The Inflammasome Activity of NLRP3 Is Independent of NEK7 in HEK293 Cells Co-Expressing ASC
Abstract
:1. Introduction
2. Results
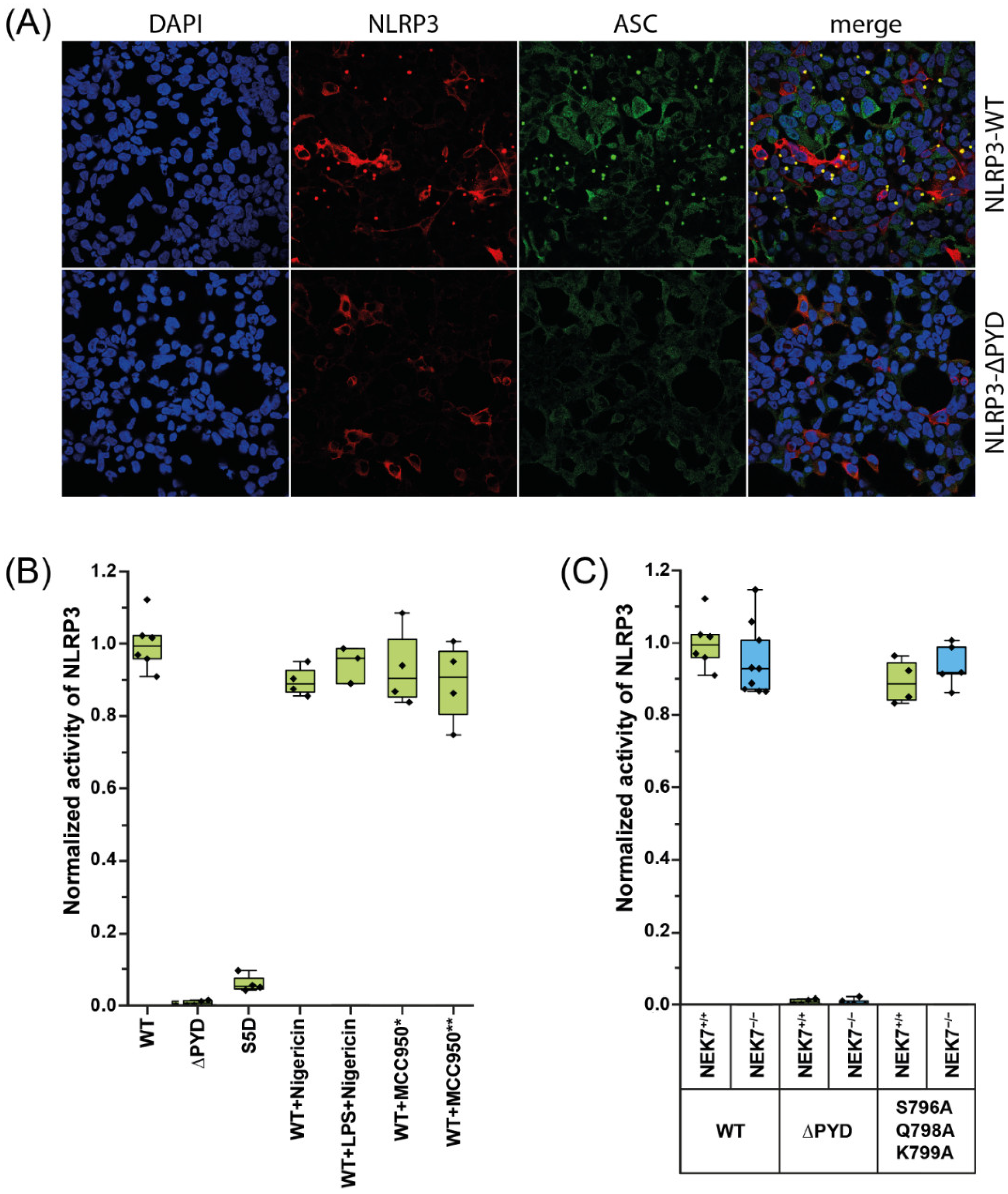
2.1. NLRP3 Is Constitutively Active in HEK293-ASC Cells Independent of NEK7
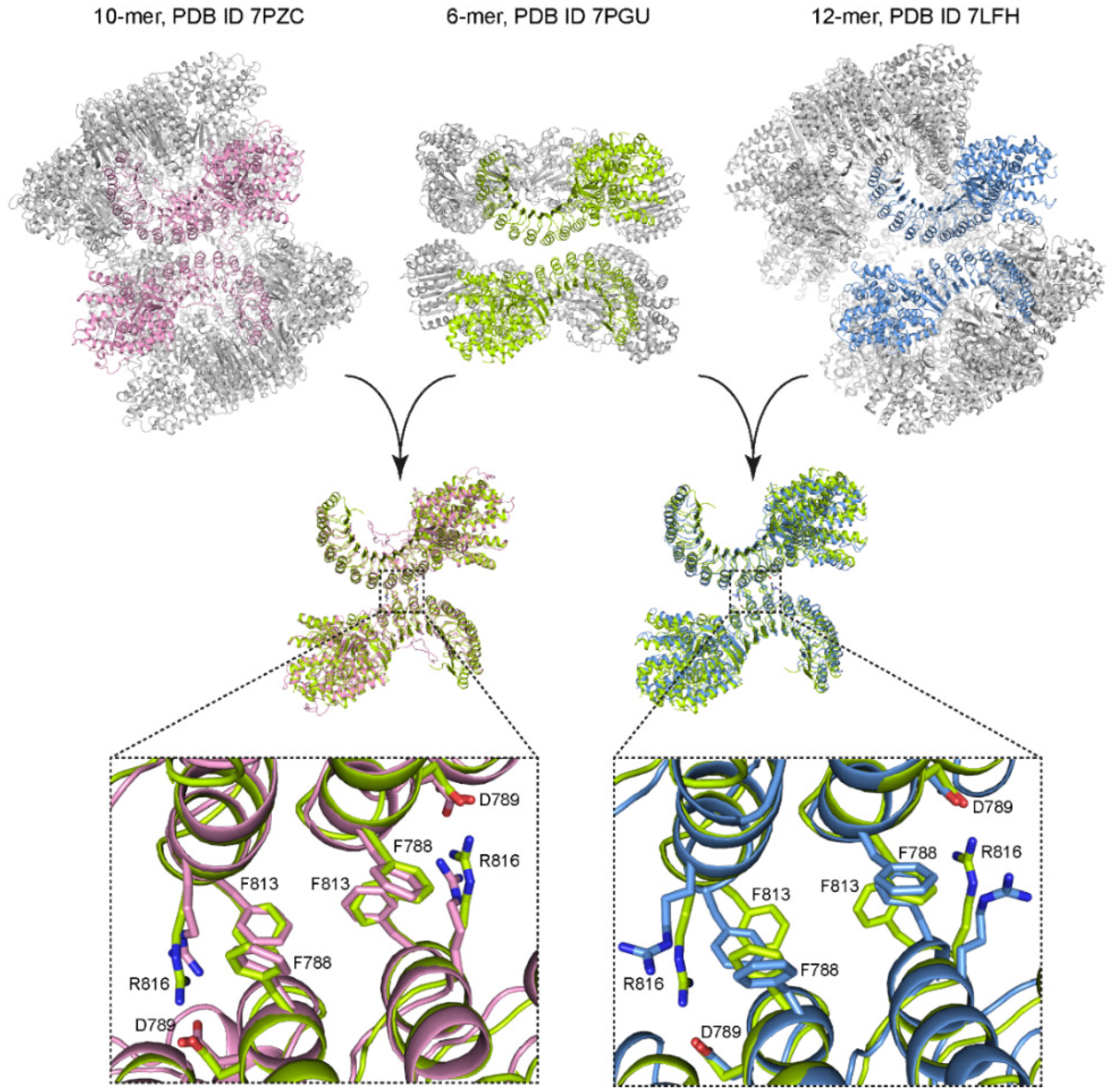
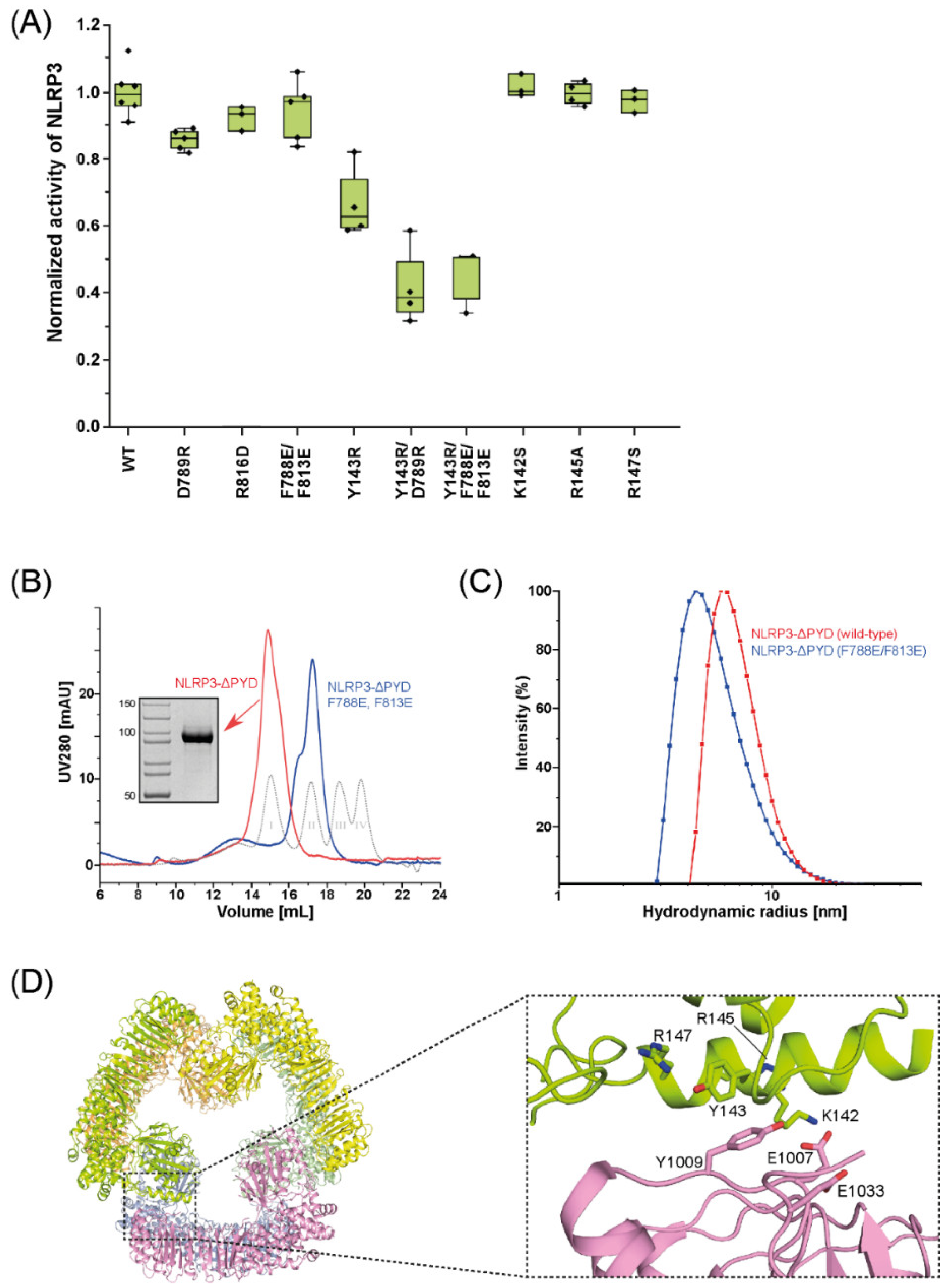
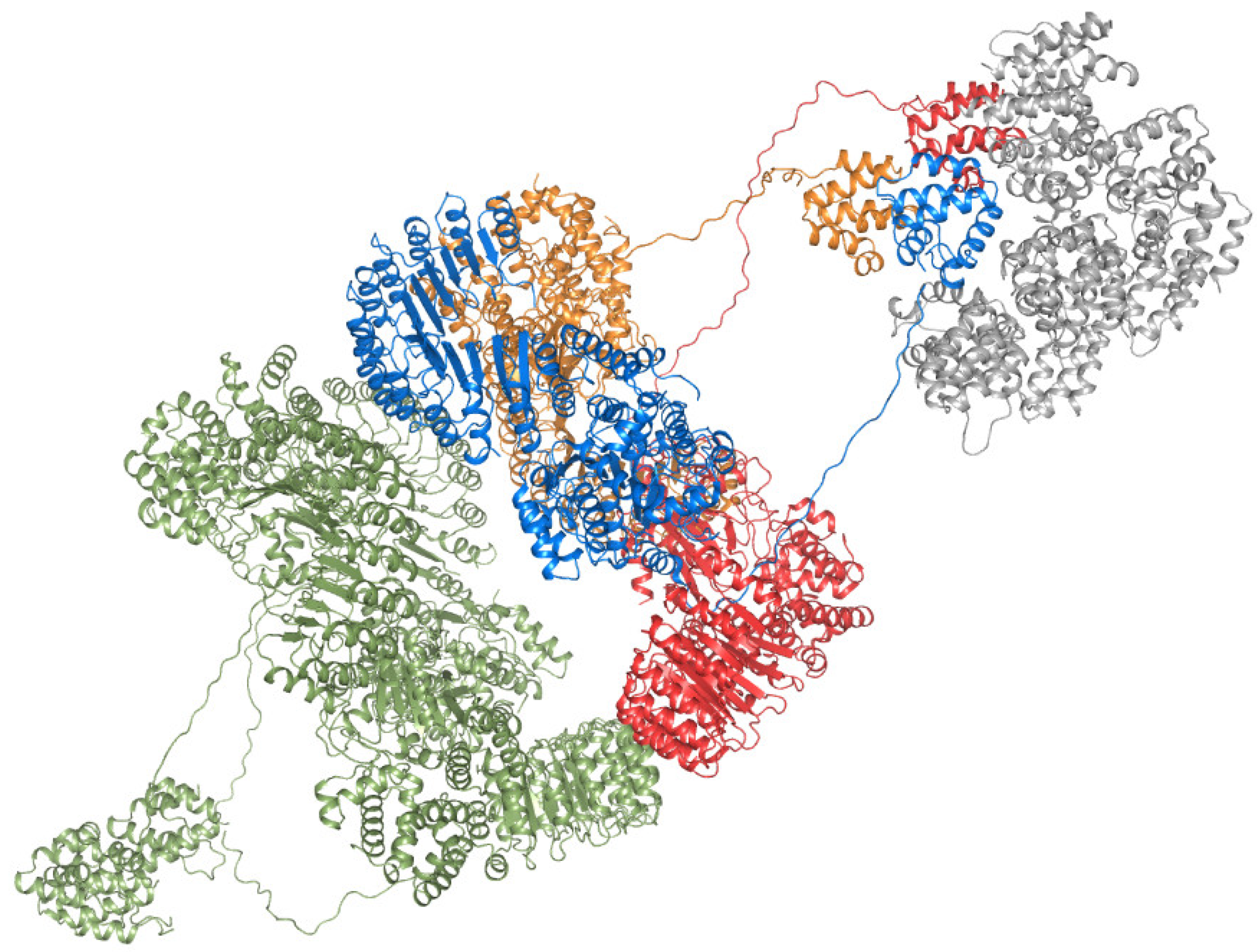
2.2. Mutations in the Interfaces of the NLRP3 Hexamer Influence NLRP3 Activity
3. Discussion
4. Materials and Methods
4.1. Cloning, Expression and Purification of Recombinant Human NLRP3
4.2. Analytical Size Exclusion Chromatography
4.3. Dynamic Light Scattering
4.4. ASC Speck Assay
4.5. CRISPR/Cas Knockout of NEK7 in HEK293-ASC Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mangan, M.S.J.; Olhava, E.J.; Roush, W.R.; Seidel, H.M.; Glick, G.D.; Latz, E. Targeting the NLRP3 Inflammasome in Inflammatory Diseases. Nat. Rev. Drug Discov. 2018, 17, 588–606. [Google Scholar] [CrossRef]
- Swanson, K.v.; Deng, M.; Ting, J.P.-Y. The NLRP3 Inflammasome: Molecular Activation and Regulation to Therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dai, Y.; Li, Q.; Chen, C.; Chen, H.; Song, Y.; Hua, F.; Zhang, Z. Beta-Amyloid Activates NLRP3 Inflammasome via TLR4 in Mouse Microglia. Neurosci. Lett. 2020, 736, 135279. