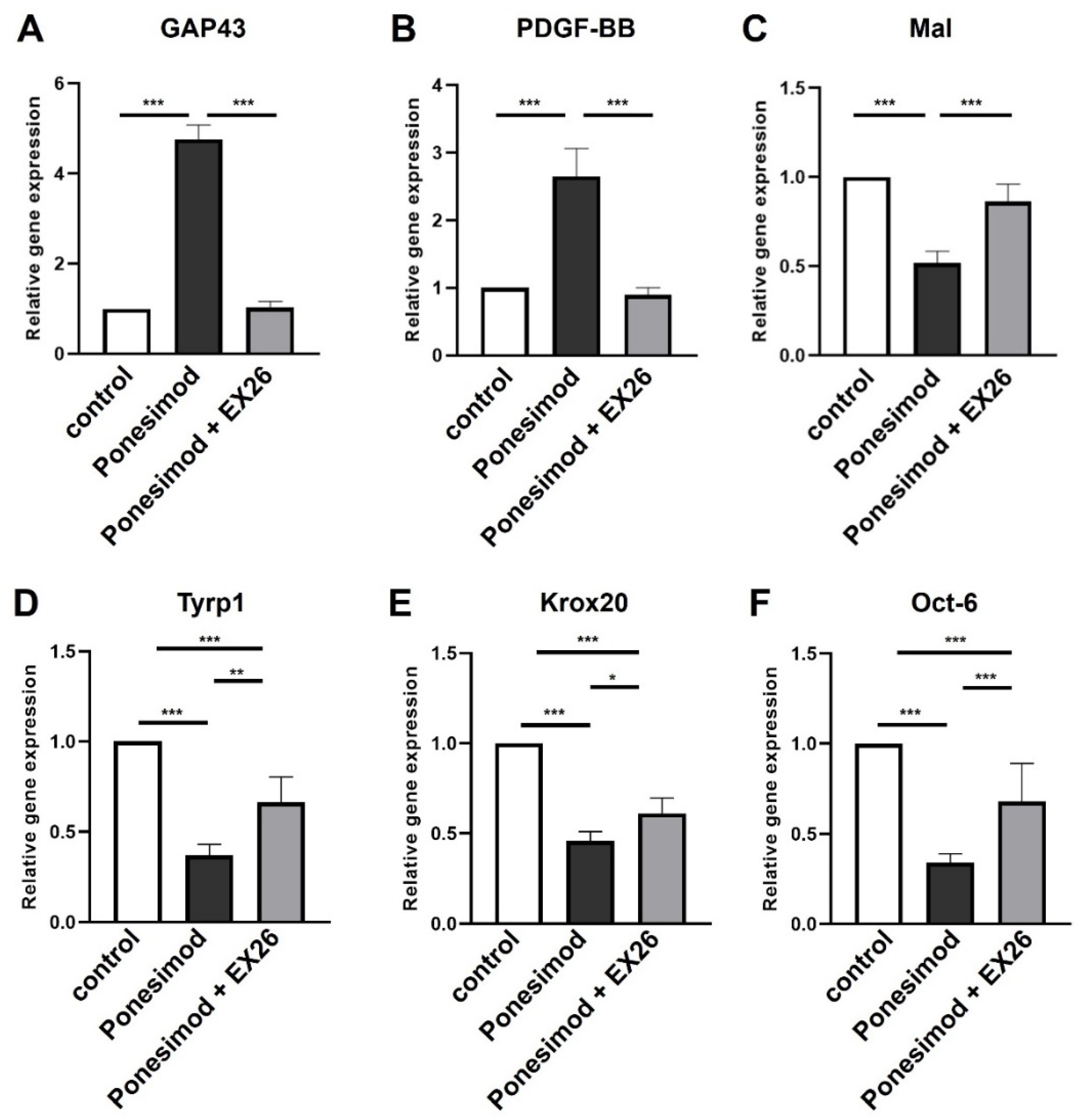
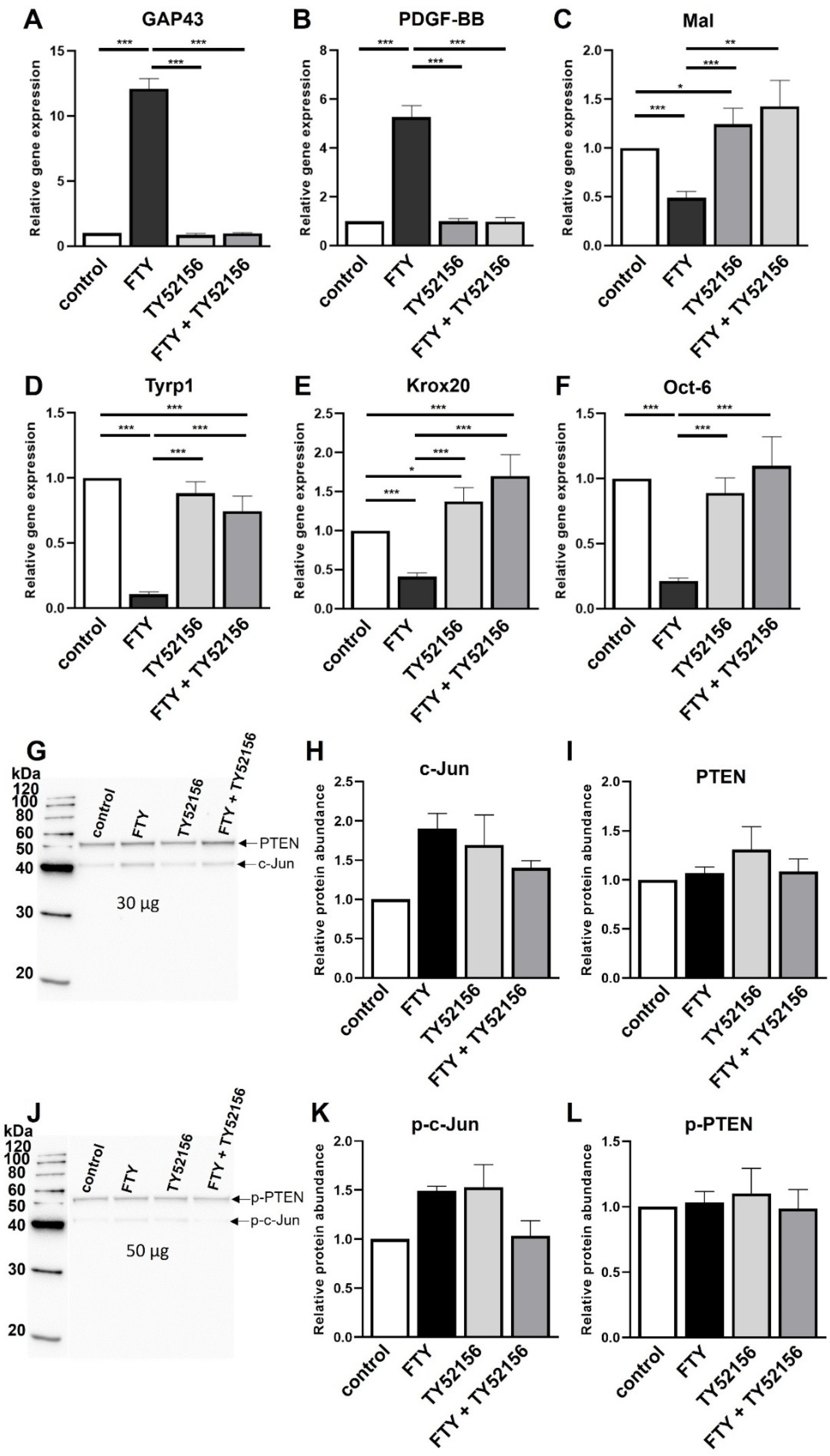
2.2. Effect of S1P1 Modulation on the Schwann Cell Phenotype
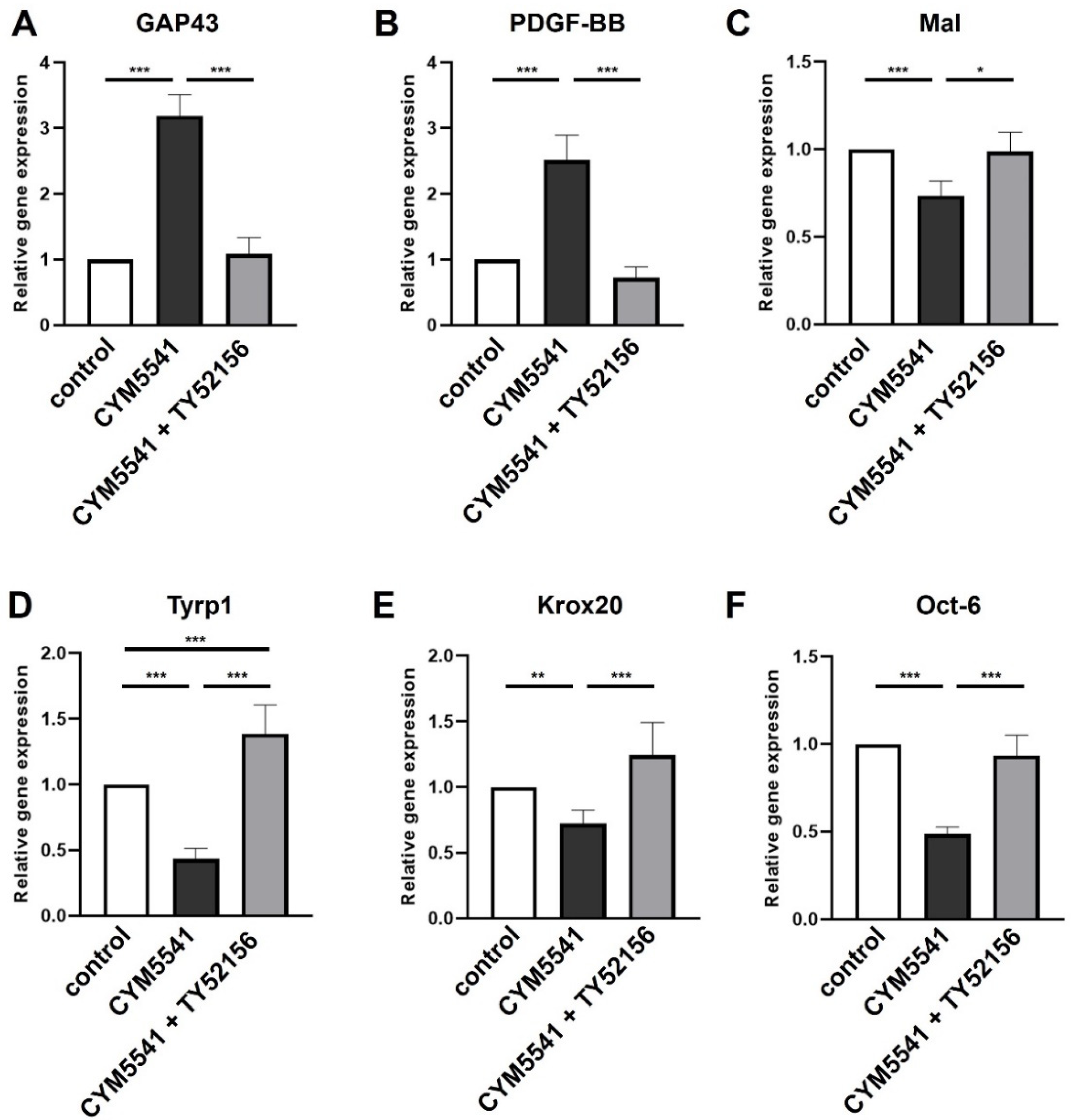
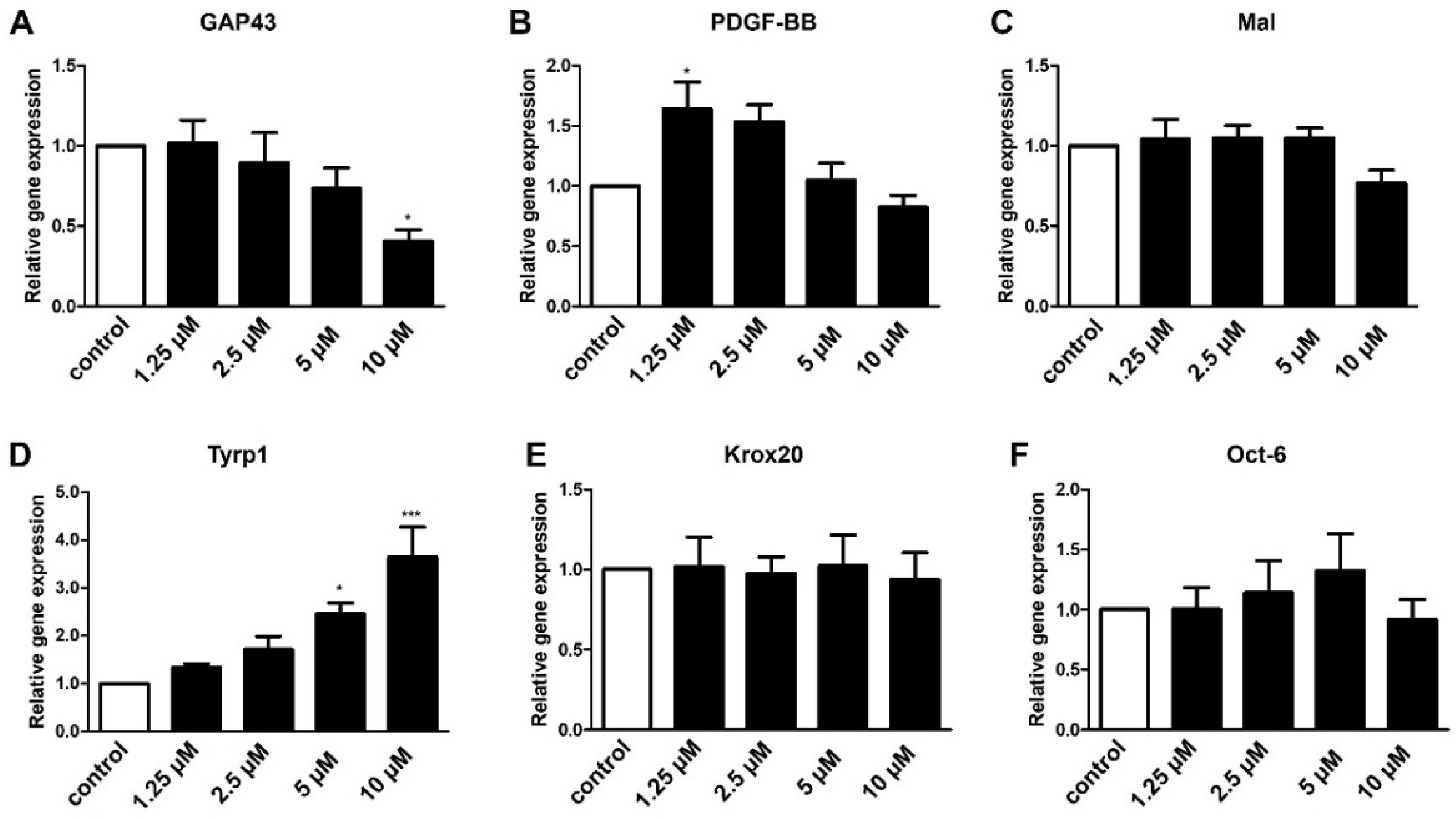
2.3. Role of S1P3 in Schwann Cell Transdifferentiation
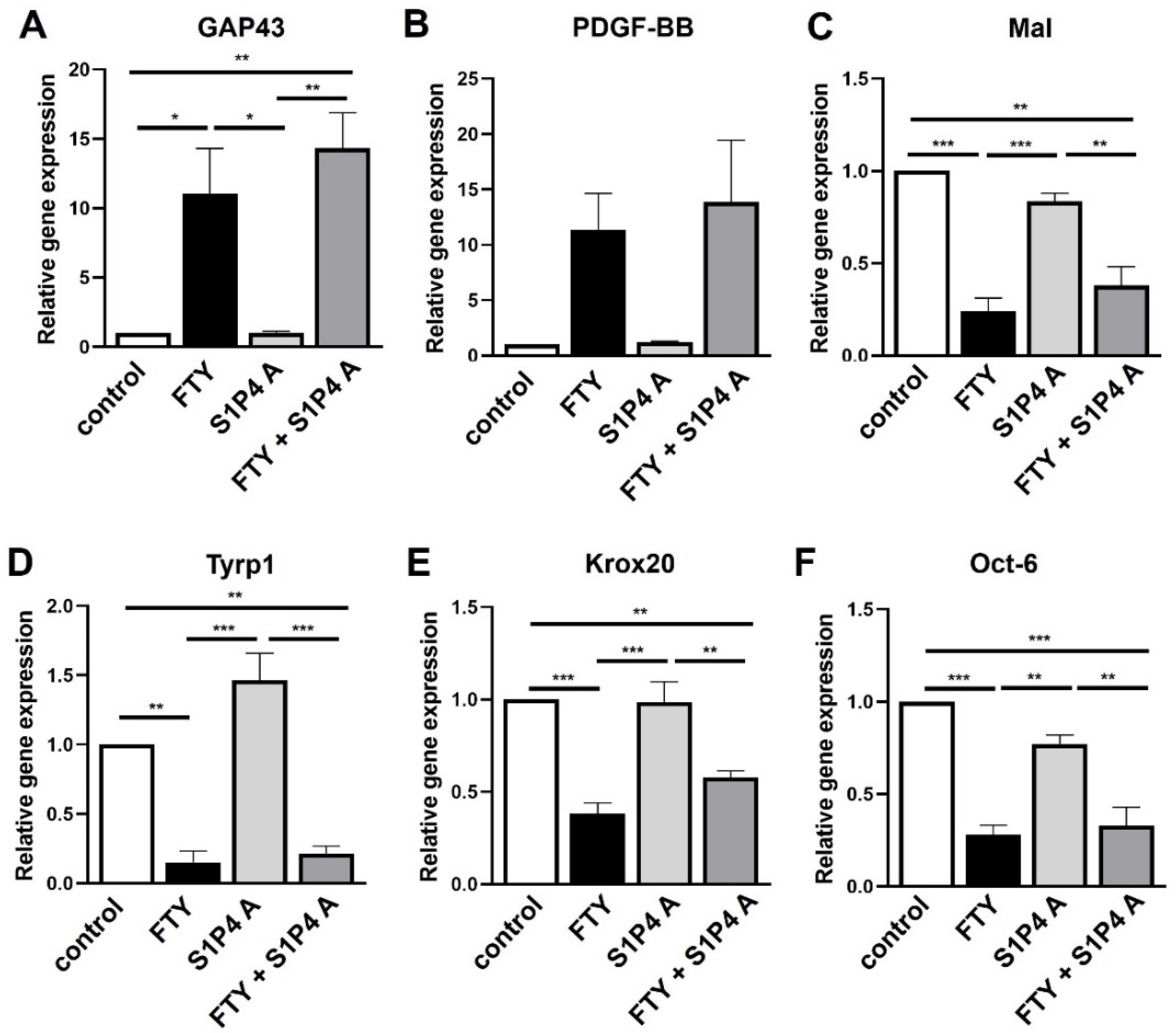
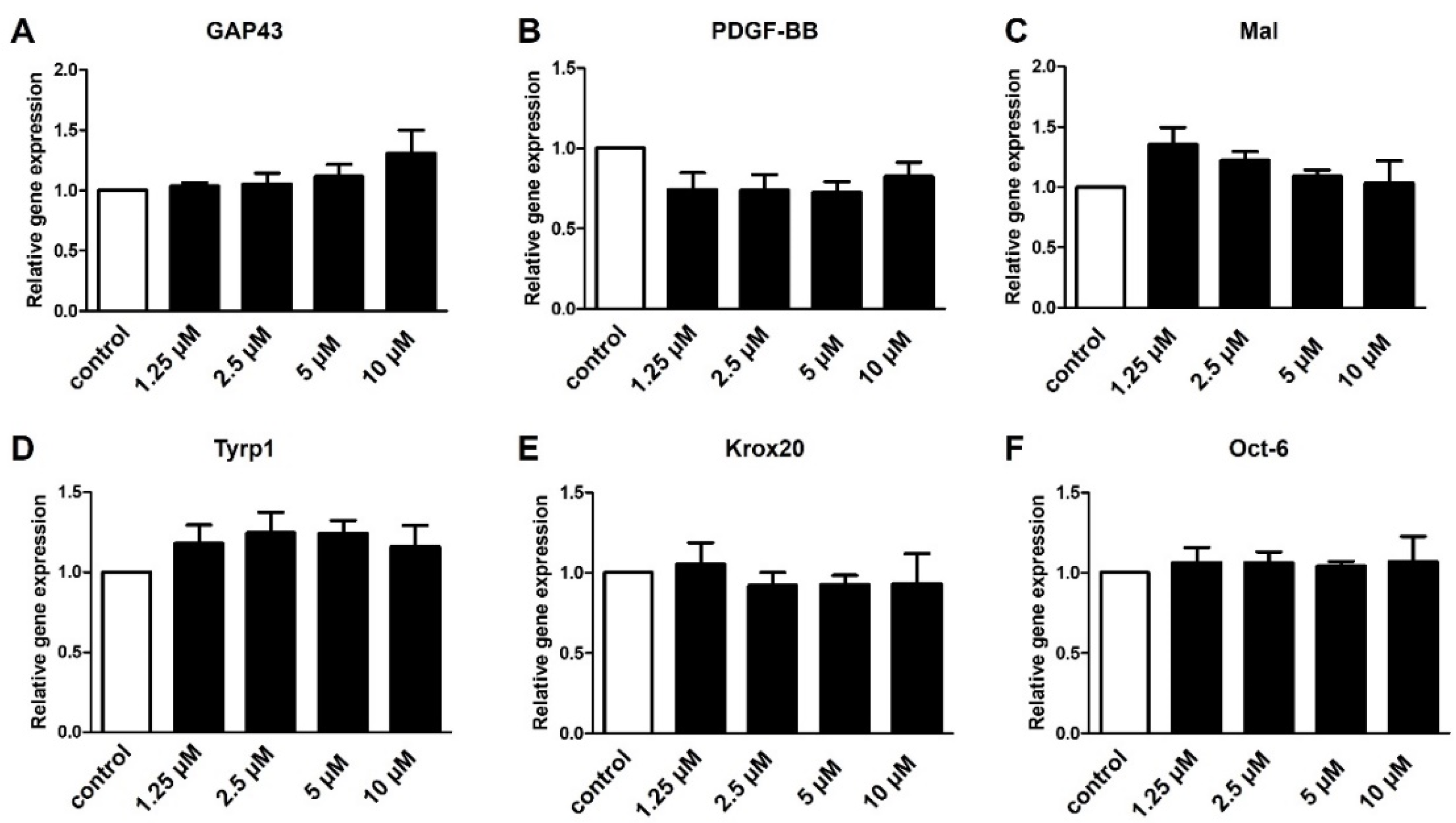
2.4. S1P4 and S1P5 Only Play a Minor Role in Schwann Cell Transdifferentiation
3. Discussion
4. Materials and Methods
4.1. Primary Neonatal Schwann Cell Culture
4.2. Modulation of S1P Receptors
4.3. Quantitative Gene Expression Analysis
4.4. Western Blot Analysis
4.5. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMSO | dimethyl sulfoxide |
| Egr2 | early growth response 2 |
| FTY720P | Fingolimod |
| GAP43 | growth associated protein 43 |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| Mal | myelin and lymphocyte protein |
| Oct-6 | octamer transcription factor 6 |
| ODC | ornithine decarboxylase |
| PDGF-BB | platelet-derived growth factor-beta |
| PNS | peripheral nervous system |
| PDL | poly-D-lysine |
| PTEN | phosphatase and tensin homolog |
| S1P | sphingosine-1-phosphate |
| S1PR | sphingosine-1-phosphate receptor |
| SC | Schwann cell |
| TGFβ | transforming growth factor-beta |
| Tyrp1 | tyrosinase-related protein 1 |
References
- Jessen, K.R.; Mirsky, R.; Lloyd, A.C. Schwann Cells: Development and Role in Nerve Repair. Cold Spring Harb. Perspect. Biol. 2015, 7, a020487. [Google Scholar] [CrossRef] [PubMed]
