Abstract
T-cell immunoglobulin and mucin domain 1 (TIM-1) has been recently identified as one of the factors involved in the internalization of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in human cells, in addition to angiotensin-converting enzyme 2 (ACE2), transmembrane serine protease 2 (TMPRSS2), neuropilin-1, and others. We hypothesized that specific microRNAs could target TIM-1, with potential implications for the management of patients suffering from coronavirus disease 2019 (COVID-19). By combining bioinformatic analyses and functional assays, we identified miR-142 as a specific regulator of TIM-1 transcription. Since TIM-1 has been implicated in the regulation of endothelial function at the level of the blood-brain barrier (BBB) and its levels have been shown to be associated with stroke and cerebral ischemia-reperfusion injury, we validated miR-142 as a functional modulator of TIM-1 in human brain microvascular endothelial cells (hBMECs). Taken together, our results indicate that miR-142 targets TIM-1, representing a novel strategy against cerebrovascular disorders, as well as systemic complications of SARS-CoV-2 and other viral infections.
1. Introduction
MicroRNAs (miRNAs, miRs) are a group of small (18–24 nucleotides) non-coding RNAs that control the post-transcriptional expression of target genes [1,2,3,4]. Specifically, miRNAs are able to recognize and bind the 3′ untranslated region (UTR) of certain genes, inhibiting their expression. Therefore, miRNAs are involved in a number of physiological and pathological processes [5,6,7,8,9,10,11,12,13,14]. We and others have identified several miRNAs involved in the regulation of endothelial function [15,16,17,18,19,20].
T-cell immunoglobulin and mucin domain 1 (TIM-1) is a type I cell-surface glycoprotein that includes four main domains: a carboxy-terminal cytoplasmic tail, a mucin domain, a single transmembrane domain, and an amino-terminal immunoglobulin (Ig)-like domain [21]. TIM-1 is expressed on endothelial cells, antigen-presenting cells, neurons and microglial cells, and proximal tubular cells in the kidney (it is also known as Kidney Injury Molecule-1; KIM-1).
TIM-1 has been shown to act as a cell receptor or entry factor for a number of viruses, including Hepatitis A (indeed, another of its nomenclatures is Hepatitis A Virus Cellular Receptor 1; HAVcR1) [22,23], Ebola virus [24,25,26], West Nile virus [26], Lassa virus [27], Marburg virus [28], Dengue virus [29,30], Japanese encephalitis virus [31], Chikungunya virus [32], and Zika virus [33,34]. Intriguingly, emerging investigations have proposed TIM-1 as a co-factor for the internalization of SARS-CoV-2 in human cells [35,36,37]. Specifically, TIM-1 has been shown to enhance a recombinant replication-competent vesicular stomatitis virus that encodes SARS-CoV-2 spike (rVSV/Spike) infection over a wide range of ACE2 concentrations [38], whereas a mutant of TIM-1 that does not bind to phosphatidylserine was unable to facilitate SARS-CoV-2 entry [39].
Since TIM-1 has been implicated in the regulation of endothelial function at the level of the blood–brain barrier (BBB), and its levels have been shown to be associated with stroke and cerebral ischemia-reperfusion injury [21,40], the main aim of this study was to identify and validate miRNAs that specifically target TIM-1 in human brain microvascular endothelial cells (hBMECs).
2. Results
2.1. miR-142 Targets TIM-1 in Different Species
By combining bioinformatic analyses and functional assays, we identified hsa-miR-142-3p (abbreviated as miR-142) as a specific and conserved regulator of TIM-1 transcription (Figure 1). We also generated a mutant construct of TIM-1 3′-UTR (TIM-1 MUT), harboring nucleotide substitutions within the predicted miR-142 binding sites of TIM-1 3′-UTR (Figure 1).

Figure 1.
Identification of miR-142 as a specific modulator of TIM-1; the complementary nucleotides between the target region of TIM-1 3′-UTR and hsa-miR-142-3p are highly conserved across different species.
2.2. Validation in Human Endothelial Cells of the Transcriptional Regulation of TIM-1 by miR-142
Preclinical assays have shown that inhibiting TIM-1 protects against cerebral ischemia-reperfusion injury [21], while in a clinical study conducted on 4591 subjects, TIM-1 has been associated with a significantly increased incidence of stroke (both ischemic stroke and all-cause stroke) [40]. On these grounds, we validated, for the first time to the best of our knowledge, miR-142 as a functional modulator of TIM-1 in hBMECs, which are generally considered to be among the most suitable cell lines to recapitulate the human BBB in vitro [41,42].
We demonstrated via luciferase assays (Figure 2) that TIM-1 is a specific molecular target of miR-142.
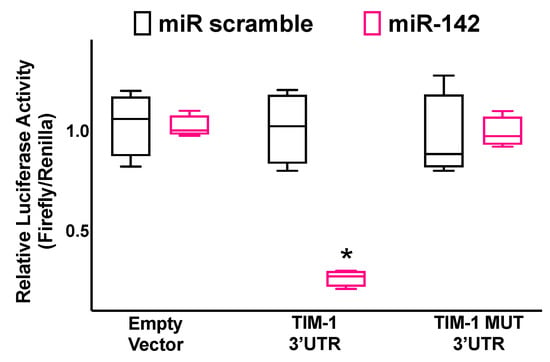
Figure 2.
Validation of TIM-1 targeting by miR-142. Luciferase activity was measured in hBMECs 48 h after transfection, using the vector without TIM-1 3′-UTR (empty vector), the vector containing the wild-type TIM-1 3′-UTR, and the vector containing a mutated TIM-1 3′-UTR (TIM-1 MUT); a non-targeting miRNA (miR scramble) was employed as a further control. All experiments were performed at least in triplicate; the box-and-whiskers graph indicates the medians and the 5th–95th percentiles; * p < 0.01 vs. miR scramble.
2.3. TIM-1 Expression Levels Are Regulated by miR-142
We observed that miR-142 was able to reduce both the mRNA levels (Figure 3; Table 1) and the protein levels (Figure 4) of TIM-1.
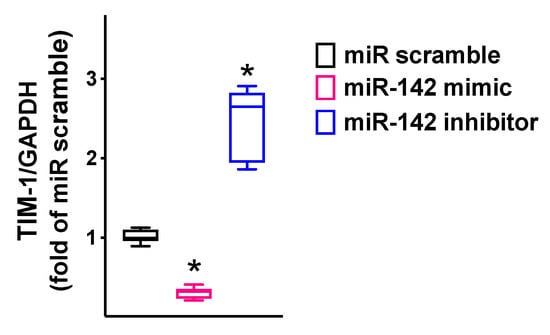
Figure 3.
TIM-1 expression in hBMECs was reduced by miR-142 and increased by miR-142 inhibitor. TIM-1 mRNA levels were measured using RT-qPCR in hBMECs transfected with miR-142 mimic, inhibitor, or scramble (negative control) for 48 h, normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). All experiments were performed at least in triplicate; the box-and-whiskers graph shows the medians and the 5th–95th percentiles; * p < 0.01 vs. miR scramble. Sequences of oligonucleotide primers are reported in Table 1.

