A Novel Pathway of Chlorimuron-Ethyl Biodegradation by Chenggangzhangella methanolivorans Strain CHL1 and Its Molecular Mechanisms
Abstract
:1. Introduction
2. Results and Discussion
2.1. Medium and Detection Conditions
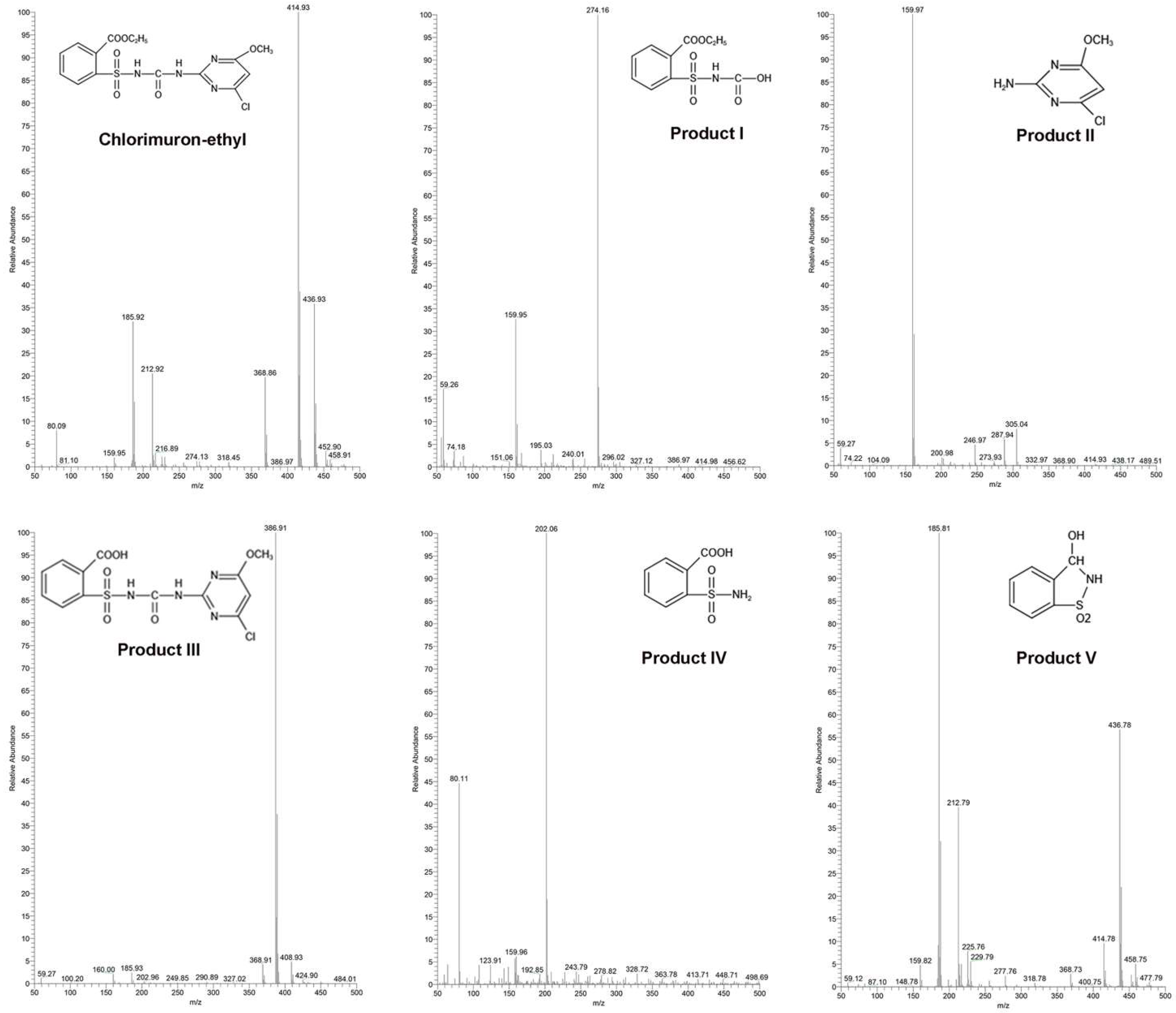
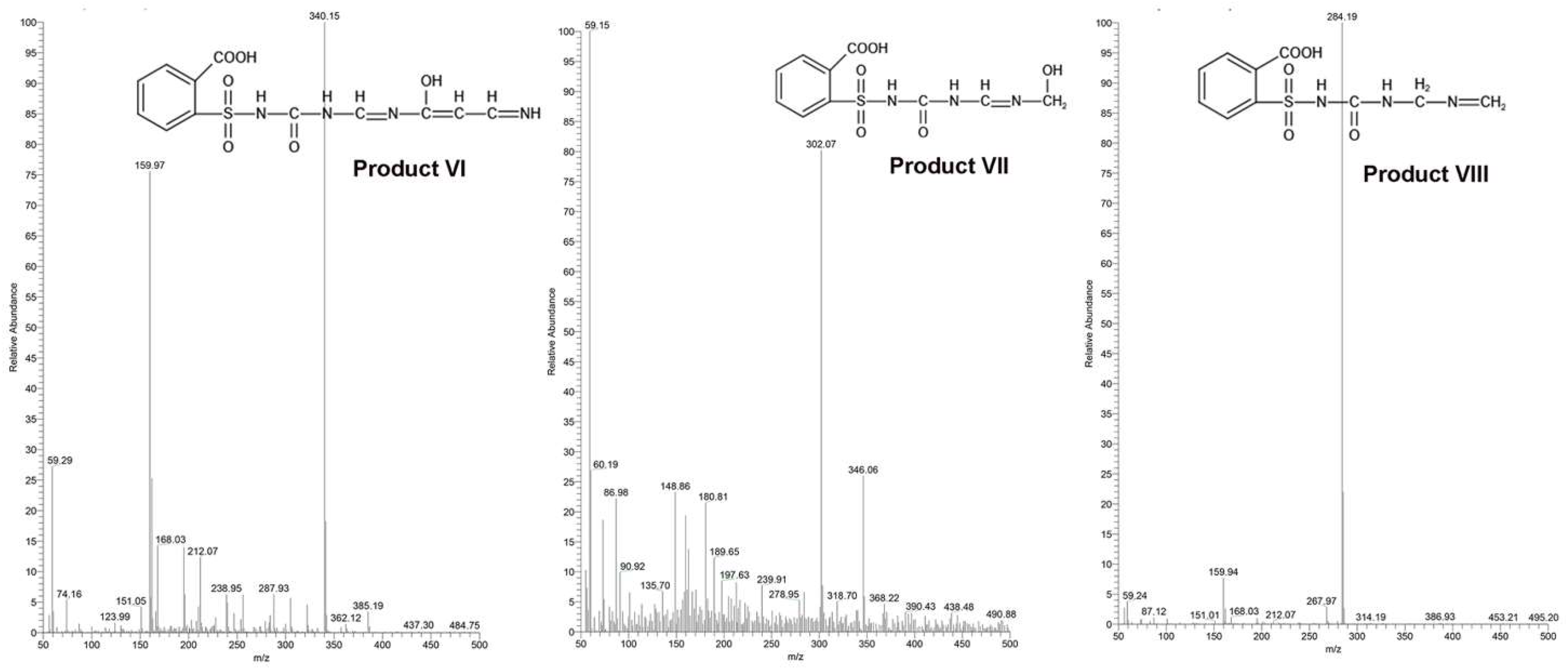
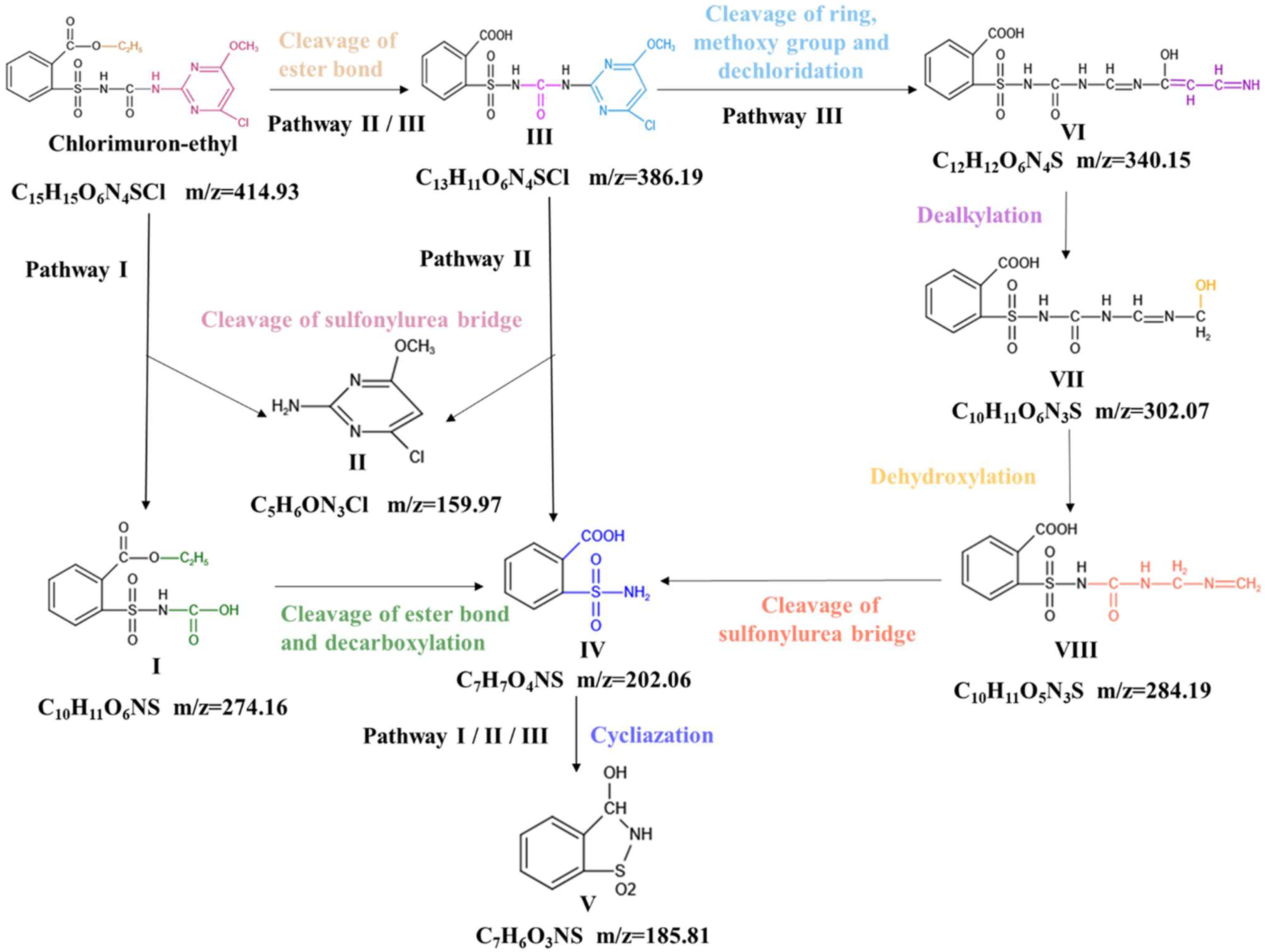
2.2. Pathways and Mechanisms Involved in Chlorimuron-Ethyl Degradation by Strain CHL1
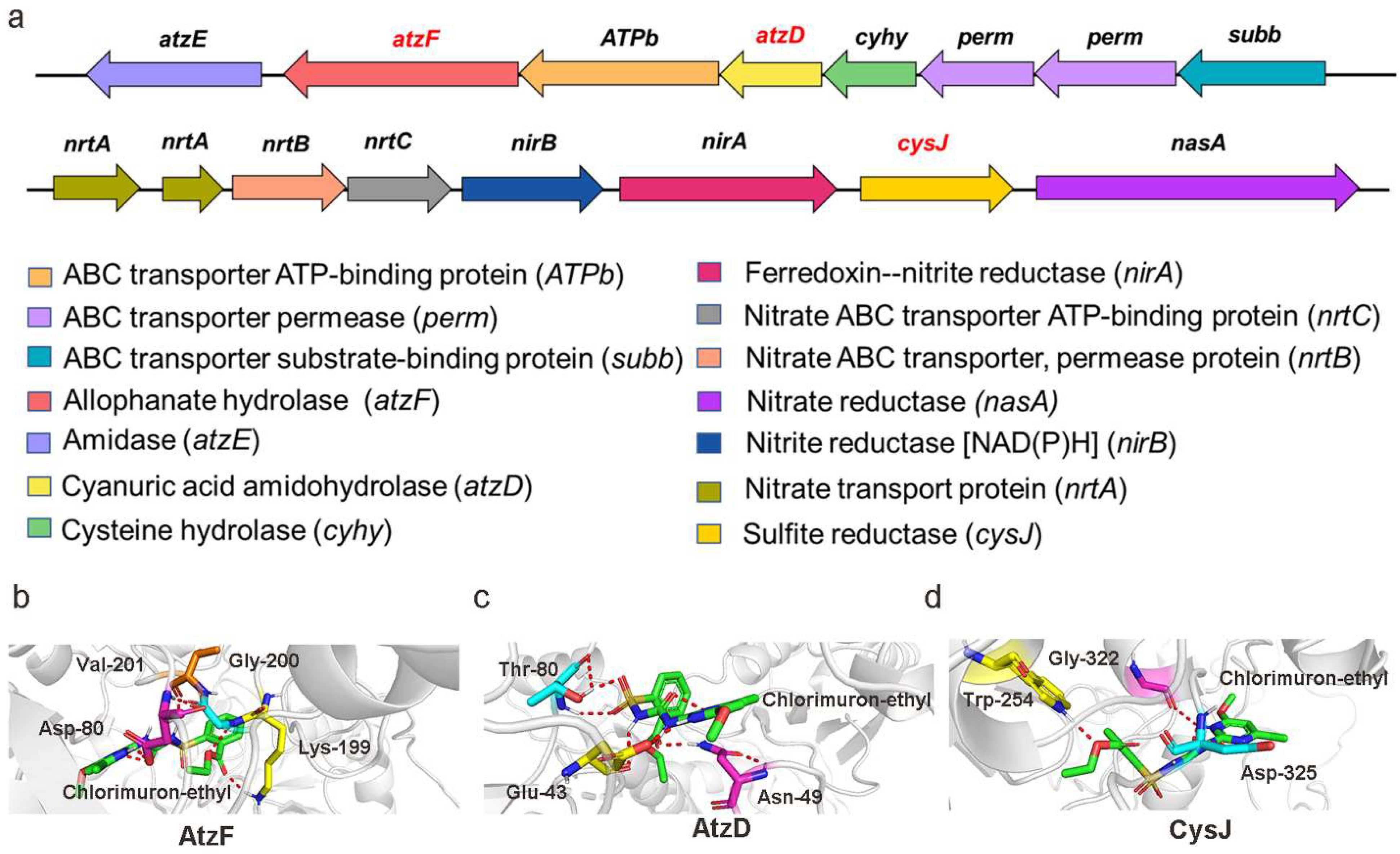
2.3. RNA-Seq Analysis of Strain CHL1 during Chlorimuron-Ethyl Degradation
2.4. Molecular Engineering
2.5. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
2.6. Structural Analysis
2.7. Predicted Mechanisms for Enzymatic Degradation of Chlorimuron-Ethyl
2.8. Schemes of the Functional Genes Involved in the Pathways Involved in Chlorimuron-Ethyl Degradation by Strain CHL1
3. Materials and Methods
3.1. Plasmids and Bacterial Strains
3.2. Medium and Detection Conditions
3.3. Pathways and Mechanisms of Chlorimuron-Ethyl Degradation
3.4. Transcriptome Sequencing
3.5. Differentially Expressed Genes (DEG) Analysis
3.6. Molecular Engineering
3.7. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) Assay
3.8. Structure and Function Prediction
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reddy, K.N.; Locke, M.A.; Wagner, S.C.; Zablotowicz, R.M.; Gaston, L.A.; Smeda, R.J. Chlorimuron ethyl sorption and desorption kinetics in soils and herbicide-desiccated cover crop residues. J. Agric. Food Chem. 1995, 43, 2752–2757. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, Q. Single and joint toxicity of chlorimuron-ethyl, cadmium, and copper acting on wheat Triticum aestivum. Ecotoxicol. Environ. Saf. 2005, 60, 169–175. [Google Scholar] [CrossRef]