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Pétrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-Associated Uric Acid Crystals Activate the NALP3 Inflammasome. Nature 2006, 440, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin Activates the Inflammasome in Response to Toxins and ATP. Nature 2006, 440, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Spel, L.; Martinon, F. Detection of Viruses by Inflammasomes. Curr. Opin. Virol. 2021, 46, 59–64. [Google Scholar] [CrossRef]
- Shimada, K.; Crother, T.R.; Karlin, J.; Dagvadorj, J.; Chiba, N.; Chen, S.; Ramanujan, V.K.; Wolf, A.J.; Vergnes, L.; Ojcius, D.M.; et al. Oxidized Mitochondrial DNA Activates the NLRP3 Inflammasome during Apoptosis. Immunity 2012, 36, 401–414. [Google Scholar] [CrossRef]
- Boucher, D.; Monteleone, M.; Coll, R.C.; Chen, K.W.; Ross, C.M.; Teo, J.L.; Gomez, G.A.; Holley, C.L.; Bierschenk, D.; Stacey, K.J.; et al. Caspase-1 Self-Cleavage Is an Intrinsic Mechanism to Terminate Inflammasome Activity. J. Exp. Med. 2018, 215, 827–840. [Google Scholar] [CrossRef]
- Cai, X.; Chen, J.; Xu, H.; Liu, S.; Jiang, Q.X.; Halfmann, R.; Chen, Z.J. Prion-like Polymerization Underlies Signal Transduction in Antiviral Immune Defense and Inflammasome Activation. Cell 2014, 156, 1207–1222. [Google Scholar] [CrossRef]