- Jessen, K.R.; Arthur-Farraj, P. Repair Schwann cell update: Adaptive reprogramming, EMT, and stemness in regenerating nerves. Glia 2019, 67, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Rigoni, M.; Negro, S. Signals Orchestrating Peripheral Nerve Repair. Cells 2020, 9, 1768. [Google Scholar] [CrossRef]
- Kumar, R.; Sinha, S.; Hagner, A.; Stykel, M.; Raharjo, E.; Singh, K.K.; Midha, R.; Biernaskie, J. Adult skin-derived precursor Schwann cells exhibit superior myelination and regeneration supportive properties compared to chronically denervated nerve-derived Schwann cells. Exp. Neurol. 2016, 278, 127–142. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, O.A.; Gordon, T. Role of chronic Schwann cell denervation in poor functional recovery after nerve injuries and experimental strategies to combat it. Neurosurgery 2009, 65 (Suppl. 4), A105–A114. [Google Scholar] [CrossRef]
- Painter, M.W.; Brosius Lutz, A.; Cheng, Y.C.; Latremoliere, A.; Duong, K.; Miller, C.M.; Posada, S.; Cobos, E.J.; Zhang, A.X.; Wagers, A.J.; et al. Diminished Schwann cell repair responses underlie age-associated impaired axonal regeneration. Neuron 2014, 83, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.; Ceballos, D.; Vilches, J.J.; Navarro, X. Influence of aging on peripheral nerve function and regeneration. J. Peripher. Nerv. Syst. 2000, 5, 191–208. [Google Scholar] [CrossRef]
- Buttner, R.; Schulz, A.; Reuter, M.; Akula, A.K.; Mindos, T.; Carlstedt, A.; Riecken, L.B.; Baader, S.L.; Bauer, R.; Morrison, H. Inflammaging impairs peripheral nerve maintenance and regeneration. Aging Cell 2018, 17, e12833. [Google Scholar] [CrossRef]
- Jessen, K.R.; Mirsky, R. The Success and Failure of the Schwann Cell Response to Nerve Injury. Front. Cell Neurosci. 2019, 13, 33. [Google Scholar] [CrossRef] [PubMed]