- Tan, H.; Xu, M.; Li, X.; Zhang, H.; Zhang, C. Effects of chlorimuron-ethyl application with or without urea fertilization on soil ammonia-oxidizing bacteria and archaea. J. Hazard. Mater. 2013, 260, 368–374. [Google Scholar] [CrossRef]
- Zawoznik, M.S.; Tomaro, M.L. Effect of chlorimuron-ethyl on Bradyrhizobium japonicum and its symbiosis with soybean. Pest Manag. Sci. 2005, 61, 1003–1008. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Zhang, C.; Li, X.; Zhang, H. Ecological risk of long-term chlorimuron-ethyl application to soil microbial community: An in situ investigation in a continuously cropped soybean field in Northeast China. Environ. Sci. Pollut. Res. 2011, 18, 407–415. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Li, X.; Su, Z.; Zhang, C.; Zhang, H. Microbial community dynamics during the bioremediation process of chlorimuron-ethyl-contaminated soil by Hansschlegelia sp. strain CHL1. PLoS ONE 2015, 10, e0117943. [Google Scholar]
- Esposito, E.; Paulillo, S.M.; Manfio, G.P. Biodegradation of the herbicide diuron in soil by indigenous actinomycetes. Chemosphere 1998, 37, 541–548. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Z.; Lu, P.; Wang, H.; Waseem Ali, S.; Li, S.P.; Huang, X. Biodegradation of the sulfonylurea herbicide chlorimuron-ethyl by the strain Pseudomonas sp. LW3. FEMS Microbiol. Lett. 2009, 296, 203–209. [Google Scholar] [CrossRef]
- Xiong, M.; Hu, Z.; Ying, Z.; Cheng, X.; Li, C. Survival of GFP-tagged Rhodococcus sp. D310-1 in chlorimuron-ethyl-contaminated soil and its effects on the indigenous microbial community. J. Hazard. Mater. 2013, 252–253, 347–354. [Google Scholar]
