Role of Muscle LIM Protein in Mechanotransduction Process
Abstract
1. Introduction
2. The Plasticity of Muscle
3. Mechanical Stress and Mechanotransduction
3.1. Mechanobiology versus Biomechanics
3.2. The Mechanical Stress
3.3. Impact of a Mechanical Stress on the Extracellular Matrix
3.4. Impact of a Mechanical Stimulation on the Intracellular Matrix
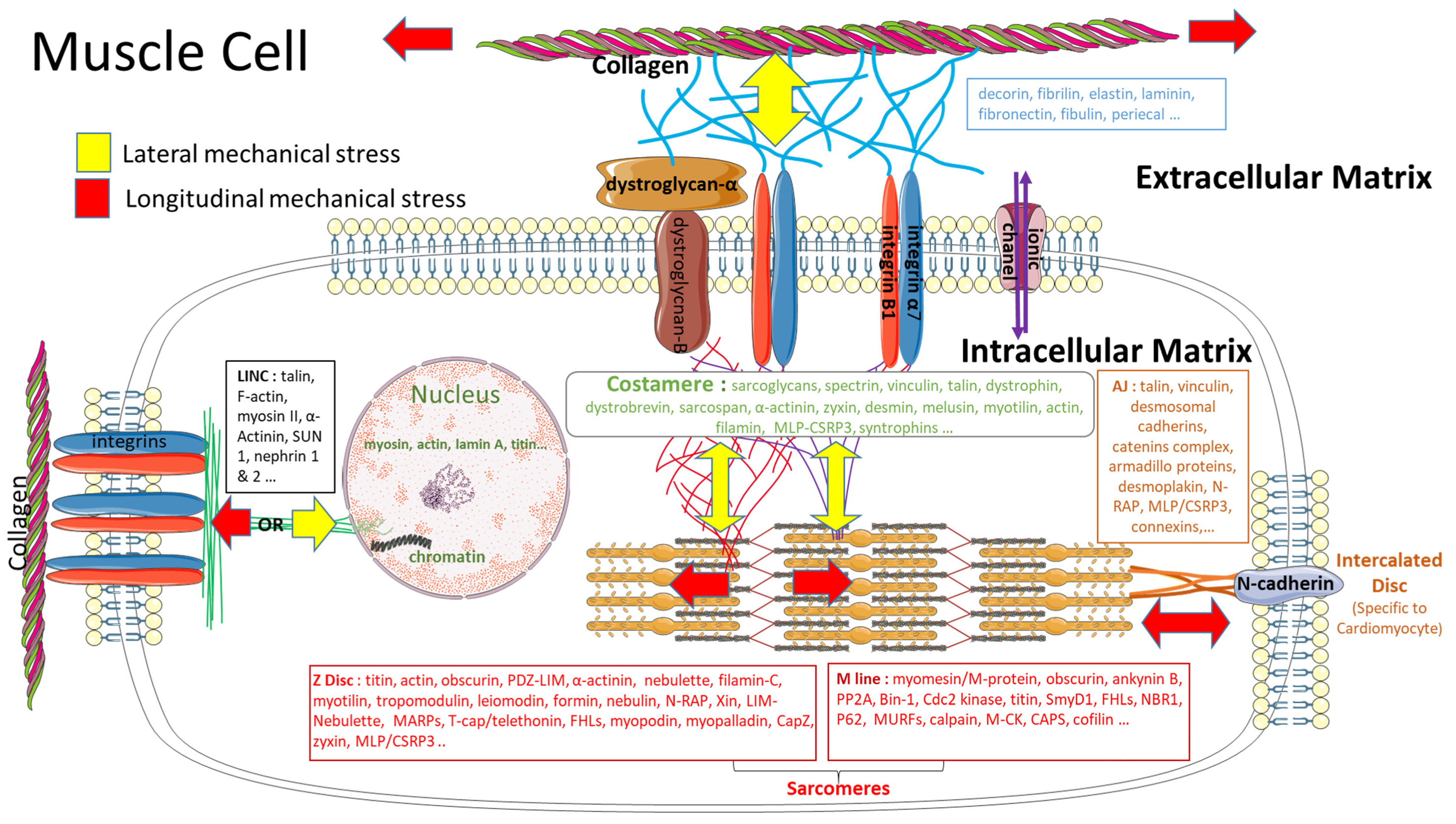
4. From Mechanosensitivity to Mechanotransduction
5. Muscle LIM Protein
5.1. MLP Isoforms
5.2. Nuclear Translocation
5.3. MLP Involvement in the Transcription Process
5.4. MLP Role in Mechanotransduction Process following Mechanical Stimulation
5.5. MLP Involvement in Human Diseases
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Argilés, J.M.; Campos, N.; Lopez-Pedrosa, J.M.; Rueda, R.; Rodriguez-Mañas, L. Skeletal Muscle Regulates Metabolism via Interorgan Crosstalk: Roles in Health and Disease. J. Am. Med. Dir. Assoc. 2016, 17, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, J.; Schneider, V.S. Space, Gravity and the Physiology of Aging: Parallel or Convergent Disciplines? A Mini-Review. Gerontology 2010, 56, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Ethgen, O.; Beaudart, C.; Buckinx, F.; Bruyère, O.; Reginster, J.Y. The Future Prevalence of Sarcopenia in Europe: A Claim for Public Health Action. Calcif. Tissue Int. 2017, 100, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Neufer, P.D.; Bamman, M.M.; Muoio, D.M.; Bouchard, C.; Cooper, D.M.; Goodpaster, B.H.; Booth, F.W.; Kohrt, W.M.; Gerszten, R.E.; Mattson, M.P.; et al. Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell Metab. 2015, 22, 4–11. [Google Scholar] [CrossRef]
- Hawley, J.A.; Hargreaves, M.; Joyner, M.J.; Zierath, J.R. Integrative Biology of Exercise. Cell 2014, 159, 738–749. [Google Scholar] [CrossRef]
- Solís, C.; Russell, B. Striated Muscle Proteins Are Regulated Both by Mechanical Deformation and by Chemical Post-Translational Modification. Biophys. Rev. 2021, 13, 679–695. [Google Scholar] [CrossRef]
- Ingber, D.E.; Wang, N.; Stamenovic, D. Tensegrity, Cellular Biophysics, and the Mechanics of Living Systems. Rep. Prog. Phys. 2014, 77, 046603. [Google Scholar] [CrossRef]
