Melatonin Inhibits hIAPP Oligomerization by Preventing β-Sheet and Hydrogen Bond Formation of the Amyloidogenic Region Revealed by Replica-Exchange Molecular Dynamics Simulation
Abstract
:1. Introduction
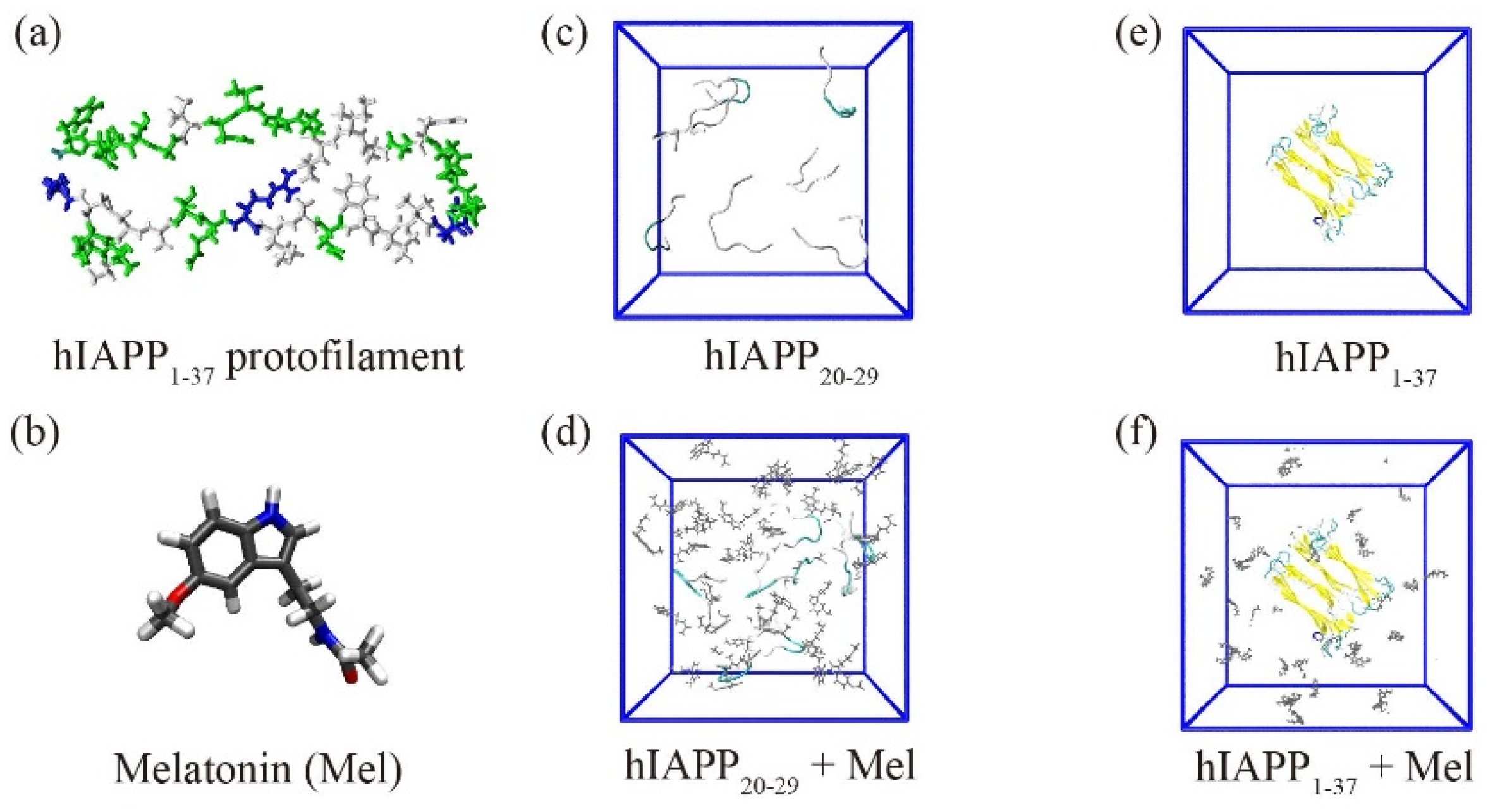
2. Results and Discussion
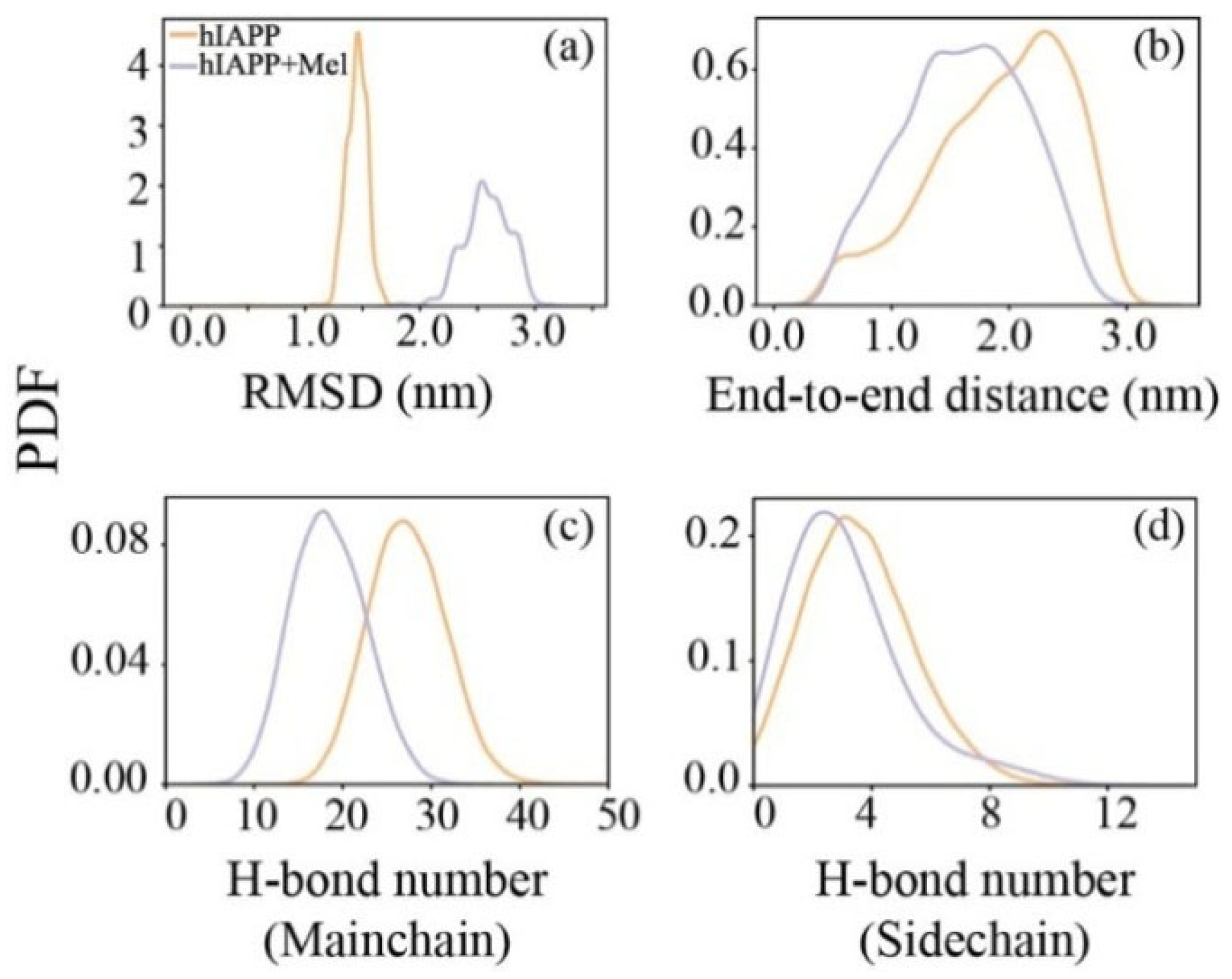