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell 2014, 156, 1193–1206. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Hara, H.; Núñez, G. Mechanism and Regulation of NLRP3 Inflammasome Activation. Trends. Biochem. Sci. 2016, 41, 1012–1021. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S.; et al. NLRP3 Activation and Mitosis Are Mutually Exclusive Events Coordinated by NEK7, a New Inflammasome Component. Nat. Immunol. 2016, 17, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A Genome-Wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.M.; Bayliss, R.; Roig, J. Mitotic Regulation by NEK Kinase Networks. Front. Cell Dev. Biol. 2017, 5, 102. [Google Scholar] [CrossRef] [PubMed]
- Fry, A.M.; O’Regan, L.; Sabir, S.R.; Bayliss, R. Cell Cycle Regulation by the NEK Family of Protein Kinases. J. Cell Sci. 2012, 125, 4423–4433. [Google Scholar] [CrossRef]
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.v.; Wu, Z.; Núñez, G.; Mao, Y.; et al. Structural Mechanism for NEK7-Licensed Activation of NLRP3 Inflammasome. Nature 2019, 570, 338–343. [Google Scholar] [CrossRef]
- Andreeva, L.; David, L.; Rawson, S.; Shen, C.; Pasricha, T.; Pelegrin, P.; Wu, H. NLRP3 Cages Revealed by Full-Length Mouse NLRP3 Structure Control Pathway Activation. Cell 2021, 184, 6299–6312. [Google Scholar] [CrossRef]
- Hochheiser, I.v.; Pilsl, M.; Hagelueken, G.; Moecking, J.; Marleaux, M.; Brinkschulte, R.; Latz, E.; Engel, C.; Geyer, M. Structure of the NLRP3 Decamer Bound to the Cytokine Release Inhibitor CRID3. Nature 2022, 604, 184–189. [Google Scholar] [CrossRef]