Table 1.
Oligonucleotide primer sequences.
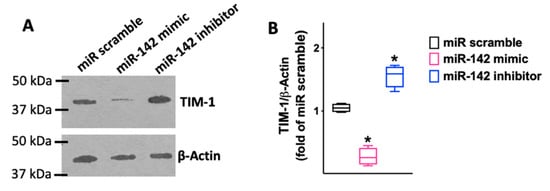
Figure 4.
The results obtained via RT-qPCR were confirmed in terms of protein levels (Biorbyt; #orb382613; Cambridge, UK), as shown in the representative immunoblots in panel (A) and their quantification in panel (B). All experiments were performed at least in triplicate; the box-and-whiskers graph indicates the medians and the 5th–95th percentiles; *: p < 0.01 vs. miR scramble.
We also confirmed that TIM-1 was expressed in other endothelial cell types, namely adult human lung microvascular endothelial cells (HMVEC-Ls) and human umbilical vein endothelial cells (HUVECs), demonstrating that its expression was significantly reduced by miR-142 (Supplementary Figure S1).
2.4. miR-142 Attenuates TIM-1-Induced Endothelial Permeability
An increased endothelial leakage represents a pathophysiological hallmark of endothelial dysfunction, especially in the settings of viral infections, including COVID-19 [43,44,45,46,47,48,49,50].
In our experimental setting, we demonstrated that miR-142 significantly attenuated endothelial permeability triggered by the main agonist of TIM-1, namely, TIMD4 [51,52] (Figure 5).
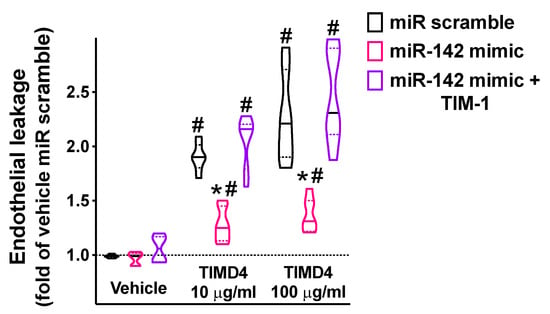
Figure 5.
The targeting of TIM-1 by miR-142 significantly reduced endothelial permeability. Endothelial leakage triggered by 48 h incubation with TIM-1 agonist TIMD4 (10 or 100 ng/mL; BioLegend San Diego, CA, USA) was measured in hBMECs transfected with miR-142 mimic, miR scramble, or by combining miR-142 mimic and TIM-1 overexpression (pcDNA3.1-TIM-1 plasmid; GenScript, Piscataway, NJ, USA). All experiments were performed at least in triplicate; in the violin plot, median (solid line) and quartiles (dotted lines) are indicated; * p < 0.01 vs. miR scramble, # p < 0.01 vs. vehicle.
Our data on endothelial leakage were further supported by the regulation of a major tight-junction protein, namely occludin [53,54,55,56], by miR-142 (Supplementary Figure S2).
3. Discussion
Our data indicate that miR-142 targets TIM-1, representing a novel potential strategy against cerebrovascular diseases, COVID-19, and other viral infections.
We established that TIM-1 is expressed in different types of endothelial cells and that miR-142 is able to significantly reduce its expression. One of the key findings of the present work is the demonstration of the functional role of miR-142 in the regulation of endothelial leakage, which represents a fundamental step in several disorders caused by different viruses, which do not have to necessarily display a definite endotheliotropism [57,58,59,60,61,62,63,64]. Moreover, many viral infections, including COVID-19, have been shown to lead to the involvement of different tissues and organs, often culminating in a systemic inflammatory response, in which the endothelium plays key roles [50,65,66,67,68]. Our results in terms of endothelial permeability are also corroborated by data showing that miR-142 can regulate the expression of Occludin, a tight-junction protein that is functionally involved in viral neuro-invasion [69].
Since TIM-1 has been shown to represent an entry co-factor for a number of viruses causing major diseases in humans [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,39,70], our findings have a major relevance in terms of public health and should enthuse further dedicated investigations exploiting miR-142 as a biomarker as well as a potential target for novel therapeutic approaches. For instance, in line with these observations, we have previously demonstrated that endothelial miRNAs can be harnessed as reliable biomarkers for cerebrovascular complications of COVID-19 [71]. Notably, the crucial role of endothelial dysfunction in the pathobiology of COVID-19, initially described by our research group [72] has been later confirmed by numerous investigators [72,73,74,75,76,77,78,79,80,81,82,83,84]. Indeed, endothelial cells have been shown to express the key co-factors involved for the internalization of SARS-CoV-2 in host cells, including angiotensin converting enzyme 2 (ACE2), transmembrane protease serine 2 (TMPRSS2), cathepsins B and D, TIM-1, neuropilin-1, and others, thereby representing a natural target of SARS-CoV-2 [83,85,86,87,88,89,90,91,92]. Furthermore, the systemic inflammatory viral reaction observed in patients affected by COVID-19 has been shown to be linked to endothelial dysfunction [93,94,95,96,97], leading to thromboembolic events [98,99,100], which represent a common feature of COVID-19 cases with a severe outcome [101,102,103].
Additional potential applications of our discovery include kidney disease, disturbances of iron metabolism, and the modulation of the immune response. Indeed, plasma levels of TIM-1 have been found to be associated with underlying tubulointerstitial and mesangial lesions and progression to kidney failure in two cohort studies of individuals with kidney diseases [104]. Preclinical investigations have demonstrated that TIM-2, the rodent homolog to TIM-1, is a binding partner to H-ferritin [105], a protein initially thought to be solely used for iron storage [106,107,108], which was later shown to serve as an iron delivery protein, secreted from the endothelial cells within the BBB as a source of iron for the brain [109]. TIM-1 can also mediate the tethering between T cells and endothelial cells in vivo and the rolling of lymphocytes on the vascular endothelium [110].
Our study is not exempt from limitations, including having performed the luciferase assays only in one cell type; nevertheless, we demonstrated that TIM-1 is indeed expressed (and regulated by miR-142) in three different types of endothelial cells, namely, hBMECs, HUVECs, and HMVEC-Ls. Further dedicated experiments are warranted to prove the effects of miR-142 in the pathophysiology of cerebrovascular events and on the actions of SARS-CoV-2 and other viruses.
4. Materials and Methods
4.1. Cell Culture and Reagents
All reagents were purchased from Millipore-Sigma (Burlington, MA, USA), unless otherwise stated. We obtained hBMECs from Neuromics (Minneapolis, MN, USA; #HEC02) [111]; these cells have been proved to be the most suitable human cell line to reproduce the BBB in vitro [112]. Adult human lung microvascular endothelial cells (HMVEC-Ls) were obtained from Lonza (Basel, Switzerland; catalog number, CC-2527) and human umbilical vein endothelial cells (HUVECs) from ThermoFisher Scientific (Waltham, MA, USA; Catalog number, #C0035C). Cells were cultured in a standard humidified atmosphere (37 °C) containing 5% CO2, as we previously described [111,113]. In some experiments, cells were transfected with pcDNA3.1-TIM-1 plasmids (GenScript, Piscataway, NJ, USA).