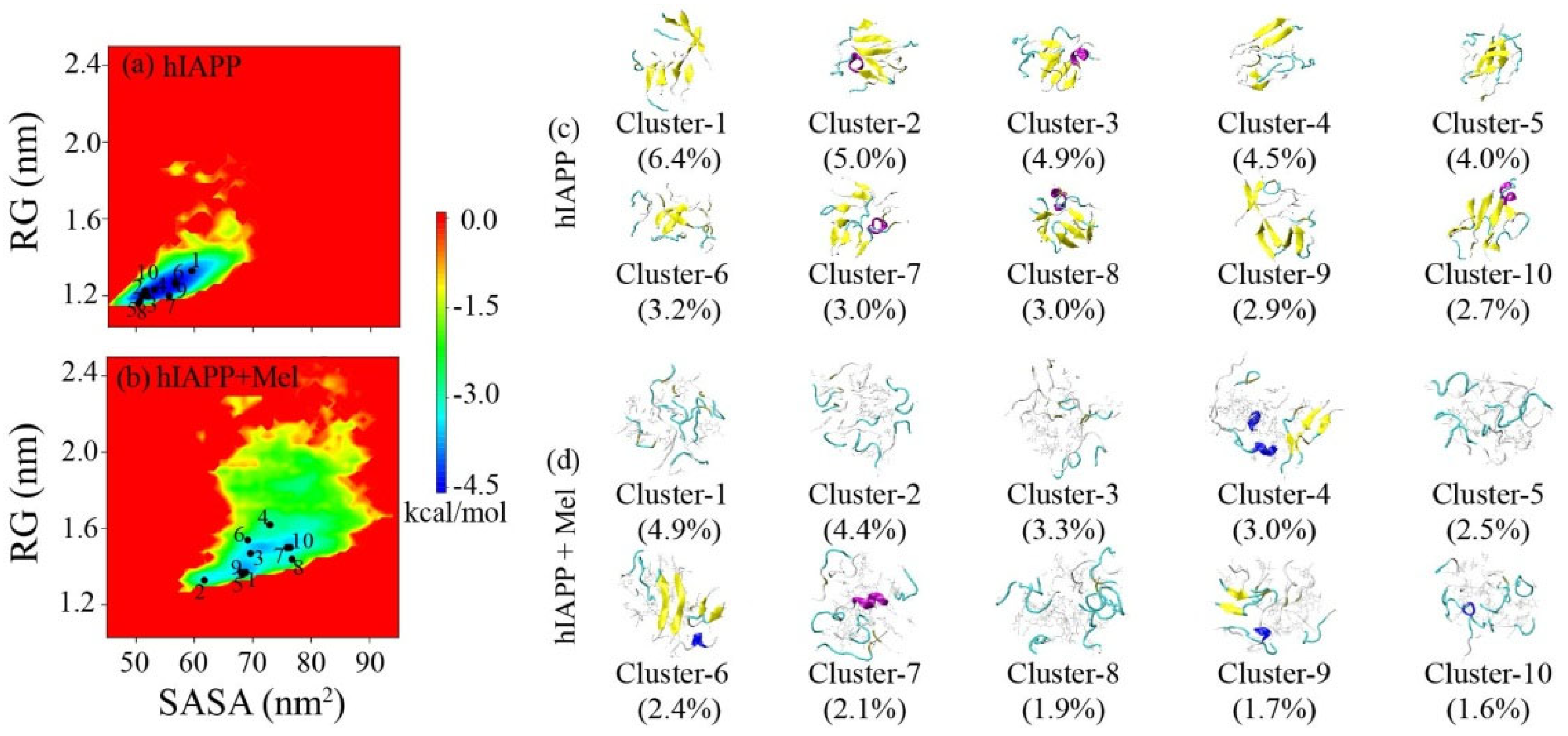
2.1. Mel Significantly Reduces β-Sheet Formation of hIAPP20–29
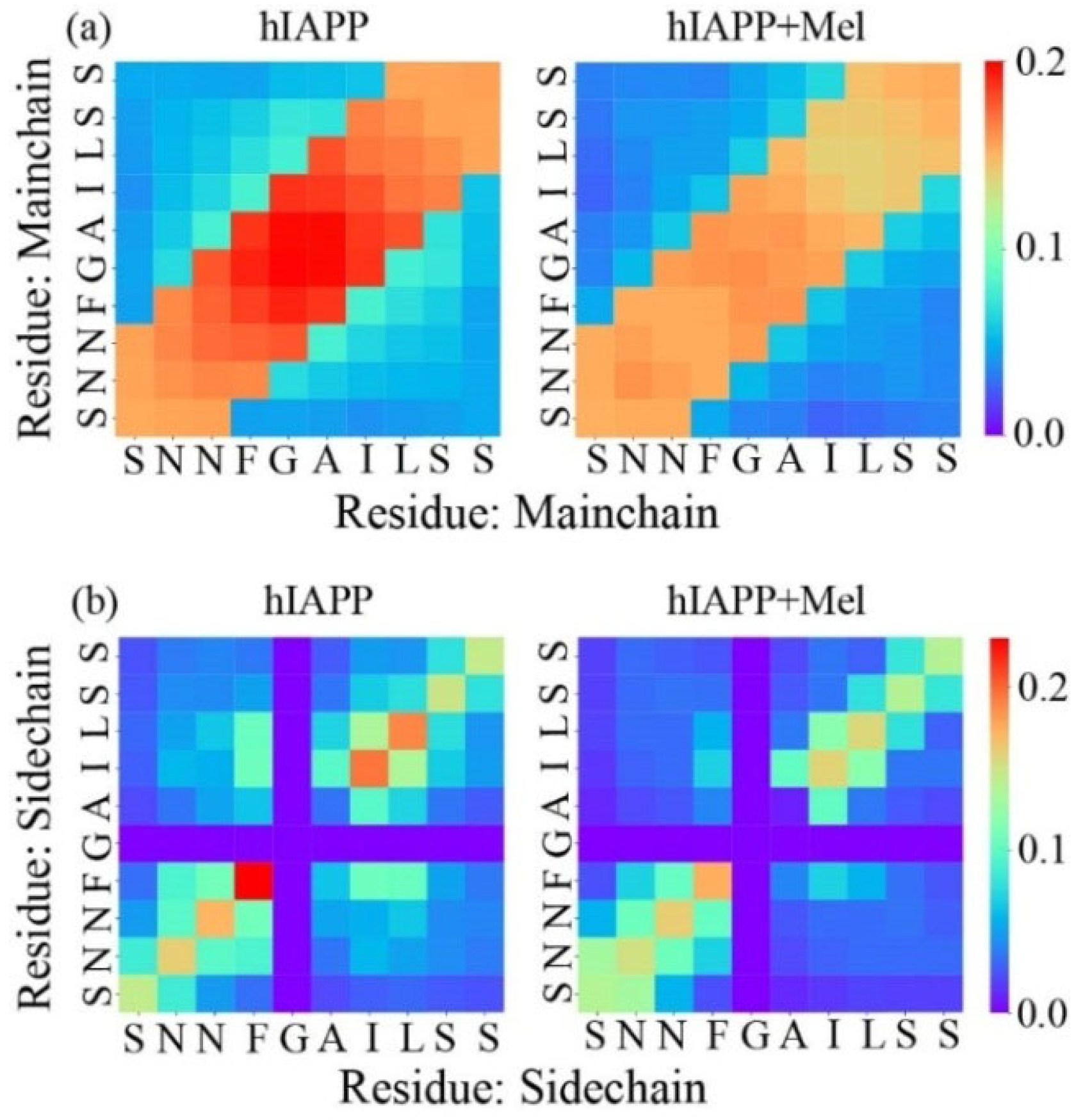
2.2. Mel Reduces Hydrogen Bond Formation of hIAPP20–29 and Transforms Peptide Conformations into Less Compacted Oligomers
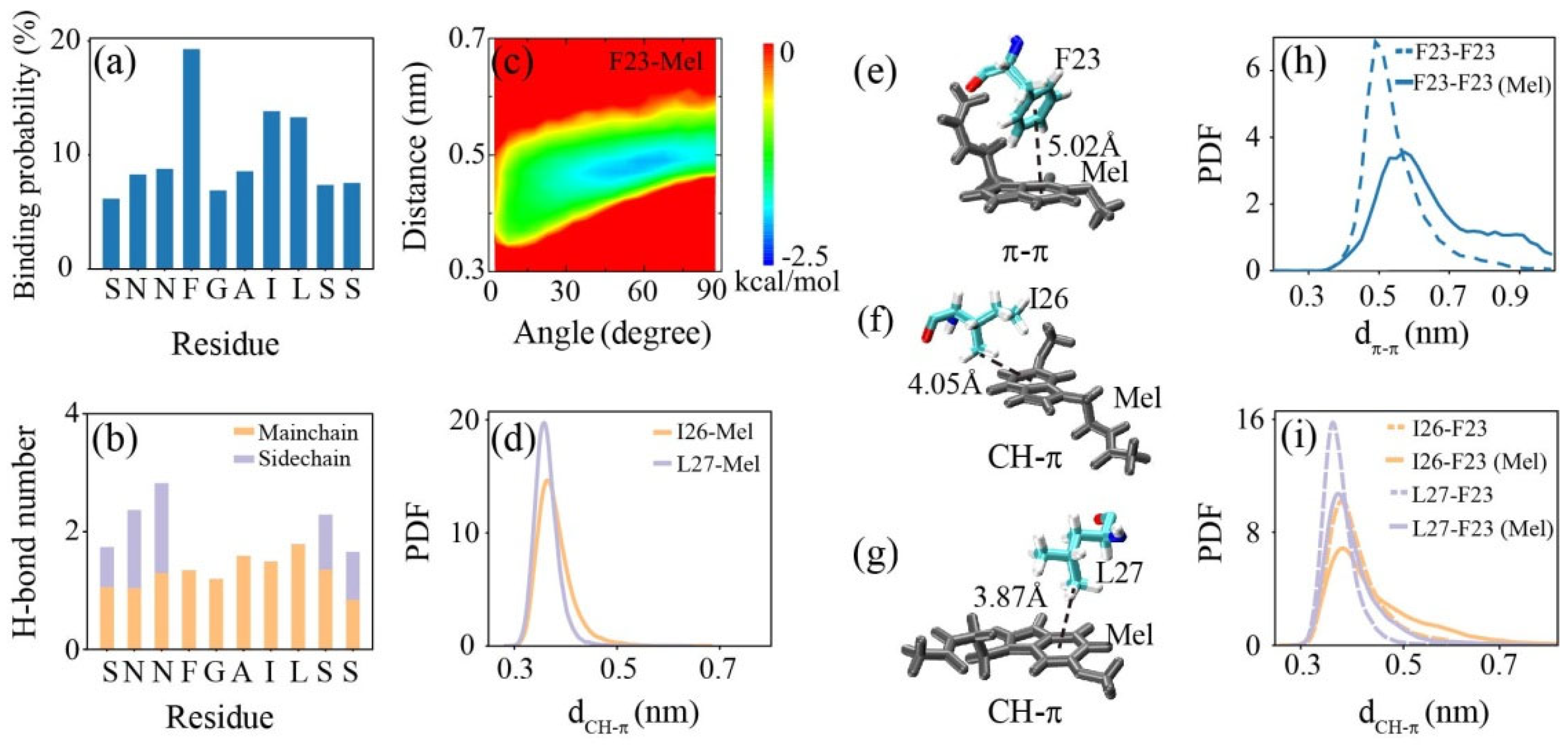
2.3. Peptide-Mel Interaction Analysis Indicates That Hydrogen Bonding, π–π, and CH–π Interactions Play Important Roles in Amyloid Inhibition
2.4. Mel Has a Slight Influence on the Structural Stability of Preformed hIAPP1–37 Fibrillar Octamer and Has a High Binding Affinity to the Amyloidogenic Region
3. Materials and Methods
3.1. Modeling hIAPP and Melatonin
3.2. Simulation Details
3.3. Analysis Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IDF Diabetes Atlas. Available online: https://www.idf.org/e-library/epidemiology-research/diabetes-atlas/159-idf-diabetes-atlas-ninth-edition-2019.html (accessed on 2 August 2022).