- Yang, L.; Li, X.; Li, X.; Su, Z.; Zhang, C.; Zhang, H. Bioremediation of chlorimuron-ethyl-contaminated soil by Hansschlegelia sp. strain CHL1 and the changes of indigenous microbial population and N-cycling function genes during the bioremediation process. J. Hazard. Mater. 2014, 274, 314–321. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, H.; Li, X.; Su, Z.; Wang, J.; Zhang, C. Isolation and characterization of Sporobolomyces sp. LF1 capable of degrading chlorimuron-ethyl. J. Environ. Sci. 2009, 21, 1253–1260. [Google Scholar] [CrossRef]
- Zang, H.; Yu, Q.; Lv, T.; Cheng, Y.; Feng, L.; Cheng, X.; Li, C. Insights into the degradation of chlorimuron-ethyl by Stenotrophomonas maltophilia D310-3. Chemosphere 2016, 144, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zang, H.; Yu, Q.; Lv, T.; Cheng, Y.; Cheng, X.; Liu, K.; Liu, W.; Xu, P.; Lan, C. Biodegradation of chlorimuron-ethyl and the associated degradation pathway by Rhodococcus sp. D310-1. Environ. Sci. Pollut. Res. 2016, 23, 8794–8805. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y. Research on the Biodegradation Mechanism and Degradation Products of Herabicide Chlorimuron-Ethyl. Ph.D. Dissertation, Northeast Agricultural University, Harbin, China, 2012. [Google Scholar]
- Zang, H.; Wang, H.; Miao, L.; Cheng, Y.; Zhang, Y.; Liu, Y.; Sun, S.; Wang, Y.; Li, C. Carboxylesterase, a de-esterification enzyme, catalyzes the degradation of chlorimuron-ethyl in Rhodococcus erythropolis D310-1. J. Hazard. Mater. 2020, 387, 121684. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, C.; Sun, F.; Zhang, Z.; Zhang, X.; Pan, H.; Sun, P.; Zhang, H. Glutathione-S-transferase (GST) catalyzes the degradation of Chlorimuron-ethyl by Klebsiella jilinsis 2N3. Sci. Total Environ. 2020, 729, 139075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hao, Q.; Zhang, Z.; Zhang, X.; Pan, H.; Zhang, J.; Zhang, H.; Sun, F. Whole genome sequencing and analysis of chlorimuron-ethyl degrading bacteria Klebsiella pneumoniae 2N3. Int. J. Mol. Sci. 2019, 20, 3053. [Google Scholar] [CrossRef]