4.2. Identification and Validation of miR-142 as a Regulator of TIM-1
To identify miRNAs targeting the 3′-UTR of TIM-1, we used TargetScanHuman 8.0, as we previously described [20,111,114,115,116,117]. To assess the effects of miR-142 on TIM-1 gene transcription, we used a luciferase reporter containing the 3′-UTR of the predicted miRNA interaction site, both wild-type and mutated, in hBMECs cells. The mutant construct of TIM-1 3′-UTR (TIM-1 MUT, as shown in Figure 1), harboring a substitution of three nucleotides within the predicted miR-142 binding sites of TIM-1 3′-UTR was obtained using the NEBaseChanger and Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA) as we previously described [111,114,116].
We transfected hBMECs with the 3′-UTR reporter plasmid (0.05 μg) and miR-142 mimic (ThermoFisher Scientific, Waltham MA, USA) or miR-142 inhibitor, as well as a non-targeting negative control (scramble), all used at a final concentration of 50 nMol/L, using Lipofectamine RNAiMAX (ThermoFisher Scientific) [111,116]. Firefly and Renilla luciferase activities were measured 48 h after transfection, using Luciferase Reporter Assay System (Promega, Madison, WI, USA), normalizing Firefly luciferase to Renilla luciferase activity [20,111,114]. The cellular expression of TIM-1 was determined using RT-qPCR as we previously described [20,114,117,118], normalizing to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The sequences of oligonucleotide primers (Merck, Darmstadt, Germany) are reported in Table 1.
4.3. Immunoblotting
Immunoblotting assays were performed as previously described and validated by our group [111,116,119]; the intensity of the bands was quantified using FIJI (“Fiji Is Just ImageJ”) software. The antibody for TIM-1 was purchased from Novus Biologicals (Centennial, CO, USA; catalog number; #NBP1-76701); the antibody for Occludin was purchased from Novus Biologicals (catalog number, #NBP1-87402); the antibody for β-Actin was purchased from abcam (Cambridge, MA, USA; catalog number, #ab8229); the antibody for GAPDH was purchased from Novus Biologicals (catalog number, #NB300-221).
4.4. Endothelial Permeability Assay
We performed the in vitro permeability assay in hBMECs as we previously described [53,111,113]. Briefly, hBMECs transfected with miR-142 mimic or miR scramble were grown on 0.4 mm fibronectin-coated transwell filters for 48 h; then, we replaced the medium in the upper well with FITC-dextran 70 kD (0.5 mg/mL in phosphate-buffered saline, PBS). Cells were stimulated in the lower well with PBS alone or PBS containing 50 ng/mL VEGF-A165 (Bio-Techne Corporation, Minneapolis, MN, USA). Endothelial permeabilization was quantified by measuring at 520 nm the fluorescence of dextran that passed in the bottom chamber through the cell monolayer [111,113,120].
4.5. Statistical Analysis
All data were expressed as means ± standard errors of the means (SEMs). The statistical analyses were carried out using GraphPad 9 (Prism, San Diego, CA, USA). Statistical significance, set at p < 0.05, was tested using the two-way ANOVA followed by Tukey–Kramer multiple comparison test or the non-parametric Mann–Whitney U test, as appropriate.
5. Conclusions
Taken together, our results indicate that miR-142 targets TIM-1, representing a novel potential strategy against cerebrovascular disease, SARS-CoV-2, and other viral infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms231810242/s1.
Author Contributions
Conceptualization, U.K., J.G., F.V. and G.S.; methodology, U.K., J.G., F.V., R.A., S.S.J. and P.M.; formal analysis, U.K., J.G. and F.V.; investigation, U.K., J.G., F.V., R.A. and A.M.; writing—original draft preparation, U.K., J.G. and F.V.; writing—review and editing, G.S. All authors have read and agreed to the published version of the manuscript.
Funding
The Santulli’s Lab is supported in part by the National Institutes of Health (NIH), i.e., National Heart, Lung, and Blood Institute (NHLBI; R01-HL159062, R01HL164772, R01-HL146691, T32-HL144456), National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; R01-DK123259, R01-DK033823), National Center for Advancing Translational Sciences (NCATS; UL1TR002556-06), with funding given to G.S.; by the Diabetes Action Research and Education Foundation (with funding given to G.S.); and by the Monique Weill-Caulier and Irma T. Hirschl Trusts (with funding given to G.S.). J.G. is supported by a postdoctoral fellowship of the American Heart Association (AHA-20POST35211151). F.V. is supported by a postdoctoral fellowship of the American Heart Association (AHA-22POST995561). S.S.J. is supported by a postdoctoral fellowship of the American Heart Association (AHA-21POST836407).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data supporting the reported results are contained within this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Santulli, G. microRNAs Distinctively Regulate Vascular Smooth Muscle and Endothelial Cells: Functional Implications in Angiogenesis, Atherosclerosis, and In-Stent Restenosis. Adv. Exp. Med. Biol. 2015, 887, 53–77. [Google Scholar] [CrossRef] [PubMed]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed]
- van der Kwast, R.; Quax, P.H.A.; Nossent, A.Y. An Emerging Role for isomiRs and the microRNA Epitranscriptome in Neovascularization. Cells 2019, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Santulli, G. Exosomal microRNA: The revolutionary endogenous Innerspace nanotechnology. Sci. Transl. Med. 2018, 10, eaav9141. [Google Scholar] [CrossRef] [PubMed]