- Fiedorova, K.; Augustynek, M.; Kubicek, J.; Kudrna, P.; Bibbo, D. Review of present method of glucose from human blood and body fluids assessment. Biosens. Bioelectron. 2022, 211, 114348. [Google Scholar] [CrossRef] [PubMed]
- Röcken, C.; Linke, R.P.; Saeger, W. Immunohistology of islet amyloid polypeptide in diabetes mellitus: Semi-quantitative studies in a post-mortem series. Virchows Arch. 1992, 421, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef] [PubMed]
- Eizirik, D.L.; Pasquali, L.; Cnop, M. Pancreatic β-cells in type 1 and type 2 diabetes mellitus: Different pathways to failure. Nat. Rev. Endocrinol. 2020, 16, 349–362. [Google Scholar] [CrossRef]
- Khemtémourian, L.; Killian, J.A.; Höppener, J.W.M.; Engel, M.F.M. Recent insights in islet amyloid polypeptide-induced membrane disruption and its role in beta-cell death in type 2 diabetes mellitus. Exp. Diabetes Res. 2008, 2008, 421287. [Google Scholar] [CrossRef]
- Westermark, G.T.; Westermark, P.; Berne, C.; Korsgren, O. Widespread amyloid deposition in transplanted human pancreatic islets. N. Engl. J. Med. 2008, 359, 977–979. [Google Scholar] [CrossRef]
- Milardi, D.; Gazit, E.; Radford, S.E.; Xu, Y.; Gallardo, R.U.; Caflisch, A.; Westermark, G.T.; Westermark, P.; Rosa, C.L.; Ramamoorthy, A. Proteostasis of islet amyloid polypeptide: A molecular perspective of risk factors and protective strategies for type II diabetes. Chem. Rev. 2021, 121, 1845–1893. [Google Scholar] [CrossRef]
- Nguyen, P.H.; Ramamoorthy, A.; Sahoo, B.R.; Zheng, J.; Faller, P.; Straub, J.E.; Dominguez, L.; Shea, J.-E.; Dokholyan, N.V.; Simone, A.D.; et al. Amyloid oligomers: A joint experimental/computational perspective on Alzheimer’s disease, Parkinson’s disease, type II diabetes, and amyotrophic lateral sclerosis. Chem. Rev. 2021, 121, 2545–2647. [Google Scholar] [CrossRef]
- Mittag, T.; Forman-Kay, J.D. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007, 17, 3–14. [Google Scholar] [CrossRef]
- Wei, G.; Xi, W.; Nussinov, R.; Ma, B. Protein ensembles: How does nature harness thermodynamic fluctuations for life? The diverse functional roles of conformational ensembles in the cell. Chem. Rev. 2016, 116, 6516–6551. [Google Scholar] [CrossRef]
- Vaiana, S.M.; Best, R.B.; Yau, W.-M.; Eaton, W.A.; Hofrichter, J. Evidence for a partially structured state of the amylin monomer. Biophys. J. 2009, 97, 2948–2957. [Google Scholar] [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008, 18, 756–764. [Google Scholar] [CrossRef]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing protein intrinsic disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef]
- Guo, A.Z.; Fluitt, A.M.; de Pablo, J.J. Early-stage human islet amyloid polypeptide aggregation: Mechanisms behind dimer formation. J. Chem. Phys. 2018, 149, 025101. [Google Scholar] [CrossRef]
- Cao, Q.; Boyer, D.R.; Sawaya, M.R.; Ge, P.; Eisenberg, D.S. Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils. Nat. Struct. Mol. Biol. 2020, 27, 653–659. [Google Scholar] [CrossRef]
- Fändrich, M. Oligomeric intermediates in amyloid formation: Structure determination and mechanisms of toxicity. J. Mol. Biol. 2012, 421, 427–440. [Google Scholar] [CrossRef]
- Benilova, I.; Karran, E.; De Strooper, B. The toxic Aβ oligomer and Alzheimer’s disease: An emperor in need of clothes. Nat. Neurosci. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Höppener, J.W.M.; Ahrén, B.; Lips, C.J.M. Islet amyloid and type 2 diabetes mellitus. N. Engl. J. Med. 2000, 343, 411–419. [Google Scholar] [CrossRef]
- Brender, J.R.; Lee, E.L.; Cavitt, M.A.; Gafni, A.; Steel, D.G.; Ramamoorthy, A. Amyloid fiber formation and membrane disruption are separate processes localized in two distinct regions of IAPP, the type-2-diabetes-related peptide. J. Am. Chem. Soc. 2008, 130, 6424–6429. [Google Scholar] [CrossRef]