- Zhang, C.; Hao, Q.; Zhang, S.; Zhang, Z.; Zhang, X.; Sun, P.; Pan, H.; Zhang, H.; Sun, F. Transcriptomic analysis of Chlorimuron-ethyl degrading bacterial strain Klebsiella jilinsis 2N3. Ecotoxicol. Environ. Saf. 2019, 183, 109581. [Google Scholar] [CrossRef]
- Yu, Z.; Gu, W.; Yang, Y.; Li, X.; Li, X.; Li, T.; Wang, J.; Su, Z.; Li, X.; Dai, Y.; et al. Whole-Genome Sequencing of a Chlorimuron-Ethyl-Degrading Strain: Chenggangzhangella methanolivorans CHL1 and Its Degrading Enzymes. Microbiol. Spectrum 2022, e01822-22. [Google Scholar] [CrossRef]
- Megadi, V.B.; Tallur, P.N.; Mulla, S.I.; Ninnekar, H.Z. Bacterial Degradation of Fungicide Captan. J. Agric. Food Chem. 2010, 58, 12863–12868. [Google Scholar] [CrossRef]
- Yang, L.; Liu, L.; Salam, N.; Xiao, M.; Kim, C.J.; Hozzein, W.N.; Park, D.; Li, W.; Zhang, H. Chenggangzhangella methanolivorans gen. Nov., sp. nov., a member of the family Methylocystaceae, transfer of Methylopila helvetica Doronina et al. 2000 to Albibacter helveticus comb. nov. and emended description of the genus Albibacter. Int. J. Syst. Evol. Microbiol. 2016, 66, 2825–2830. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Li, X.; Wang, H.; Su, Z.; Wang, X.; Zhang, H. Dynamic changes in microbial communities during the bioremediation of herbicide (chlorimuron-ethyl and atrazine) contaminated soils by combined degrading bacteria. PLoS ONE 2018, 13, e0194753. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, H.; Wang, J.; Li, X.; Li, X.; Su, Z. High bioremediation potential of strain Chenggangzhangella methanolivorans CHL1 for soil polluted with metsulfuron-methyl or tribenuron-methyl in a pot experiment. Environ. Sci. Pollut. Res. 2021, 28, 4731–4738. [Google Scholar] [CrossRef]