- Godlewski, J.; Lenart, J.; Salinska, E. MicroRNA in Brain pathology: Neurodegeneration the Other Side of the Brain Cancer. Noncoding RNA 2019, 5, 20. [Google Scholar] [CrossRef]
- Wronska, A.; Kurkowska-Jastrzebska, I.; Santulli, G. Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol. 2015, 213, 60–83. [Google Scholar] [CrossRef]
- Wong, W.K.M.; Sorensen, A.E.; Joglekar, M.V.; Hardikar, A.A.; Dalgaard, L.T. Non-Coding RNA in Pancreas and beta-Cell Development. Noncoding RNA 2018, 4, 41. [Google Scholar] [CrossRef]
- Dama, E.; Melocchi, V.; Mazzarelli, F.; Colangelo, T.; Cuttano, R.; Di Candia, L.; Ferretti, G.M.; Taurchini, M.; Graziano, P.; Bianchi, F. Non-Coding RNAs as Prognostic Biomarkers: A miRNA Signature Specific for Aggressive Early-Stage Lung Adenocarcinomas. Noncoding RNA 2020, 6, 48. [Google Scholar] [CrossRef]
- Creemers, E.E.; Tijsen, A.J.; Pinto, Y.M. Circulating microRNAs: Novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012, 110, 483–495. [Google Scholar] [CrossRef]
- Fay, E.J.; Langlois, R.A. MicroRNA-Attenuated Virus Vaccines. Noncoding RNA 2018, 4, 25. [Google Scholar] [CrossRef]
- Bar, C.; Chatterjee, S.; Falcao Pires, I.; Rodrigues, P.; Sluijter, J.P.G.; Boon, R.A.; Nevado, R.M.; Andres, V.; Sansonetti, M.; de Windt, L.; et al. Non-coding RNAs: Update on mechanisms and therapeutic targets from the ESC Working Groups of Myocardial Function and Cellular Biology of the Heart. Cardiovasc. Res. 2020, 116, 1805–1819. [Google Scholar] [CrossRef] [PubMed]
- Slota, J.A.; Booth, S.A. MicroRNAs in Neuroinflammation: Implications in Disease Pathogenesis, Biomarker Discovery and Therapeutic Applications. Noncoding RNA 2019, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect Clin. Res. 2016, 7, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Santulli, G. MicroRNA: From Molecular Biology to Clinical Practice; Santulli, G., Ed.; Springer Nature: New York, NY, USA, 2016. [Google Scholar]
- Yang, Y.; Liu, Y.; Li, Y.; Chen, Z.; Xiong, Y.; Zhou, T.; Tao, W.; Xu, F.; Yang, H.; Yla-Herttuala, S.; et al. MicroRNA-15b Targets VEGF and Inhibits Angiogenesis in Proliferative Diabetic Retinopathy. J. Clin. Endocrinol. Metab. 2020, 105, 3404–3415. [Google Scholar] [CrossRef]
- Widlansky, M.E.; Jensen, D.M.; Wang, J.; Liu, Y.; Geurts, A.M.; Kriegel, A.J.; Liu, P.; Ying, R.; Zhang, G.; Casati, M.; et al. miR-29 contributes to normal endothelial function and can restore it in cardiometabolic disorders. EMBO Mol. Med. 2018, 10, e8046. [Google Scholar] [CrossRef]
- Santulli, G. MicroRNAs and Endothelial (Dys) Function. J. Cell Physiol. 2016, 231, 1638–1644. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Rafehi, H.; El-Osta, A. HDAC Inhibition in Vascular Endothelial Cells Regulates the Expression of ncRNAs. Noncoding RNA 2016, 2, 4. [Google Scholar] [CrossRef]
- Santulli, G.; Wronska, A.; Uryu, K.; Diacovo, T.G.; Gao, M.; Marx, S.O.; Kitajewski, J.; Chilton, J.M.; Akat, K.M.; Tuschl, T.; et al. A selective microRNA-based strategy inhibits restenosis while preserving endothelial function. J. Clin. Investig. 2014, 124, 4102–4114. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, L.; Chen, M.; Liu, L.; Pei, A.; Zhang, R.; Gan, S.; Zhu, S. Inhibition of T cell immunoglobulin and mucin-1 (TIM-1) protects against cerebral ischemia-reperfusion injury. Cell Commun. Signal. 2019, 17, 103. [Google Scholar] [CrossRef]
- Costafreda, M.I.; Abbasi, A.; Lu, H.; Kaplan, G. Exosome mimicry by a HAVCR1-NPC1 pathway of endosomal fusion mediates hepatitis A virus infection. Nat. Microbiol. 2020, 5, 1096–1106. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, M.; Agnihotry, S.; Srivastava, A.; Bolia, R.; Yachha, S.K.; Aggarwal, R. Relationship of Severity of Hepatitis A with Polymorphisms in Hepatitis A Virus Cellular Receptor 1 (HAVCR1) Gene. Ann. Hepatol. 2018, 17, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, X.; Hu, L.; Zhang, Y.; Zheng, H.; Wu, H.; Wang, J.; Luo, L.; Xiao, H.; Qiao, C.; et al. TIM-1 Augments Cellular Entry of Ebola Virus Species and Mutants, Which Is Blocked by Recombinant TIM-1 Protein. Microbiol. Spectr. 2022, 10, e0221221. [Google Scholar] [CrossRef]
- Dragovich, M.A.; Fortoul, N.; Jagota, A.; Zhang, W.; Schutt, K.; Xu, Y.; Sanabria, M.; Moyer, D.M., Jr.; Moller-Tank, S.; Maury, W.; et al. Biomechanical characterization of TIM protein-mediated Ebola virus-host cell adhesion. Sci. Rep. 2019, 9, 267. [Google Scholar] [CrossRef]
- Richard, A.S.; Zhang, A.; Park, S.J.; Farzan, M.; Zong, M.; Choe, H. Virion-associated phosphatidylethanolamine promotes TIM1-mediated infection by Ebola, dengue, and West Nile viruses. Proc. Natl. Acad. Sci. USA 2015, 112, 14682–14687. [Google Scholar] [CrossRef]
- Brouillette, R.B.; Phillips, E.K.; Patel, R.; Mahauad-Fernandez, W.; Moller-Tank, S.; Rogers, K.J.; Dillard, J.A.; Cooney, A.L.; Martinez-Sobrido, L.; Okeoma, C.; et al. TIM-1 Mediates Dystroglycan-Independent Entry of Lassa Virus. J. Virol. 2018, 92, e00093-18. [Google Scholar] [CrossRef]
- Kondratowicz, A.S.; Lennemann, N.J.; Sinn, P.L.; Davey, R.A.; Hunt, C.L.; Moller-Tank, S.; Meyerholz, D.K.; Rennert, P.; Mullins, R.F.; Brindley, M.; et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 2011, 108, 8426–8431. [Google Scholar] [CrossRef]
- Dejarnac, O.; Hafirassou, M.L.; Chazal, M.; Versapuech, M.; Gaillard, J.; Perera-Lecoin, M.; Umana-Diaz, C.; Bonnet-Madin, L.; Carnec, X.; Tinevez, J.Y.; et al. TIM-1 Ubiquitination Mediates Dengue Virus Entry. Cell Rep. 2018, 23, 1779–1793. [Google Scholar] [CrossRef] [PubMed]