- Buchanan, L.E.; Dunkelberger, E.B.; Tran, H.Q.; Cheng, P.-N.; Chiu, C.-C.; Cao, P.; Raleigh, D.P.; de Pablo, J.J.; Nowick, J.S.; Zanni, M.T. Mechanism of IAPP amyloid fibril formation involves an intermediate with a transient β-sheet. Proc. Natl. Acad. Sci. USA 2013, 110, 19285–19290. [Google Scholar] [CrossRef] [Green Version]
- Porat, Y.; Mazor, Y.; Efrat, S.; Gazit, E. Inhibition of islet amyloid polypeptide fibril formation: A potential role for heteroaromatic interactions. Biochemistry 2004, 43, 14454–14462. [Google Scholar] [CrossRef] [PubMed]
- Scherzer-Attali, R.; Shaltiel-Karyo, R.; Adalist, Y.; Segal, D.; Gazit, E. Generic inhibition of amyloidogenic proteins by two naphthoquinone-tryptophan hybrid molecules. Proteins 2012, 80, 1962–1973. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Guo, J.; Jiao, P.; Liu, H.; Yao, X. Exploring the influence of EGCG on the β-sheet-rich oligomers of human islet amyloid polypeptide (hIAPP1–37) and identifying its possible binding sites from molecular dynamics simulation. PLoS ONE 2014, 9, e94796. [Google Scholar] [CrossRef] [PubMed]
- Franko, A.; Rodriguez Camargo, D.C.; Böddrich, A.; Garg, D.; Rodriguez Camargo, A.; Rathkolb, B.; Janik, D.; Aichler, M.; Feuchtinger, A.; Neff, F.; et al. Epigallocatechin gallate (EGCG) reduces the intensity of pancreatic amyloid fibrils in human islet amyloid polypeptide (hIAPP) transgenic mice. Sci. Rep. 2018, 8, 1116. [Google Scholar] [CrossRef] [PubMed]
- Kakinen, A.; Adamcik, J.; Wang, B.; Ge, X.; Mezzenga, R.; Davis, T.P.; Ding, F.; Ke, P.C. Nanoscale inhibition of polymorphic and ambidextrous IAPP amyloid aggregation with small molecules. Nano Res. 2018, 11, 3636–3647. [Google Scholar] [CrossRef]
- Chaari, A. Inhibition of human islet amyloid polypeptide aggregation and cellular toxicity by oleuropein and derivatives from olive oil. Int. J. Biol. Macromol. 2020, 162, 284–300. [Google Scholar] [CrossRef]
- Floris, S.; Fais, A.; Medda, R.; Pintus, F.; Piras, A.; Kumar, A.; Kuś, P.M.; Westermark, G.T.; Era, B. Washingtonia filifera seed extracts inhibit the islet amyloid polypeptide fibrils formations and α-amylase and α-glucosidase activity. J. Enzym. Inhib. Med. Chem. 2021, 36, 517–524. [Google Scholar] [CrossRef]
- Wu, L.; Velander, P.; Brown, A.M.; Wang, Y.; Liu, D.; Bevan, D.R.; Zhang, S.; Xu, B. Rosmarinic acid potently detoxifies amylin amyloid and ameliorates diabetic pathology in a transgenicrat model of type 2 diabetes. ACS Pharmacol. Transl. Sci. 2021, 4, 1322–1337. [Google Scholar] [CrossRef]
- Tang, Y.; Liu, Y.; Zhang, Y.; Zhang, D.; Gong, X.; Zheng, J. Repurposing a cardiovascular disease drug of cloridarol as hIAPP inhibitor. ACS Chem. Neurosci. 2021, 12, 1419–1427. [Google Scholar] [CrossRef]
- Nedumpully-Govindan, P.; Gurzov, E.N.; Chen, P.; Pilkington, E.H.; Stanley, W.J.; Litwak, S.A.; Davis, T.P.; Ke, P.C.; Ding, F. Graphene oxide inhibits hIAPP amyloid fibrillation and toxicity in insulin-producing NIT-1 cells. Phys. Chem. Chem. Phys. 2016, 18, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Koppel, K.; Tang, H.; Javed, I.; Parsa, M.; Mortimer, M.; Davis, T.P.; Lin, S.; Chaffee, A.L.; Ding, F.; Ke, P.C. Elevated amyloidoses of human IAPP and amyloid beta by lipopolysaccharide and their mitigation by carbon quantum dots. Nanoscale 2020, 12, 12317–12328. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.C.; Pilkington, E.H.; Sun, Y.; Javed, I.; Kakinen, A.; Peng, G.; Ding, F.; Davis, T.P. Mitigation of amyloidosis with nanomaterials. Adv. Mater. 2020, 32, 1901690. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Pithadia, A.S.; Bhat, J.; Bera, S.; Midya, A.; Fierke, C.A.; Ramamoorthy, A.; Bhunia, A. Self-assembly of a nine-residue amyloid-forming peptide fragment of SARS corona virus E-protein: Mechanism of self aggregation and amyloid-inhibition of hIAPP. Biochemistry 2015, 54, 2249–2261. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, Q.; Ren, M.; Zhang, A.; Liu, H.; Yao, X. Molecular dynamics simulation on the inhibition mechanism of peptide-based inhibitor of islet amyloid polypeptide (IAPP) to islet amyloid polypeptide (IAPP22-28) oligomers. Chem. Biol. Drug. Des. 2017, 90, 31–39. [Google Scholar] [CrossRef]