- Ohto, U.; Kamitsukasa, Y.; Ishida, H.; Zhang, Z.; Murakami, K.; Hirama, C.; Maekawa, S.; Shimizu, T. Structural Basis for the Oligomerization-Mediated Regulation of NLRP3 Inflammasome Activation. Proc. Natl. Acad. Sci. USA 2022, 119, e2121353119. [Google Scholar] [CrossRef]
- Zito, G.; Buscetta, M.; Cimino, M.; Dino, P.; Bucchieri, F.; Cipollina, C. Cellular Models and Assays to Study NLRP3 Inflammasome Biology. Int. J. Mol. Sci. 2020, 21, 4294. [Google Scholar] [CrossRef]
- Chen, S.; Chi, Z.; Wang, D. Reconstitution System of NLRP3 Inflammasome in HEK293T Cells. Methods Mol. Biol. 2022, 2459, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Murray, A.; Beutler, B. Reconstruction of the Mouse Inflammasome System in HEK293T Cells. Bio-Protocol 2016, 6, e1986. [Google Scholar] [CrossRef] [PubMed]
- Compan, V.; López-Castejón, G. Functional Reconstruction of NLRs in HEK293 Cells. Methods Mol. Biol. 2016, 1417, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Stutz, A.; Kolbe, C.-C.; Stahl, R.; Horvath, G.L.; Franklin, B.S.; van Ray, O.; Brinkschulte, R.; Geyer, M.; Meissner, F.; Latz, E. NLRP3 Inflammasome Assembly Is Regulated by Phosphorylation of the Pyrin Domain. J. Exp. Med. 2017, 214, 1725–1736. [Google Scholar] [CrossRef] [PubMed]