- Zhu, S.; Qiu, J.; Wang, H.; Wang, X.; Jin, W.; Zhang, Y.; Zhang, C.; Hu, G.; He, J.; Hong, Q. Cloning and expression of the carbaryl hydrolase gene mcba and the identification of a key amino acid necessary for carbaryl hydrolysis. J. Hazard. Mater. 2018, 344, 1126–1135. [Google Scholar] [CrossRef]
- Shapir, N.; Sadowsky, M.J.; Wackett, L.P. Purification and characterization of allophanate hydrolase (AtzF) from Pseudomonas sp. strain ADP. J. Bacteriol. 2005, 187, 3731–3738. [Google Scholar] [CrossRef]
- Karns, J.S. Gene sequence and properties of an s-triazine ring-cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol 1999, 65, 3512–3517. [Google Scholar] [CrossRef]
- Harder, P.A.; O’Keefe, D.P.; Romesser, J.A.; Leto, K.J.; Omer, C.A. Isolation and characterization of Streptomyces griseolus deletion mutants affected in cytochrome P-450-mediated herbicide metabolism. Mol. Gen. Genet. 1991, 227, 238–244. [Google Scholar] [CrossRef]
- Holland, I.B. ABC transporters, mechanisms and biology: An overview. Essays Biochem. 2011, 50, 1–17. [Google Scholar]
- Hang, B.; Hong, Q.; Xie, X.; Huang, X.; Wang, C.; He, J.; Li, S. SulE, a sulfonylurea herbicide de-esterification esterase from Hansschlegelia zhihuaiae S113. Appl. Environ. Microbiol. 2012, 78, 1962–1968. [Google Scholar] [CrossRef]
- Yang, L.; Li, X.; Li, X.; Su, Z.; Zhang, C.; Xu, M.; Zhang, H. Improved stability and enhanced efficiency to degrade chlorimuron-ethyl by the entrapment of esterase SulE in cross-linked poly (γ-glutamic acid)/gelatin hydrogel. J. Hazard. Mater. 2015, 287, 287–295. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, H.; Fu, X.; Li, X.; Guo, Q.; Yang, T.; Li, X. Immobilization of esterase SulE in cross-linked gelatin/chitosan and its application in remediating soils polluted with tribenuron-methyl and metsulfuron-methyl. Process Biochem. 2020, 98, 217–223. [Google Scholar] [CrossRef]