- Abioye, R.O.; Okagu, O.D.; Udenigwe, C.C. Disaggregation of islet amyloid polypeptide fibrils as a potential anti-fibrillation mechanism of tetrapeptide TNGQ. Int. J. Mol. Sci. 2022, 23, 1972. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.-H.; Kim, J.-H. Role and therapeutic potential of melatonin in the central nervous system and cancers. Cancers 2020, 12, 1567. [Google Scholar] [CrossRef]
- Serrano, E.; Venegas, C.; Escames, G.; Sánchez-Muñoz, C.; Zabala, M.; Puertas, A.; de Haro, T.; Gutierrez, A.; Castillo, M.; Acuna-Castroviejo, D. Antioxidant defence and inflammatory response in professional road cyclists during a 4-day competition. J. Sports Sci. 2010, 28, 1047–1056. [Google Scholar] [CrossRef]
- Escames, G.; Ozturk, G.; Baño-Otálora, B.; Pozo, M.J.; Madrid, J.A.; Reiter, R.J.; Serrano, E.; Concepción, M.; Acuña-Castroviejo, D. Exercise and melatonin in humans: Reciprocal benefits. J. Pineal Res. 2012, 52, 1–11. [Google Scholar] [CrossRef]
- Yang, X.; Zou, D.; Tang, S.; Fan, T.; Su, H.; Hu, R.; Zhou, Q.; Gui, S.; Zuo, L.; Wang, Y. Ameliorative effect of melatonin against increased intestinal permeability in diabetic rats: Possible involvement of MLCK-dependent MLC phosphorylation. Mol. Cell. Biochem. 2016, 416, 23–32. [Google Scholar] [CrossRef]
- Ergenc, M.; Ozacmak, H.S.; Turan, I.; Ozacmak, V.H. Melatonin reverses depressive and anxiety like-behaviours induced by diabetes: Involvement of oxidative stress, age, rage and S100B levels in the hippocampus and prefrontal cortex of rats. Arch. Physiol. Biochem. 2022, 128, 402–410. [Google Scholar] [CrossRef]
- Costes, S.; Boss, M.; Thomas, A.P.; Matveyenko, A.V. Activation of melatonin signaling promotes β-cell survival and function. Mol. Endocrinol. 2015, 29, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Jung, E.-M.; Yoo, Y.-M.; Jeung, E.-B. Melatonin influences the expression and oligomerization of amylin in rat INS-1E cells. J. Physiol. Pharmacol. 2019, 70, 695–703. [Google Scholar]
- Aarabi, M.H.; Mirhashemi, S.M. To estimate effective antiamyloidogenic property of melatonin and fisetin and their actions to destabilize amyloid fibrils. Pak. J. Pharm. Sci. 2017, 30, 1589–1593. [Google Scholar] [PubMed]
- King, K.M.; Bevan, D.R.; Brown, A.M. Molecular dynamics simulations indicate aromaticity as a key factor in the inhibition of IAPP((20–29)) aggregation. ACS Chem. Neurosci. 2022, 13, 1615–1626. [Google Scholar] [CrossRef]
- Bai, C.; Lin, D.; Mo, Y.; Lei, J.; Sun, Y.; Xie, L.; Yang, X.; Wei, G. Influence of fullerenol on hIAPP aggregation: Amyloid inhibition and mechanistic aspects. Phys. Chem. Chem. Phys. 2019, 21, 4022–4031. [Google Scholar] [CrossRef]
- Kamihira-Ishijima, M.; Nakazawa, H.; Kira, A.; Naito, A.; Nakayama, T. Inhibitory mechanism of pancreatic amyloid fibril formation: Formation of the complex between tea catechins and the fragment of residues 22–27. Biochemistry 2012, 51, 10167–10174. [Google Scholar] [CrossRef]
- Zhu, L.; Gong, Y.; Lju, H.; Sun, G.; Zhang, Q.; Qian, Z. Mechanisms of melatonin binding and destabilizing the protofilament and filament of tau R3–R4 domains revealed by molecular dynamics simulation. Phys. Chem. Chem. Phys. 2021, 23, 20615–20626. [Google Scholar] [CrossRef]
- Levy, M.; Porat, Y.; Bacharach, E.; Shalev, D.E.; Gazit, E. Phenolsulfonphthalein, but not phenolphthalein, inhibits amyloid fibril formation: Implications for the modulation of amyloid self-assembly. Biochemistry 2008, 47, 5896–5904. [Google Scholar] [CrossRef]
- Kumaki, Y.; Kawano, K.; Hikichi, K.; Matsumoto, T.; Matsushima, N. A circular loop of the 16-residue repeating unit in ice nucleation protein. Biochem. Biophys. Res. Commun. 2008, 371, 5–9. [Google Scholar] [CrossRef]