- Buljan, M.; Ciuffa, R.; van Drogen, A.; Vichalkovski, A.; Mehnert, M.; Rosenberger, G.; Lee, S.; Varjosalo, M.; Pernas, L.E.; Spegg, V.; et al. Kinase Interaction Network Expands Functional and Disease Roles of Human Kinases. Mol. Cell 2020, 79, 504–520.e9. [Google Scholar] [CrossRef] [PubMed]
- O’Regan, L.; Fry, A.M. The Nek6 and Nek7 Protein Kinases Are Required for Robust Mitotic Spindle Formation and Cytokinesis. Mol. Cell Biol. 2009, 29, 3975–3990. [Google Scholar] [CrossRef]
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Núñez, G. NEK7 Is an Essential Mediator of NLRP3 Activation Downstream of Potassium Efflux. Nature 2016, 530, 354–357. [Google Scholar] [CrossRef]
- Niu, T.; de Rosny, C.; Chautard, S.; Rey, A.; Patoli, D.; Groslambert, M.; Cosson, C.; Lagrange, B.; Zhang, Z.; Visvikis, O.; et al. NLRP3 Phosphorylation in Its LRR Domain Critically Regulates Inflammasome Assembly. Nat. Commun. 2021, 12, 5862. [Google Scholar] [CrossRef]
- Chen, J.; Chen, Z.J. PtdIns4P on Dispersed Trans-Golgi Network Mediates NLRP3 Inflammasome Activation. Nature 2018, 564, 71–76. [Google Scholar] [CrossRef]
- Kelley, N.; Jeltema, D.; Duan, Y.; He, Y. The NLRP3 Inflammasome: An Overview of Mechanisms of Activation and Regulation. Int. J. Mol. Sci. 2019, 20, 3328. [Google Scholar] [CrossRef] [Green Version]