- Stierli, S.; Imperatore, V.; Lloyd, A.C. Schwann cell plasticity-roles in tissue homeostasis, regeneration, and disease. Glia 2019, 67, 2203–2215. [Google Scholar] [CrossRef] [PubMed]
- Wagstaff, L.J.; Gomez-Sanchez, J.A.; Fazal, S.V.; Otto, G.W.; Kilpatrick, A.M.; Michael, K.; Wong, L.Y.N.; Ma, K.H.; Turmaine, M.; Svaren, J.; et al. Failures of nerve regeneration caused by aging or chronic denervation are rescued by restoring Schwann cell c-Jun. Elife 2021, 10, e62232. [Google Scholar] [CrossRef] [PubMed]
- Kohama, T.; Olivera, A.; Edsall, L.; Nagiec, M.M.; Dickson, R.; Spiegel, S. Molecular cloning and functional characterization of murine sphingosine kinase. J. Biol. Chem. 1998, 273, 23722–23728. [Google Scholar] [CrossRef] [PubMed]
- Olivera, A.; Spiegel, S. Sphingosine-1-phosphate as second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature 1993, 365, 557–560. [Google Scholar] [CrossRef] [PubMed]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, S.; Milstien, S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat. Rev. Immunol. 2011, 11, 403–415. [Google Scholar] [CrossRef]
- Heinen, A.; Beyer, F.; Tzekova, N.; Hartung, H.P.; Küry, P. Fingolimod induces the transition to a nerve regeneration promoting Schwann cell phenotype. Exp. Neurol. 2015, 271, 25–35. [Google Scholar] [CrossRef]
- Arthur-Farraj, P.J.; Latouche, M.; Wilton, D.K.; Quintes, S.; Chabrol, E.; Banerjee, A.; Woodhoo, A.; Jenkins, B.; Rahman, M.; Turmaine, M.; et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron 2012, 75, 633–647. [Google Scholar] [CrossRef]