- Schiaffino, S.; Reggiani, C.; Akimoto, T.; Blaauw, B. Molecular Mechanisms of Skeletal Muscle Hypertrophy. J. Neuromuscul. Dis. 2021, 8, 169–183. [Google Scholar] [CrossRef]
- Buyandelger, B.; Ng, K.-E.; Miocic, S.; Piotrowska, I.; Gunkel, S.; Ku, C.-H.; Knöll, R. MLP (Muscle LIM Protein) as a Stress Sensor in the Heart. Pflügers Arch.-Eur. J. Physiol. 2011, 462, 135–142. [Google Scholar] [CrossRef]
- Boateng, S.Y.; Senyo, S.E.; Qi, L.; Goldspink, P.H.; Russell, B. Myocyte Remodeling in Response to Hypertrophic Stimuli Requires Nucleocytoplasmic Shuttling of Muscle LIM Protein. J. Mol. Cell. Cardiol. 2009, 47, 426–435. [Google Scholar] [CrossRef]
- Soltow, Q.A.; Zeanah, E.H.; Lira, V.A.; Criswell, D.S. Cessation of Cyclic Stretch Induces Atrophy of C2C12 Myotubes. Biochem. Biophys. Res. Commun. 2013, 434, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.K.; Verlengia, R.; Bueno Junior, C.R. Mechanotransduction Pathways in Skeletal Muscle Hypertrophy. J. Recept. Signal Transduct. Res. 2012, 32, 42–44. [Google Scholar] [CrossRef]
- Bernardo, B.C.; McMullen, J.R. Molecular Aspects of Exercise-Induced Cardiac Remodeling. Cardiol. Clin. 2016, 34, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Pette, D.; Staron, R.S. Transitions of Muscle Fiber Phenotypic Profiles. Histochem. Cell Biol. 2001, 115, 359–372. [Google Scholar] [CrossRef]
- Davies, K.J.A. Adaptive Homeostasis. Mol. Aspects Med. 2016, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. Overcompensation Stimulation: A Mechanism for Hormetic Effects. Crit. Rev. Toxicol. 2001, 31, 425–470. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Mattson, M.P. How Does Hormesis Impact Biology, Toxicology, and Medicine? NPJ Aging Mech. Dis. 2017, 3, 13. [Google Scholar] [CrossRef]
- Pikosky, M.A.; Gaine, P.C.; Martin, W.F.; Grabarz, K.C.; Ferrando, A.A.; Wolfe, R.R.; Rodriguez, N.R. Aerobic Exercise Training Increases Skeletal Muscle Protein Turnover in Healthy Adults at Rest. J. Nutr. 2006, 136, 379–383. [Google Scholar] [CrossRef]
- Demangel, R.; Treffel, L.; Py, G.; Brioche, T.; Pagano, A.F.; Bareille, M.-P.; Beck, A.; Pessemesse, L.; Candau, R.; Gharib, C.; et al. Early Structural and Functional Signature of 3-Day Human Skeletal Muscle Disuse Using the Dry Immersion Model. J. Physiol. 2017, 595, 4301–4315. [Google Scholar] [CrossRef]