- Sogawa, K.; Minoura, K.; In, Y.; Ishida, T.; Taniguchi, T.; Tomoo, K. CH-π interaction in VQIVYK sequence elucidated by NMR spectroscopy is essential for PHF formation of tau: CH-π interaction in VQIVYK sequence. Biopolymers 2014, 102, 288–295. [Google Scholar] [CrossRef]
- Lao, Z.; Chen, Y.; Tang, Y.; Wei, G. Molecular dynamics simulations reveal the inhibitory mechanism of dopamine against human islet amyloid polypeptide (hIAPP) aggregation and its destabilization effect on hIAPP protofibrils. ACS Chem. Neurosci. 2019, 10, 4151–4159. [Google Scholar] [CrossRef] [PubMed]
- Luca, S.; Yau, W.-M.; Leapman, R.; Tycko, R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid-state NMR. Biochemistry 2007, 46, 13505–13522. [Google Scholar] [CrossRef] [PubMed]
- Schüttelkopf, A.W.; van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Molnar, L.F.; Jung, Y.; Kussmann, J.; Ochsenfeld, C.; Brown, S.T.; Gilbert, A.T.B.; Slipchenko, L.V.; Levchenko, S.V.; O’Neill, D.P.; et al. Advances in methods and algorithms in a modern quantum chemistry program package. Phys. Chem. Chem. Phys. 2006, 8, 3172–3191. [Google Scholar] [CrossRef]
- Schmidt, K.; Baldridge, K.K.; Boatz, J.A.; Elbert, S.T.; Gordon, M.S.; Jensen, J.H.; Koseki, S.; Matsunaga, N.; Nguyen, K.A.; Su, S.; et al. GAMESS-MW. J. Comput. Chem. 1993, 14, 1347–1363. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Moore, S.; Sonar, K.; Bharadwaj, P.; Deplazes, E.; Mancera, R. Characterisation of the structure and oligomerisation of islet amyloid polypeptides (IAPP): A review of molecular dynamics simulation studies. Molecules 2018, 23, 2142. [Google Scholar] [CrossRef]
- Shaw, D.E.; Maragakis, P.; Lindorff-Larsen, K.; Piana, S.; Dror, R.O.; Eastwood, M.P.; Bank, J.A.; Jumper, J.M.; Salmon, J.K.; Shan, Y.; et al. Atomic-level characterization of the structural dynamics of proteins. Science 2010, 330, 341–346. [Google Scholar] [CrossRef]
- Piana, S.; Lindorff-Larsen, K.; Shaw, D.E. How robust are protein folding simulations with respect to force field parameterization? Biophys. J. 2011, 100, L47–L49. [Google Scholar] [CrossRef]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [Green Version]
- Lindorff-Larsen, K.; Piana, S.; Palmo, K.; Maragakis, P.; Klepeis, J.L.; Dror, R.O.; Shaw, D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 2010, 78, 1950–1958. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Kabsch, W.; Sander, C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983, 22, 2577–2637. [Google Scholar] [CrossRef]
- Daura, X.; Gademann, K.; Jaun, B.; Seebach, D.; van Gunsteren, W.F.; Mark, A.E. Peptide folding: When simulation meets experiment. Angew. Chem. Int. Ed. 1999, 38, 236–240. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Zhu, X.; Song, X.; Zhang, Q.; Qian, Z. Melatonin Inhibits hIAPP Oligomerization by Preventing β-Sheet and Hydrogen Bond Formation of the Amyloidogenic Region Revealed by Replica-Exchange Molecular Dynamics Simulation. Int. J. Mol. Sci. 2022, 23, 10264. https://doi.org/10.3390/ijms231810264
Wang G, Zhu X, Song X, Zhang Q, Qian Z. Melatonin Inhibits hIAPP Oligomerization by Preventing β-Sheet and Hydrogen Bond Formation of the Amyloidogenic Region Revealed by Replica-Exchange Molecular Dynamics Simulation. International Journal of Molecular Sciences. 2022; 23(18):10264. https://doi.org/10.3390/ijms231810264
Chicago/Turabian StyleWang, Gang, Xinyi Zhu, Xiaona Song, Qingwen Zhang, and Zhenyu Qian. 2022. "Melatonin Inhibits hIAPP Oligomerization by Preventing β-Sheet and Hydrogen Bond Formation of the Amyloidogenic Region Revealed by Replica-Exchange Molecular Dynamics Simulation" International Journal of Molecular Sciences 23, no. 18: 10264. https://doi.org/10.3390/ijms231810264
APA StyleWang, G., Zhu, X., Song, X., Zhang, Q., & Qian, Z. (2022). Melatonin Inhibits hIAPP Oligomerization by Preventing β-Sheet and Hydrogen Bond Formation of the Amyloidogenic Region Revealed by Replica-Exchange Molecular Dynamics Simulation. International Journal of Molecular Sciences, 23(18), 10264. https://doi.org/10.3390/ijms231810264