- Parkinson, D.B.; Bhaskaran, A.; Arthur-Farraj, P.; Noon, L.A.; Woodhoo, A.; Lloyd, A.C.; Feltri, M.L.; Wrabetz, L.; Behrens, A.; Mirsky, R.; et al. c-Jun is a negative regulator of myelination. J. Cell Biol. 2008, 181, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Schira, J.; Heinen, A.; Poschmann, G.; Ziegler, B.; Hartung, H.P.; Stühler, K.; Küry, P. Secretome analysis of nerve repair mediating Schwann cells reveals Smad-dependent trophism. FASEB J. 2019, 33, 4703–4715. [Google Scholar] [CrossRef]
- Rogister, B.; Delree, P.; Leprince, P.; Martin, D.; Sadzot, C.; Malgrange, B.; Munaut, C.; Rigo, J.M.; Lefebvre, P.P.; Octave, J.N.; et al. Transforming growth factor beta as a neuronoglial signal during peripheral nervous system response to injury. J. Neurosci. Res. 1993, 34, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, O.A.; Gordon, T. Transforming growth factor-beta and forskolin attenuate the adverse effects of long-term Schwann cell denervation on peripheral nerve regeneration in vivo. Glia 2002, 37, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Clements, M.P.; Byrne, E.; Camarillo Guerrero, L.F.; Cattin, A.L.; Zakka, L.; Ashraf, A.; Burden, J.J.; Khadayate, S.; Lloyd, A.C.; Marguerat, S.; et al. The Wound Microenvironment Reprograms Schwann Cells to Invasive Mesenchymal-like Cells to Drive Peripheral Nerve Regeneration. Neuron 2017, 96, 98–114.e7. [Google Scholar] [CrossRef]
- Zeng, Y.; Yao, X.; Chen, L.; Yan, Z.; Liu, J.; Zhang, Y.; Feng, T.; Wu, J.; Liu, X. Sphingosine-1-phosphate induced epithelial-mesenchymal transition of hepatocellular carcinoma via an MMP-7/ syndecan-1/TGF-beta autocrine loop. Oncotarget 2016, 7, 63324–63337. [Google Scholar] [CrossRef]