- Baldwin, K.M.; Haddad, F.; Pandorf, C.E.; Roy, R.R.; Edgerton, V.R. Alterations in Muscle Mass and Contractile Phenotype in Response to Unloading Models: Role of Transcriptional/Pretranslational Mechanisms. Front. Physiol. 2013, 4, 284. [Google Scholar] [CrossRef]
- Anastasi, G.; Cutroneo, G.; Santoro, G.; Arco, A.; Rizzo, G.; Bramanti, P.; Rinaldi, C.; Sidoti, A.; Amato, A.; Favaloro, A. Costameric Proteins in Human Skeletal Muscle during Muscular Inactivity. J. Anat. 2008, 213, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Bogdanis, G.C. Effects of Physical Activity and Inactivity on Muscle Fatigue. Front. Physiol. 2012, 3, 142. [Google Scholar] [CrossRef] [PubMed]
- Hackney, K.J.; Ploutz-Snyder, L.L. Unilateral Lower Limb Suspension: Integrative Physiological Knowledge from the Past 20 Years (1991–2011). Eur. J. Appl. Physiol. 2012, 112, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Narici, M.V.; Erskine, R.M.; Seynnes, O.R.; Rittweger, J.; Pišot, R.; Šimunič, B.; Flück, M. Costamere Remodeling with Muscle Loading and Unloading in Healthy Young Men. J. Anat. 2013, 223, 525–536. [Google Scholar] [CrossRef]
- Akpulat, U.; Onbaşılar, İ.; Kocaefe, Y.Ç. Tenotomy Immobilization as a Model to Investigate Skeletal Muscle Fibrosis (with Emphasis on Secreted Frizzled-Related Protein 2). Physiol. Genom. 2016, 48, 397–408. [Google Scholar] [CrossRef]
- Dirks-Naylor, A.J.; Lennon-Edwards, S. Cellular and Molecular Mechanisms of Apoptosis in Age-Related Muscle Atrophy. Curr. Aging Sci. 2011, 4, 269–278. [Google Scholar] [CrossRef]
- Power, G.A.; Dalton, B.H.; Rice, C.L. Human Neuromuscular Structure and Function in Old Age: A Brief Review. J. Sport Health Sci. 2013, 2, 215–226. [Google Scholar] [CrossRef]
- Landi, F.; Calvani, R.; Cesari, M.; Tosato, M.; Martone, A.M.; Ortolani, E.; Savera, G.; Salini, S.; Sisto, A.N.; Picca, A.; et al. Sarcopenia: An Overview on Current Definitions, Diagnosis and Treatment. Curr. Protein Pept. Sci. 2017, 19, 633–638. [Google Scholar] [CrossRef]
- Phymedexp. Abrégé des Protéines Musculaires. Available online: https://u1046.edu.umontpellier.fr/163-2/abrege-des-proteines-musculaires/ (accessed on 30 August 2019).