- Meertens, L.; Carnec, X.; Lecoin, M.P.; Ramdasi, R.; Guivel-Benhassine, F.; Lew, E.; Lemke, G.; Schwartz, O.; Amara, A. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 2012, 12, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Niu, J.; Jiang, Y.; Xu, H.; Zhao, C.; Zhou, G.; Chen, P.; Cao, R. TIM-1 Promotes Japanese Encephalitis Virus Entry and Infection. Viruses 2018, 10, 630. [Google Scholar] [CrossRef]
- Kirui, J.; Abidine, Y.; Lenman, A.; Islam, K.; Gwon, Y.D.; Lasswitz, L.; Evander, M.; Bally, M.; Gerold, G. The Phosphatidylserine Receptor TIM-1 Enhances Authentic Chikungunya Virus Cell Entry. Cells 2021, 10, 1828. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, G.M.; Samogim, A.P.; Parise, P.L.; Venceslau, E.M.; Guida, J.P.S.; Japecanga, R.R.; Amorim, M.R.; Toledo-Teixeira, D.A.; Forato, J.; Consonni, S.R.; et al. TAM and TIM receptors mRNA expression in Zika virus infected placentas. Placenta 2020, 101, 204–207. [Google Scholar] [CrossRef]
- Giraldo, M.I.; Xia, H.; Aguilera-Aguirre, L.; Hage, A.; van Tol, S.; Shan, C.; Xie, X.; Sturdevant, G.L.; Robertson, S.J.; McNally, K.L.; et al. Envelope protein ubiquitination drives entry and pathogenesis of Zika virus. Nature 2020, 585, 414–419. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, Y.; Zeng, X.; Chen, H.; Chen, Y.; Yang, D.; Shen, Z.; Wang, X.; Liu, X.; Xiong, M.; et al. Kidney injury molecule-1 is a potential receptor for SARS-CoV-2. J. Mol. Cell Biol. 2021, 13, 185–196. [Google Scholar] [CrossRef]
- Mori, Y.; Fink, C.; Ichimura, T.; Sako, K.; Mori, M.; Lee, N.N.; Aschauer, P.; Padmanabha Das, K.M.; Hong, S.; Song, M.; et al. KIM-1/TIM-1 is a Receptor for SARS-CoV-2 in Lung and Kidney. medRxiv 2022. [Google Scholar] [CrossRef]
- Baggen, J.; Vanstreels, E.; Jansen, S.; Daelemans, D. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021, 6, 1219–1232. [Google Scholar] [CrossRef]
- Havranek, K.E.; Jimenez, A.R.; Acciani, M.D.; Lay Mendoza, M.F.; Reyes Ballista, J.M.; Diaz, D.A.; Brindley, M.A. SARS-CoV-2 Spike Alterations Enhance Pseudoparticle Titers and Replication-Competent VSV-SARS-CoV-2 Virus. Viruses 2020, 12, 1465. [Google Scholar] [CrossRef]
- Bohan, D.; Van Ert, H.; Ruggio, N.; Rogers, K.J.; Badreddine, M.; Aguilar Briseno, J.A.; Elliff, J.M.; Rojas Chavez, R.A.; Gao, B.; Stokowy, T.; et al. Phosphatidylserine receptors enhance SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009743. [Google Scholar] [CrossRef]
- Song, L.; Sun, J.; Soderholm, M.; Melander, O.; Orho-Melander, M.; Nilsson, J.; Borne, Y.; Engstrom, G. Association of TIM-1 (T-Cell Immunoglobulin and Mucin Domain 1) With Incidence of Stroke. Arter. Thromb. Vasc. Biol. 2020, 40, 1777–1786. [Google Scholar] [CrossRef]
- Helms, H.C.; Abbott, N.J.; Burek, M.; Cecchelli, R.; Couraud, P.O.; Deli, M.A.; Forster, C.; Galla, H.J.; Romero, I.A.; Shusta, E.V.; et al. In Vitro models of the blood-brain barrier: An overview of commonly used brain endothelial cell culture models and guidelines for their use. J. Cereb. Blood Flow Metab. 2016, 36, 862–890. [Google Scholar] [CrossRef]
- Ahn, S.I.; Sei, Y.J.; Park, H.J.; Kim, J.; Ryu, Y.; Choi, J.J.; Sung, H.J.; MacDonald, T.J.; Levey, A.I.; Kim, Y. Microengineered human blood-brain barrier platform for understanding nanoparticle transport mechanisms. Nat. Commun. 2020, 11, 175. [Google Scholar] [CrossRef]
- Huang, L.Y.; Stuart, C.; Takeda, K.; D’Agnillo, F.; Golding, B. Poly(I:C) Induces Human Lung Endothelial Barrier Dysfunction by Disrupting Tight Junction Expression of Claudin-5. PLoS ONE 2016, 11, e0160875. [Google Scholar] [CrossRef]
- Puerta-Guardo, H.; Glasner, D.R.; Harris, E. Dengue Virus NS1 Disrupts the Endothelial Glycocalyx, Leading to Hyperpermeability. PLoS Pathog. 2016, 12, e1005738. [Google Scholar] [CrossRef]
- Han, T.; Lai, Y.; Jiang, Y.; Liu, X.; Li, D. Influenza A virus infects pulmonary microvascular endothelial cells leading to microvascular leakage and release of pro-inflammatory cytokines. PeerJ 2021, 9, e11892. [Google Scholar] [CrossRef]
- Gulati, A. Vascular Endothelium and Hypovolemic Shock. Curr. Vasc. Pharm. 2016, 14, 187–195. [Google Scholar] [CrossRef]
- Trung, D.T.; Wills, B. Systemic vascular leakage associated with dengue infections-the clinical perspective. Curr. Top. Microbiol. Immunol. 2010, 338, 57–66. [Google Scholar] [CrossRef]
- Terajima, M.; Hayasaka, D.; Maeda, K.; Ennis, F.A. Immunopathogenesis of hantavirus pulmonary syndrome and hemorrhagic fever with renal syndrome: Do CD8+ T cells trigger capillary leakage in viral hemorrhagic fevers? Immunol. Lett. 2007, 113, 117–120. [Google Scholar] [CrossRef]
- Srikiatkhachorn, A.; Spiropoulou, C.F. Vascular events in viral hemorrhagic fevers: A comparative study of dengue and hantaviruses. Cell Tissue Res. 2014, 355, 621–633. [Google Scholar] [CrossRef][Green Version]
- Robles, J.P.; Zamora, M.; Adan-Castro, E.; Siqueiros-Marquez, L.; Martinez de la Escalera, G.; Clapp, C. The spike protein of SARS-CoV-2 induces endothelial inflammation through integrin alpha5beta1 and NF-kappaB signaling. J. Biol. Chem. 2022, 298, 101695. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L.; Liang, X.; Liu, X.; Zhao, Y.; Ma, C.; Gao, L. Tim-4 in Health and Disease: Friend or Foe? Front. Immunol. 2020, 11, 537. [Google Scholar] [CrossRef]