- Milara, J.; Navarro, R.; Juan, G.; Peiro, T.; Serrano, A.; Ramon, M.; Morcillo, E.; Cortijo, J. Sphingosine-1-phosphate is increased in patients with idiopathic pulmonary fibrosis and mediates epithelial to mesenchymal transition. Thorax 2012, 67, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Davis, M.D.; Heise, C.E.; Albert, R.; Cottens, S.; Hof, R.; Bruns, C.; Prieschl, E.; Baumruker, T.; Hiestand, P.; et al. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 2002, 277, 21453–21457. [Google Scholar] [CrossRef] [PubMed]
- Mandala, S.; Hajdu, R.; Bergstrom, J.; Quackenbush, E.; Xie, J.; Milligan, J.; Thornton, R.; Shei, G.J.; Card, D.; Keohane, C.; et al. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 2002, 296, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Jung, C.G.; Dukala, D.; Bae, H.; Kakazu, R.; Wollmann, R.; Soliven, B. Fingolimod and related compounds in a spontaneous autoimmune polyneuropathy. J. Neuroimmunol. 2009, 214, 93–100. [Google Scholar] [CrossRef][Green Version]
- Köhne, A.; Stettner, M.; Jangouk, P.; Dehmel, T.; Hartung, H.P.; Lehmann, H.C.; Kieseier, B.C. Fingolimod impedes Schwann cell-mediated myelination: Implications for the treatment of immune neuropathies? Arch. Neurol. 2012, 69, 1280–1289. [Google Scholar] [CrossRef]
- Brockes, J.P.; Fields, K.L.; Raff, M.C. Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve. Brain Res. 1979, 165, 105–118. [Google Scholar] [CrossRef]