- Dekker, C.; Mattes, H.; Wright, M.; Boettcher, A.; Hinniger, A.; Hughes, N.; Kapps-Fouthier, S.; Eder, J.; Erbel, P.; Stiefl, N.; et al. Crystal Structure of NLRP3 NACHT Domain With an Inhibitor Defines Mechanism of Inflammasome Inhibition. J. Mol. Biol. 2021, 433, 167309. [Google Scholar] [CrossRef] [PubMed]
- Gambin, Y.; Giles, N.; O’Carroll, A.; Polinkovsky, M.; Hunter, D.; Sierecki, E. Single-Molecule Fluorescence Reveals the Oligomerization and Folding Steps Driving the Prion-like Behavior of ASC. J. Mol. Biol. 2018, 430, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Sušjan, P.; Roškar, S.; Hafner-Bratkovič, I. The Mechanism of NLRP3 Inflammasome Initiation: Trimerization but Not Dimerization of the NLRP3 Pyrin Domain Induces Robust Activation of IL-1β. Biochem. Biophys. Res. Commun. 2017, 483, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Tapia-Abellán, A.; Angosto-Bazarra, D.; Alarcón-Vila, C.; Baños, M.C.; Hafner-Bratkovič, I.; Oliva, B.; Pelegrín, P. Sensing Low Intracellular Potassium by NLRP3 Results in a Stable Open Structure That Promotes Inflammasome Activation. Sci. Adv. 2021, 7, eabf4468. [Google Scholar] [CrossRef]
- Ito, S.; Hara, Y.; Kubota, T. CARD8 Is a Negative Regulator for NLRP3 Inflammasome, but Mutant NLRP3 in Cryopyrin-Associated Periodic Syndromes Escapes the Restriction. Arthritis Res. Ther. 2014, 16, R52. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Ran, F.; Hsu, P.; Wright, J.; Agarwala, V.; Scott, D.