- Santiago, C.; Ballesteros, A.; Martinez-Munoz, L.; Mellado, M.; Kaplan, G.G.; Freeman, G.J.; Casasnovas, J.M. Structures of T cell immunoglobulin mucin protein 4 show a metal-Ion-dependent ligand binding site where phosphatidylserine binds. Immunity 2007, 27, 941–951. [Google Scholar] [CrossRef]
- Mone, P.; Varzideh, F.; Jankauskas, S.S.; Pansini, A.; Lombardi, A.; Frullone, S.; Santulli, G. SGLT2 Inhibition via Empagliflozin Improves Endothelial Function and Reduces Mitochondrial Oxidative Stress: Insights From Frail Hypertensive and Diabetic Patients. Hypertension 2022, 79, 1633–1643. [Google Scholar] [CrossRef]
- Yuan, S.; Liu, K.J.; Qi, Z. Occludin regulation of blood-brain barrier and potential therapeutic target in ischemic stroke. Brain Circ. 2020, 6, 152–162. [Google Scholar] [CrossRef]
- Bauer, H.C.; Krizbai, I.A.; Bauer, H.; Traweger, A. “You Shall Not Pass”-tight junctions of the blood brain barrier. Front. Neurosci. 2014, 8, 392. [Google Scholar] [CrossRef]
- Dallasta, L.M.; Pisarov, L.A.; Esplen, J.E.; Werley, J.V.; Moses, A.V.; Nelson, J.A.; Achim, C.L. Blood-brain barrier tight junction disruption in human immunodeficiency virus-1 encephalitis. Am. J. Pathol. 1999, 155, 1915–1927. [Google Scholar] [CrossRef]
- Mackow, E.R.; Gorbunova, E.E.; Gavrilovskaya, I.N. Endothelial cell dysfunction in viral hemorrhage and edema. Front. Microbiol. 2014, 5, 733. [Google Scholar] [CrossRef]
- Chanthick, C.; Suttitheptumrong, A.; Rawarak, N.; Pattanakitsakul, S.N. Transcytosis Involvement in Transport System and Endothelial Permeability of Vascular Leakage during Dengue Virus Infection. Viruses 2018, 10, 69. [Google Scholar] [CrossRef]
- Dalrymple, N.A.; Mackow, E.R. Virus interactions with endothelial cell receptors: Implications for viral pathogenesis. Curr. Opin. Virol. 2014, 7, 134–140. [Google Scholar] [CrossRef]
- Suwarto, S.; Sasmono, R.T.; Sinto, R.; Ibrahim, E.; Suryamin, M. Association of Endothelial Glycocalyx and Tight and Adherens Junctions with Severity of Plasma Leakage in Dengue Infection. J. Infect. Dis. 2017, 215, 992–999. [Google Scholar] [CrossRef]
- Vaheri, A.; Strandin, T.; Hepojoki, J.; Sironen, T.; Henttonen, H.; Makela, S.; Mustonen, J. Uncovering the mysteries of hantavirus infections. Nat. Rev. Microbiol. 2013, 11, 539–550. [Google Scholar] [CrossRef]
- Zheng, B.; Wang, H.; Cui, G.; Guo, Q.; Si, L.; Yan, H.; Fang, D.; Jiang, L.; Jiang, Z.; Zhou, J. ERG-Associated lncRNA (ERGAL) Promotes the Stability and Integrity of Vascular Endothelial Barrier during Dengue Viral Infection via Interaction With miR-183-5p. Front. Cell Infect. Microbiol. 2020, 10, 477. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Kroeze, E.; Fouchier, R.A.M.; Kuiken, T. Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect. Dis. 2014, 14, 57–69. [Google Scholar] [CrossRef]
- Rathore, A.P.; Mantri, C.K.; Aman, S.A.; Syenina, A.; Ooi, J.; Jagaraj, C.J.; Goh, C.C.; Tissera, H.; Wilder-Smith, A.; Ng, L.G.; et al. Dengue virus-elicited tryptase induces endothelial permeability and shock. J. Clin. Investig. 2019, 129, 4180–4193. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka-Tojo, M. Endothelial glycocalyx damage as a systemic inflammatory microvascular endotheliopathy in COVID-19. Biomed. J. 2020, 43, 399–413. [Google Scholar] [CrossRef]
- Pan, P.; Li, G.; Shen, M.; Yu, Z.; Ge, W.; Lao, Z.; Fan, Y.; Chen, K.; Ding, Z.; Wang, W.; et al. DENV NS1 and MMP-9 cooperate to induce vascular leakage by altering endothelial cell adhesion and tight junction. PLoS Pathog. 2021, 17, e1008603. [Google Scholar] [CrossRef]
- Tramontini Gomes de Sousa Cardozo, F.; Baimukanova, G.; Lanteri, M.C.; Keating, S.M.; Moraes Ferreira, F.; Heitman, J.; Pannuti, C.S.; Pati, S.; Romano, C.M.; Cerdeira Sabino, E. Serum from dengue virus-infected patients with and without plasma leakage differentially affects endothelial cells barrier function in vitro. PLoS ONE 2017, 12, e0178820. [Google Scholar] [CrossRef]
- Tundup, S.; Kandasamy, M.; Perez, J.T.; Mena, N.; Steel, J.; Nagy, T.; Albrecht, R.A.; Manicassamy, B. Endothelial cell tropism is a determinant of H5N1 pathogenesis in mammalian species. PLoS Pathog. 2017, 13, e1006270. [Google Scholar] [CrossRef]
- Chen, Z.; Li, G. Immune response and blood-brain barrier dysfunction during viral neuroinvasion. Innate Immun. 2021, 27, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Moller-Tank, S.; Kondratowicz, A.S.; Davey, R.A.; Rennert, P.D.; Maury, W. Role of the phosphatidylserine receptor TIM-1 in enveloped-virus entry. J. Virol. 2013, 87, 8327–8341. [Google Scholar] [CrossRef]
- Gambardella, J.; Coppola, A.; Izzo, R.; Fiorentino, G.; Trimarco, B.; Santulli, G. Role of endothelial miR-24 in COVID-19 cerebrovascular events. Crit. Care 2021, 25, 306. [Google Scholar] [CrossRef]
- Sardu, C.; Gambardella, J.; Morelli, M.B.; Wang, X.; Marfella, R.; Santulli, G. Hypertension, Thrombosis, Kidney Failure, and Diabetes: Is COVID-19 an Endothelial Disease? A Comprehensive Evaluation of Clinical and Basic Evidence. J. Clin. Med. 2020, 9, 1417. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Liu, F.; Blair, R.; Wang, C.; Yang, H.; Mudd, J.; Currey, J.M.; Iwanaga, N.; He, J.; Mi, R.; et al. Endothelial cell infection and dysfunction, immune activation in severe COVID-19. Theranostics 2021, 11, 8076–8091. [Google Scholar] [CrossRef] [PubMed]
- Libby, P.; Luscher, T. COVID-19 is, in the end, an endothelial disease. Eur. Heart J. 2020, 41, 3038–3044. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.X.; Tyagi, T.; Jain, K.; Gu, V.W.; Lee, S.H.; Hwa, J.M.; Kwan, J.M.; Krause, D.S.; Lee, A.I.; Halene, S.; et al. Thrombocytopathy and endotheliopathy: Crucial contributors to COVID-19 thromboinflammation. Nat. Rev. Cardiol. 2020, 18, 194–209. [Google Scholar] [CrossRef]