- Heinen, A.; Kremer, D.; Göttle, P.; Kruse, F.; Hasse, B.; Lehmann, H.; Hartung, H.P.; Küry, P. The cyclin-dependent kinase inhibitor p57kip2 is a negative regulator of Schwann cell differentiation and in vitro myelination. Proc. Natl. Acad. Sci. USA 2008, 105, 8748–8753. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiang, P.; Chew, W.S.; Torta, F.; Bandla, A.; Lopez, V.; Seow, W.L.; Lam, B.W.S.; Chang, J.K.; Wong, P.; et al. Activation of sphingosine 1-phosphate receptor 2 attenuates chemotherapy-induced neuropathy. J. Biol. Chem. 2020, 295, 1143–1152. [Google Scholar] [CrossRef]
- Cahalan, S.M.; Gonzalez-Cabrera, P.J.; Nguyen, N.; Guerrero, M.; Cisar, E.A.; Leaf, N.B.; Brown, S.J.; Roberts, E.; Rosen, H. Sphingosine 1-phosphate receptor 1 (S1P(1)) upregulation and amelioration of experimental autoimmune encephalomyelitis by an S1P(1) antagonist. Mol. Pharmacol. 2013, 83, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Takasugi, H.; Ohnuma, S.; Koide, Y.; Sakurai, A.; Takeda, S.; Hasegawa, T.; Sasamori, J.; Konno, T.; Hayashi, K.; et al. Sphingosine 1-phosphate (S1P) regulates vascular contraction via S1P3 receptor: Investigation based on a new S1P3 receptor antagonist. Mol. Pharmacol. 2010, 77, 704–713. [Google Scholar] [CrossRef]
- Patmanathan, S.N.; Wang, W.; Yap, L.F.; Herr, D.R.; Paterson, I.C. Mechanisms of sphingosine 1-phosphate receptor signalling in cancer. Cell. Signal. 2017, 34, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Jo, E.; Bhhatarai, B.; Repetto, E.; Guerrero, M.; Riley, S.; Brown, S.J.; Kohno, Y.; Roberts, E.; Schurer, S.C.; Rosen, H. Novel selective allosteric and bitopic ligands for the S1P(3) receptor. ACS Chem. Biol. 2012, 7, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Lucaciu, A.; Brunkhorst, R.; Pfeilschifter, J.M.; Pfeilschifter, W.; Subburayalu, J. The S1P-S1PR Axis in Neurological Disorders-Insights into Current and Future Therapeutic Perspectives. Cells 2020, 9, 1515. [Google Scholar] [CrossRef] [PubMed]
- Meyer Zu Reckendorf, S.; Brand, C.; Pedro, M.T.; Hegler, J.; Schilling, C.S.; Lerner, R.; Bindila, L.; Antoniadis, G.; Knoll, B. Lipid metabolism adaptations are reduced in human compared to murine Schwann cells following injury. Nat. Commun. 2020, 11, 2123. [Google Scholar] [CrossRef]
- Barkovits, K.; Pfeiffer, K.; Eggers, B.; Marcus, K. Protein Quantification Using the “Rapid Western Blot” Approach. Methods Mol. Biol. 2021, 2228, 29–39. [Google Scholar] [PubMed]
- Diller, T.; Thompson, J.; Steer, B. Biological validation of a novel process and product for quantitating western blots. J. Biotechnol. 2021, 326, 52–60. [Google Scholar] [CrossRef] [PubMed]
| Gene | Primer Sequence |
|---|---|
| GAP43 | fwd: CCG GAG GAT AAG GCT CAT AAG G rev: TTG TTA TGT GTC CAC GGA AGC T |
| GAPDH | fwd: GAA CGG GAA GCT CAC TGG C rev: GCA TGT CAG ATC CAC AAC GG |
| Krox20 | fwd: TTT TTC CAT CTC CGT GCC A rev: GAA CGG CTT TCG ATC AGG G |
| Mal | fwd: AGG AGG CCT TTG GTT ATC CC rev: GCA AAT GGC AGA TTT GGG TAC |
| Oct-6 | fwd: GGC ACC CTC TAC GGT AAT GTG T rev: TTG AGC AGC GGT TTG AGC T |
| ODC | fwd: GGT TCC AGA GGC CAA ACA TC rev: GTT GCC ACA TTG ACC GTG AC |
| PDGF-BB | fwd: GTT CGG ACG GTG CGA ATC rev: GTG TGC TTA AAC TTT CGG TGC TT |
| S1P1 | fwd: AAA TGC CCC AAC GGA GAC T rev: TCC ATG CCC GGG ATG AT |
| S1P2 | fwd: GGT CAA GCT CTA CGG CAG TGA rev: AAG AGG CCC CAA TGA GCA T |