- Fan, B.; Rosen, B.P. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J. Biol. Chem. 2002, 277, 46987–46992. [Google Scholar] [CrossRef]
- Steiger, S.; Astier, C.; Sandmann, G. Substrate specificity of the expressed carotenoid 3,4-desaturase from Rubrivivax gelatinosus reveals the detailed reaction sequence to spheroidene and spirilloxanthin. Biochem. J. 2000, 349, 635–640. [Google Scholar] [CrossRef]
- Hiseni, A.; Arends, I.; Otten, L.G. Biochemical characterization of the carotenoid 1,2-hydratases (CrtC) from Rubrivivax gelatinosus and Thiocapsa roseopersicina. Appl. Microbiol. Biotechnol. 2011, 91, 1029–1036. [Google Scholar] [CrossRef] [Green Version]
- van der Werf, M.J.; Swarts, H.J.; De Bont, J.A.M. Rhodococcus erythropolis DCL14 contains a novel degradation pathway for limonene. Appl. Environ. Microbiol. 1999, 65, 2092–2102. [Google Scholar] [CrossRef]
- Benzi, M.; Robotti, E.; Gianotti, V. Study on the photodegradation of amidosulfuron in aqueous solutions by LC-MS/MS. Environ. Sci. Pollut. Res. 2013, 20, 9034–9043. [Google Scholar] [CrossRef]
- Jin, M.; Yu, X.; Chen, X.; Zeng, R. Pseudomonas putida IOFA1 transcriptome profiling reveals a metabolic pathway involved in formaldehyde degradation. Process Biochem. 2016, 51, 220–228. [Google Scholar] [CrossRef]
- Bruchmann, S.; Muthukumarasamy, U.; Pohl, S.; Preusse, M.; Bielecka, A.; Nicolai, T.; Hamann, I.; Hillert, R.; Kola, A.; Gastmeier, P.; et al. Deep transcriptome profiling of clinical klebsiella pneumoniae isolates reveals strain and sequence type-specific adaptation. Environ. Microbiol. 2015, 17, 4690–4710. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, Z.; Hu, Z.; Wang, W.; Xiong, J. Morphology and molecular analysis of Moesziomyces antarcticus isolated from the blood samples of a Chinese patient. Front. Microbiol. 2019, 10, 254. [Google Scholar] [CrossRef]