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Brinkman, E.; van Steensel, B. Rapid Quantitative Evaluation of CRISPR Genome Editing by TIDE and TIDER. Methods Mol. Biol. 2019, 1961, 29–44. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Machtens, D.A.; Bresch, I.P.; Eberhage, J.; Reubold, T.F.; Eschenburg, S. The Inflammasome Activity of NLRP3 Is Independent of NEK7 in HEK293 Cells Co-Expressing ASC. Int. J. Mol. Sci. 2022, 23, 10269. https://doi.org/10.3390/ijms231810269
Machtens DA, Bresch IP, Eberhage J, Reubold TF, Eschenburg S. The Inflammasome Activity of NLRP3 Is Independent of NEK7 in HEK293 Cells Co-Expressing ASC. International Journal of Molecular Sciences. 2022; 23(18):10269. https://doi.org/10.3390/ijms231810269
Chicago/Turabian StyleMachtens, Dominik Alexander, Ian Philipp Bresch, Jan Eberhage, Thomas Frank Reubold, and Susanne Eschenburg. 2022. "The Inflammasome Activity of NLRP3 Is Independent of NEK7 in HEK293 Cells Co-Expressing ASC" International Journal of Molecular Sciences 23, no. 18: 10269. https://doi.org/10.3390/ijms231810269
APA StyleMachtens, D. A., Bresch, I. P., Eberhage, J., Reubold, T. F., & Eschenburg, S. (2022). The Inflammasome Activity of NLRP3 Is Independent of NEK7 in HEK293 Cells Co-Expressing ASC. International Journal of Molecular Sciences, 23(18), 10269. https://doi.org/10.3390/ijms231810269