| S1P3 | fwd: TCC TCA ACT CGG CCA TGA A rev: ACG CCG CAT CTC TTT GCT |
| S1P4 | fwd: TGA GGC CCA GGG ACA GTT T rev: GGC TCT CGC ATC TTG AAG CT |
| S1P5 | fwd: TTG GCT GTG TGC GCT TTC rev: GGT GCA TGG AAG CGA GGA T |
| Tyrp1 | fwd: CTT CGT CAG GGC CTT GGA rev: GCA ATG ACA AAT TGA GGG TGA GT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schira-Heinen, J.; Wang, L.; Akgün, S.; Blum, S.; Ziegler, B.; Heinen, A.; Hartung, H.-P.; Küry, P. Modulation of Specific Sphingosine-1-Phosphate Receptors Augments a Repair Mediating Schwann Cell Phenotype. Int. J. Mol. Sci. 2022, 23, 10311. https://doi.org/10.3390/ijms231810311
Schira-Heinen J, Wang L, Akgün S, Blum S, Ziegler B, Heinen A, Hartung H-P, Küry P. Modulation of Specific Sphingosine-1-Phosphate Receptors Augments a Repair Mediating Schwann Cell Phenotype. International Journal of Molecular Sciences. 2022; 23(18):10311. https://doi.org/10.3390/ijms231810311
Chicago/Turabian StyleSchira-Heinen, Jessica, Luzhou Wang, Seda Akgün, Sofia Blum, Brigida Ziegler, André Heinen, Hans-Peter Hartung, and Patrick Küry. 2022. "Modulation of Specific Sphingosine-1-Phosphate Receptors Augments a Repair Mediating Schwann Cell Phenotype" International Journal of Molecular Sciences 23, no. 18: 10311. https://doi.org/10.3390/ijms231810311
APA StyleSchira-Heinen, J., Wang, L., Akgün, S., Blum, S., Ziegler, B., Heinen, A., Hartung, H.-P., & Küry, P. (2022). Modulation of Specific Sphingosine-1-Phosphate Receptors Augments a Repair Mediating Schwann Cell Phenotype. International Journal of Molecular Sciences, 23(18), 10311. https://doi.org/10.3390/ijms231810311