- Zhan, Y.; Guo, S. Three-dimensional (3D) structure prediction and function analysis of the chitin-binding domain 3 protein HD73-3189 from Bacillus thuringiensis HD73. Biomed. Mater. Eng. 2015, 26, S2019–S2024. [Google Scholar] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Unzue, A.; Zhao, H.; Lolli, G.; Dong, J.; Zhu, J. The “gatekeeper” residue influences the mode of binding of acetyl indoles to bromodomains. J. Med. Chem. 2016, 59, 3087–3097. [Google Scholar] [CrossRef]
- Guo, L.; Fang, W.; Guo, L.; Yao, C.; Zhao, Y.; Ge, F.; Dai, Y. Biodegradation of the neonicotinoid insecticide acetamiprid by actinomycetes Streptomyces canus CGMCC 13662 and characterization of the novel nitrile hydratase involved. J. Agric. Food Chem. 2019, 67, 5922–5931. [Google Scholar] [CrossRef]
- Liu, X.; Luo, Y.; Mohamed, O.A.; Liu, D.; Wei, G. Global transcriptome analysis of Mesorhizobium alhagi CCNWXJ12-2 under salt stress. BMC Microbiol. 2014, 14, 319. [Google Scholar] [CrossRef]
- Kristoffersen, S.M.; Haase, C.; Weil, M.R.; Passalacqua, K.D.; Niazi, F.; Hutchison, S.K.; Desany, B.; Kolstø, A.; Tourasse, N.J.; Read, T.D. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol. 2012, 13, R30. [Google Scholar] [CrossRef]
- Ran, S.; Liu, B.; Jiang, W.; Sun, Z.; Liang, J. Transcriptome analysis of Enterococcus faecalis in response to alkaline stress. Front. Microbiol. 2015, 6, 795. [Google Scholar] [CrossRef] [PubMed]
- Thomason, L.C.; Sawitzke, J.A.; Li, X.; Costantino, N.; Court, D.L. Recombineering: Genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. 2014, 78, 1–24. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Annotation | Log2FC | KEGG | ||