- Perea Polak, A.; Romero Madrid, B.; Garcia Ocana, P.P.; Lomena Alvarez, G.; Martinez Pilar, L.; Gomez-Moyano, E. Complement-mediated thrombogenic vasculopathy in COVID-19. Int J. Derm. 2020, 60, 229–232. [Google Scholar] [CrossRef]
- Yin, J.; Wang, S.; Liu, Y.; Chen, J.; Li, D.; Xu, T. Coronary microvascular dysfunction pathophysiology in COVID-19. Microcirculation 2021, 28, e12718. [Google Scholar] [CrossRef]
- Teuwen, L.A.; Geldhof, V.; Pasut, A.; Carmeliet, P. COVID-19: The vasculature unleashed. Nat. Rev. Immunol. 2020, 20, 389–391. [Google Scholar] [CrossRef]
- Kelliher, S.; Weiss, L.; Cullivan, S.; O’Rourke, E.; Murphy, C.A.; Toolan, S.; Lennon, A.; Szklanna, P.B.; Comer, S.P.; Macleod, H.; et al. Non-severe COVID-19 is associated with endothelial damage and hypercoagulability despite pharmacological thromboprophylaxis. J. Thromb. Haemost. 2022, 20, 1008–1014. [Google Scholar] [CrossRef]
- Mesquida, J.; Caballer, A.; Cortese, L.; Vila, C.; Karadeniz, U.; Pagliazzi, M.; Zanoletti, M.; Pacheco, A.P.; Castro, P.; Garcia-de-Acilu, M.; et al. Peripheral microcirculatory alterations are associated with the severity of acute respiratory distress syndrome in COVID-19 patients admitted to intermediate respiratory and intensive care units. Crit. Care 2021, 25, 381. [Google Scholar] [CrossRef]
- Schmaier, A.A.; Pajares Hurtado, G.M.; Manickas-Hill, Z.J.; Sack, K.D.; Chen, S.M.; Bhambhani, V.; Quadir, J.; Nath, A.K.; Collier, A.Y.; Ngo, D.; et al. Tie2 activation protects against prothrombotic endothelial dysfunction in COVID-19. JCI Insight 2021, 6, e151527. [Google Scholar] [CrossRef]
- Otifi, H.M.; Adiga, B.K. Endothelial Dysfunction in COVID-19. Am. J. Med. Sci. 2022, 363, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Tang, K.; Levin, M.; Irfan, O.; Morris, S.K.; Wilson, K.; Klein, J.D.; Bhutta, Z.A. COVID-19 and multisystem inflammatory syndrome in children and adolescents. Lancet Infect Dis. 2020, 20, e276–e288. [Google Scholar] [CrossRef]
- Amraei, R.; Xia, C.; Olejnik, J.; White, M.R.; Napoleon, M.A.; Lotfollahzadeh, S.; Hauser, B.M.; Schmidt, A.G.; Chitalia, V.; Muhlberger, E.; et al. Extracellular vimentin is an attachment factor that facilitates SARS-CoV-2 entry into human endothelial cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2113874119. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, J.; Laurence, J. Long COVID endotheliopathy: Hypothesized mechanisms and potential therapeutic approaches. J. Clin. Investig. 2022, 132, e161167. [Google Scholar] [CrossRef]
- Gambardella, J.; Santulli, G. What is linking COVID-19 and endothelial dysfunction? Updates on nanomedicine and bioengineering from the 2020 AHA Scientific Sessions. Eur. Heart J. (Cardiovasc. Pharmacother.) 2021, 7, e2–e3. [Google Scholar] [CrossRef]
- Tharappel, A.M.; Samrat, S.K.; Li, Z.; Li, H. Targeting Crucial Host Factors of SARS-CoV-2. ACS Infect. Dis. 2020, 6, 2844–2865. [Google Scholar] [CrossRef]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Kaur, S.; Tripathi, D.M.; Yadav, A. The Enigma of Endothelium in COVID-19. Front. Physiol. 2020, 11, 989. [Google Scholar] [CrossRef]
- Oxford, A.E.; Halla, F.; Robertson, E.B.; Morrison, B.E. Endothelial Cell Contributions to COVID-19. Pathogens 2020, 9, 785. [Google Scholar] [CrossRef]
- Ahmetaj-Shala, B.; Vaja, R.; Atanur, S.S.; George, P.M.; Kirkby, N.S.; Mitchell, J.A. Cardiorenal Tissues Express SARS-CoV-2 Entry Genes and Basigin (BSG/CD147) Increases With Age in Endothelial Cells. JACC Basic Transl. Sci. 2020, 5, 1111–1123. [Google Scholar] [CrossRef]
- Roberts, K.A.; Colley, L.; Agbaedeng, T.A.; Ellison-Hughes, G.M.; Ross, M.D. Vascular Manifestations of COVID-19-Thromboembolism and Microvascular Dysfunction. Front. Cardiovasc. Med. 2020, 7, 598400. [Google Scholar] [CrossRef] [PubMed]
- Perico, L.; Benigni, A.; Casiraghi, F.; Ng, L.F.P.; Renia, L.; Remuzzi, G. Immunity, endothelial injury and complement-induced coagulopathy in COVID-19. Nat. Rev. Nephrol. 2020, 17, 46–64. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.; Raju, S.; Wu, R.; Ching, C.; Veitch, S.; Rathnakumar, K.; Boudreau, E.; Howe, K.L.; Fish, J.E. Overcoming Barriers: The Endothelium As a Linchpin of Coronavirus Disease 2019 Pathogenesis? Arter. Thromb. Vasc. Biol. 2020, 40, 1818–1829. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Matthay, M.A.; Calfee, C.S. Is a “Cytokine Storm” Relevant to COVID-19? JAMA Intern. Med. 2020, 180, 1152–1154. [Google Scholar] [CrossRef]
- Kang, S.; Tanaka, T.; Inoue, H.; Ono, C.; Hashimoto, S.; Kioi, Y.; Matsumoto, H.; Matsuura, H.; Matsubara, T.; Shimizu, K.; et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc. Natl. Acad. Sci. USA 2020, 117, 22351–22356. [Google Scholar] [CrossRef]
- Fraser, D.D.; Patterson, E.K.; Slessarev, M.; Gill, S.E.; Martin, C.; Daley, M.; Miller, M.R.; Patel, M.A.; Dos Santos, C.C.; Bosma, K.J.; et al. Endothelial Injury and Glycocalyx Degradation in Critically Ill Coronavirus Disease 2019 Patients: Implications for Microvascular Platelet Aggregation. Crit. Care Explor. 2020, 2, e0194. [Google Scholar] [CrossRef]
- Malas, M.B.; Naazie, I.N.; Elsayed, N.; Mathlouthi, A.; Marmor, R.; Clary, B. Thromboembolism risk of COVID-19 is high and associated with a higher risk of mortality: A systematic review and meta-analysis. eClinicalMedicine 2020, 29, 100639. [Google Scholar] [CrossRef]