- Nigro, V.; Piluso, G. Spectrum of Muscular Dystrophies Associated with Sarcolemmal-Protein Genetic Defects. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2015, 1852, 585–593. [Google Scholar] [CrossRef]
- Sweeney, M.; Yiu, A.; Lyon, A.R. Cardiac Atrophy and Heart Failure in Cancer. Card. Fail. Rev. 2017, 3, 62–65. [Google Scholar] [CrossRef]
- Tan, P.M.; Buchholz, K.S.; Omens, J.H.; McCulloch, A.D.; Saucerman, J.J. Predictive Model Identifies Key Network Regulators of Cardiomyocyte Mechano-Signaling. PLOS Comput. Biol. 2017, 13, e1005854. [Google Scholar] [CrossRef] [PubMed]
- Heineke, J.; Molkentin, J.D. Regulation of Cardiac Hypertrophy by Intracellular Signalling Pathways. Nat. Rev. Mol. Cell Biol. 2006, 7, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Katz, A.M. The “Modern” View of Heart Failure: How Did We Get Here? Circ. Heart Fail. 2008, 1, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.M.; Hoffman, J.R.; Stout, J.R.; Fukuda, D.H.; Willoughby, D.S. Intramuscular Anabolic Signaling and Endocrine Response Following Resistance Exercise: Implications for Muscle Hypertrophy. Sports Med. 2016, 46, 671–685. [Google Scholar] [CrossRef]
- Egerman, M.A.; Glass, D.J. Signaling Pathways Controlling Skeletal Muscle Mass. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 59–68. [Google Scholar] [CrossRef]
- Frost, R.A.; Lang, C.H. Protein Kinase B/Akt: A Nexus of Growth Factor and Cytokine Signaling in Determining Muscle Mass. J. Appl. Physiol. 2007, 103, 378–387. [Google Scholar] [CrossRef]
- Sandri, M. Signaling in Muscle Atrophy and Hypertrophy. Physiology 2008, 23, 160–170. [Google Scholar] [CrossRef]
- Khalilimeybodi, A.; Daneshmehr, A.; Sharif Kashani, B. Ca2+-Dependent Calcineurin/NFAT Signaling in β-Adrenergic-Induced Cardiac Hypertrophy. Gen. Physiol. Biophys. 2018, 37, 41–56. [Google Scholar] [CrossRef]
- Fink, J.; Schoenfeld, B.J.; Nakazato, K. The Role of Hormones in Muscle Hypertrophy. Phys. Sportsmed. 2018, 46, 129–134. [Google Scholar] [CrossRef]
- Dillmann, W. Cardiac Hypertrophy and Thyroid Hormone Signaling. Heart Fail. Rev. 2010, 15, 125–132. [Google Scholar] [CrossRef]
- Alli, N.S.; Yang, E.C.; Miyake, T.; Aziz, A.; Collins-Hooper, H.; Patel, K.; McDermott, J.C. Signal-Dependent Fra-2 Regulation in Skeletal Muscle Reserve and Satellite Cells. Cell Death Dis. 2013, 4, e692. [Google Scholar] [CrossRef] [PubMed]
- Proud, C.G. Ras, PI3-Kinase and MTOR Signaling in Cardiac Hypertrophy. Cardiovasc. Res. 2004, 63, 403–413. [Google Scholar] [CrossRef]
- Hindi, S.M.; Sato, S.; Xiong, G.; Bohnert, K.R.; Gibb, A.A.; Gallot, Y.S.; McMillan, J.D.; Hill, B.G.; Uchida, S.; Kumar, A. TAK1 Regulates Skeletal Muscle Mass and Mitochondrial Function. JCI Insight 2018, 3, e98441. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Cholewa, J.M.; Zhao, Y.; Yang, Y.; Shang, H.; Jiang, H.; Su, Q.; Zanchi, N.E. A Potential Strategy for Counteracting Age-Related Sarcopenia: Preliminary Evidence of Combined Exercise Training and Leucine Supplementation. Food Funct. 2017, 8, 4528–4538. [Google Scholar] [CrossRef]
- Wang, J.; Liew, O.W.; Richards, A.M.; Chen, Y.-T. Overview of MicroRNAs in Cardiac Hypertrophy, Fibrosis, and Apoptosis. Int. J. Mol. Sci. 2016, 17, 749. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, J.R.; Mongue-Din, H.; Eaton, P.; Shah, A.M. Redox Signaling in Cardiac Physiology and Pathology. Circ. Res. 2012, 111, 1091–1106. [Google Scholar] [CrossRef]
- Takahashi, K.; Matsuda, Y.; Naruse, K. Mechanosensitive Ion Channels. AIMS Biophys. 2016, 3, 63–74. [Google Scholar] [CrossRef]
- Xia, L.; Cheung, K.-K.; Yeung, S.S.; Yeung, E.W. The Involvement of Transient Receptor Potential Canonical Type 1 in Skeletal Muscle Regrowth after Unloading-Induced Atrophy. J. Physiol. 2016, 592, 3111–3126. [Google Scholar] [CrossRef]
- De, R.; Zemel, A.; Safran, S.A. Theoretical Concepts and Models of Cellular Mechanosensing. Methods Cell Biol. 2010, 98, 143–175. [Google Scholar] [CrossRef]