|---|---|---|---|---|---|
| Sample A | Sample B | Sample C | |||
| atzF | Allophanate hydrolase | 0.23 | 0.82 * | 1.77 * | Atrazine degradation |
| atzD | Cyanuric acid amidohydrolase | −0.65 | 1.74 * | 2.03 * | Atrazine degradation |
| cysJ | Sulfite reductase | 0.06 | 1.17 * | 2.57 * | Sulfur metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Dai, Y.; Li, T.; Gu, W.; Yang, Y.; Li, X.; Peng, P.; Yang, L.; Li, X.; Wang, J.; et al. A Novel Pathway of Chlorimuron-Ethyl Biodegradation by Chenggangzhangella methanolivorans Strain CHL1 and Its Molecular Mechanisms. Int. J. Mol. Sci. 2022, 23, 9890. https://doi.org/10.3390/ijms23179890
Yu Z, Dai Y, Li T, Gu W, Yang Y, Li X, Peng P, Yang L, Li X, Wang J, et al. A Novel Pathway of Chlorimuron-Ethyl Biodegradation by Chenggangzhangella methanolivorans Strain CHL1 and Its Molecular Mechanisms. International Journal of Molecular Sciences. 2022; 23(17):9890. https://doi.org/10.3390/ijms23179890
Chicago/Turabian StyleYu, Zhixiong, Yumeng Dai, Tingting Li, Wu Gu, Yi Yang, Xiang Li, Pai Peng, Lijie Yang, Xinyu Li, Jian Wang, and et al. 2022. "A Novel Pathway of Chlorimuron-Ethyl Biodegradation by Chenggangzhangella methanolivorans Strain CHL1 and Its Molecular Mechanisms" International Journal of Molecular Sciences 23, no. 17: 9890. https://doi.org/10.3390/ijms23179890
APA StyleYu, Z., Dai, Y., Li, T., Gu, W., Yang, Y., Li, X., Peng, P., Yang, L., Li, X., Wang, J., Su, Z., Li, X., Xu, M., & Zhang, H. (2022). A Novel Pathway of Chlorimuron-Ethyl Biodegradation by Chenggangzhangella methanolivorans Strain CHL1 and Its Molecular Mechanisms. International Journal of Molecular Sciences, 23(17), 9890. https://doi.org/10.3390/ijms23179890