- Piazza, G.; Morrow, D.A. Diagnosis, Management, and Pathophysiology of Arterial and Venous Thrombosis in COVID-19. JAMA 2020, 324, 2548. [Google Scholar] [CrossRef]
- Pillai, P.; Joseph, J.P.; Fadzillah, N.H.M.; Mahmod, M. COVID-19 and Major Organ Thromboembolism: Manifestations in Neurovascular and Cardiovascular Systems. J. Stroke Cereb. Dis. 2020, 30, 105427. [Google Scholar] [CrossRef]
- Mondal, S.; Quintili, A.L.; Karamchandani, K.; Bose, S. Thromboembolic disease in COVID-19 patients: A brief narrative review. J. Intensive Care 2020, 8, 70. [Google Scholar] [CrossRef]
- Bilaloglu, S.; Aphinyanaphongs, Y.; Jones, S.; Iturrate, E.; Hochman, J.; Berger, J.S. Thrombosis in Hospitalized Patients With COVID-19 in a New York City Health System. JAMA 2020, 324, 799–801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Feng, X.; Zhang, D.; Jiang, C.; Mei, H.; Wang, J.; Zhang, C.; Li, H.; Xia, X.; Kong, S.; et al. Deep Vein Thrombosis in Hospitalized Patients With COVID-19 in Wuhan, China: Prevalence, Risk Factors, and Outcome. Circulation 2020, 142, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, I.M.; Srivastava, A.; Sabbisetti, V.; McMahon, G.M.; He, J.; Chen, J.; Kusek, J.W.; Taliercio, J.; Ricardo, A.C.; Hsu, C.Y.; et al. Plasma Kidney Injury Molecule 1 in CKD: Findings From the Boston Kidney Biopsy Cohort and CRIC Studies. Am. J. Kidney Dis. 2021, 79, 231–243.e1. [Google Scholar] [CrossRef] [PubMed]
- Todorich, B.; Zhang, X.; Slagle-Webb, B.; Seaman, W.E.; Connor, J.R. Tim-2 is the receptor for H-ferritin on oligodendrocytes. J. Neurochem. 2008, 107, 1495–1505. [Google Scholar] [CrossRef]
- Mesquita, G.; Silva, T.; Gomes, A.C.; Oliveira, P.F.; Alves, M.G.; Fernandes, R.; Almeida, A.A.; Moreira, A.C.; Gomes, M.S. H-Ferritin is essential for macrophages’ capacity to store or detoxify exogenously added iron. Sci. Rep. 2020, 10, 3061. [Google Scholar] [CrossRef]
- Li, L.; Fang, C.J.; Ryan, J.C.; Niemi, E.C.; Lebron, J.A.; Bjorkman, P.J.; Arase, H.; Torti, F.M.; Torti, S.V.; Nakamura, M.C.; et al. Binding and uptake of H-ferritin are mediated by human transferrin receptor-1. Proc. Natl. Acad. Sci. USA 2010, 107, 3505–3510. [Google Scholar] [CrossRef]
- Ferreira, C.; Santambrogio, P.; Martin, M.E.; Andrieu, V.; Feldmann, G.; Henin, D.; Beaumont, C. H ferritin knockout mice: A model of hyperferritinemia in the absence of iron overload. Blood 2001, 98, 525–532. [Google Scholar] [CrossRef]
- Chiou, B.; Neal, E.H.; Bowman, A.B.; Lippmann, E.S.; Simpson, I.A.; Connor, J.R. Endothelial cells are critical regulators of iron transport in a model of the human blood-brain barrier. J. Cereb. Blood Flow Metab. 2019, 39, 2117–2131. [Google Scholar] [CrossRef]
- Angiari, S.; Donnarumma, T.; Rossi, B.; Dusi, S.; Pietronigro, E.; Zenaro, E.; Della Bianca, V.; Toffali, L.; Piacentino, G.; Budui, S.; et al. TIM-1 glycoprotein binds the adhesion receptor P-selectin and mediates T cell trafficking during inflammation and autoimmunity. Immunity 2014, 40, 542–553. [Google Scholar] [CrossRef]
- Mone, P.; Gambardella, J.; Wang, X.; Jankauskas, S.S.; Matarese, A.; Santulli, G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Noncoding RNA 2021, 7, 9. [Google Scholar] [CrossRef]
- Eigenmann, D.E.; Xue, G.; Kim, K.S.; Moses, A.V.; Hamburger, M.; Oufir, M. Comparative study of four immortalized human brain capillary endothelial cell lines, hCMEC/D3, hBMEC, TY10, and BB19, and optimization of culture conditions, for an in vitro blood-brain barrier model for drug permeability studies. Fluids Barriers CNS 2013, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Mone, P.; Gambardella, J.; Pansini, A.; de Donato, A.; Martinelli, G.; Boccalone, E.; Matarese, A.; Frullone, S.; Santulli, G. Cognitive Impairment in Frail Hypertensive Elderly Patients: Role of Hyperglycemia. Cells 2021, 10, 2115. [Google Scholar] [CrossRef] [PubMed]
- Matarese, A.; Gambardella, J.; Lombardi, A.; Wang, X.; Santulli, G. miR-7 Regulates GLP-1-Mediated Insulin Release by Targeting beta-Arrestin 1. Cells 2020, 9, 1621. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Morelli, M.B.; Matarese, A.; Sardu, C.; Santulli, G. Cardiomyocyte-derived exosomal microRNA-92a mediates post-ischemic myofibroblast activation both in vitro and ex vivo. ESC Heart Fail. 2020, 7, 284–288. [Google Scholar] [CrossRef]
- Matarese, A.; Gambardella, J.; Sardu, C.; Santulli, G. miR-98 Regulates TMPRSS2 Expression in Human Endothelial Cells: Key Implications for COVID-19. Biomedicines 2020, 8, 462. [Google Scholar] [CrossRef]
- Morelli, M.B.; Shu, J.; Sardu, C.; Matarese, A.; Santulli, G. Cardiosomal microRNAs Are Essential in Post-Infarction Myofibroblast Phenoconversion. Int. J. Mol. Sci. 2019, 21, 201. [Google Scholar] [CrossRef]
- Tang, X.H.; Gambardella, J.; Jankauskas, S.; Wang, X.; Santulli, G.; Gudas, L.J.; Levi, R. A Retinoic Acid Receptor beta 2 Agonist Improves Cardiac Function in a Heart Failure Model. J. Pharmacol. Exp. Ther. 2021, 379, 182–190. [Google Scholar] [CrossRef]
- Santulli, G.; Xie, W.; Reiken, S.R.; Marks, A.R. Mitochondrial calcium overload is a key determinant in heart failure. Proc. Natl. Acad. Sci. USA 2015, 112, 11389–11394. [Google Scholar] [CrossRef]
- Gambardella, J.; Sorriento, D.; Bova, M.; Rusciano, M.; Loffredo, S.; Wang, X.; Petraroli, A.; Carucci, L.; Mormile, I.; Oliveti, M.; et al. Role of Endothelial G Protein-Coupled Receptor Kinase 2 in Angioedema. Hypertension 2020, 76, 1625–1636. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).