- Rindom, E.; Vissing, K. Mechanosensitive Molecular Networks Involved in Transducing Resistance Exercise-Signals into Muscle Protein Accretion. Front. Physiol. 2016, 7, 547. [Google Scholar] [CrossRef][Green Version]
- Bernardo, B.C.; Weeks, K.L.; Pretorius, L.; McMullen, J.R. Molecular Distinction between Physiological and Pathological Cardiac Hypertrophy: Experimental Findings and Therapeutic Strategies. Pharmacol. Ther. 2010, 128, 191–227. [Google Scholar] [CrossRef] [PubMed]
- Gundersen, K. Excitation-Transcription Coupling in Skeletal Muscle: The Molecular Pathways of Exercise. Biol. Rev. Camb. Philos. Soc. 2011, 86, 564–600. [Google Scholar] [CrossRef]
- Silver, F.H.; Siperko, L.M. Mechanosensing and Mechanochemical Transduction: How Is Mechanical Energy Sensed and Converted into Chemical Energy in an Extracellular Matrix? Crit. Rev. Biomed. Eng. 2003, 31, 255–331. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Kamm, R.D.; Lee, R.T. Cell Mechanics and Mechanotransduction: Pathways, Probes, and Physiology. Am. J. Physiol.-Cell Physiol. 2004, 287, C1–C11. [Google Scholar] [CrossRef] [PubMed]
- Ingber, D.E. Tensegrity-Based Mechanosensing from Macro to Micro. Prog. Biophys. Mol. Biol. 2008, 97, 163–179. [Google Scholar] [CrossRef] [PubMed]
- Lyon, R.C.; Zanella, F.; Omens, J.H.; Sheikh, F. Mechanotransduction in Cardiac Hypertrophy and Failure. Circ. Res. 2015, 116, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Jansen, K.A.; Donato, D.M.; Balcioglu, H.E.; Schmidt, T.; Danen, E.H.J.; Koenderink, G.H. A Guide to Mechanobiology: Where Biology and Physics Meet. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2015, 1853, 3043–3052. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.L.; Kersh, M.E.; Kilbreath, S.; Knothe Tate, M. Establishing the Basis for Mechanobiology-Based Physical Therapy Protocols to Potentiate Cellular Healing and Tissue Regeneration. Front. Physiol. 2017, 8, 303. [Google Scholar] [CrossRef]
- Goubel, F.; Lensel-corbeil, G. Biomécanique: Éléments de Biomécnique Musculaire, 2nd ed.; STAPS; Masson: Paris, France, 2003. [Google Scholar]
- Herzog, W.; Powers, K.; Johnston, K.; Duvall, M. A New Paradigm for Muscle Contraction. Front. Physiol. 2015, 6, 174. [Google Scholar] [CrossRef]
- DuFort, C.C.; Paszek, M.J.; Weaver, V.M. Balancing Forces: Architectural Control of Mechanotransduction. Nat. Rev. Mol. Cell Biol. 2011, 12, 308–319. [Google Scholar] [CrossRef]
- Hughes, D.C.; Wallace, M.A.; Baar, K. Effects of Aging, Exercise, and Disease on Force Transfer in Skeletal Muscle. Am. J. Physiol. Endocrinol. Metab. 2015, 309, E1–E10. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, B.L.; Dunn, A.R.; Weis, W.I.; Nelson, W.J. Mechano-Transduction: From Molecules to Tissues. PLoS Biol. 2014, 12, e1001996. [Google Scholar] [CrossRef] [PubMed]
- Petriz, B.A.; Gomes, C.P.C.; Almeida, J.A.; de Oliveira, G.P.; Ribeiro, F.M.; Pereira, R.W.; Franco, O.L. The Effects of Acute and Chronic Exercise on Skeletal Muscle Proteome. J. Cell. Physiol. 2017, 232, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Germain, A.; Bourzac, C.; Pichon, C.; Portier, H.; Pallu, S.; Germain, P. Impact of Treadmill Interval Running on the Appearance of Zinc Finger Protein FHL2 in Bone Marrow Cells in a Rat Model: A Pilot Study. Life 2022, 12, 528. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.; Zhang, M.; Wang, Y.; Yu, L.; Zhao, T.; Qiu, X.; Wang, L. Mechanical Stretch Regulates MicroRNA Expression Profile via NF-ΚB Activation in C2C12 Myoblasts. Mol. Med. Rep. 2016, 14, 5084–5092. [Google Scholar] [CrossRef] [PubMed]
- Juffer, P.; Bakker, A.D.; Klein-Nulend, J.; Jaspers, R.T. Mechanical Loading by Fluid Shear Stress of Myotube Glycocalyx Stimulates Growth Factor Expression and Nitric Oxide Production. Cell Biochem. Biophys. 2014, 69, 411–419. [Google Scholar] [CrossRef]
- Tsimbouri, P.M. Adult Stem Cell Responses to Nanostimuli. J. Funct. Biomater. 2015, 6, 598–622. [Google Scholar] [CrossRef]
- Zhao, W.; Hanson, L.; Lou, H.-Y.; Akamatsu, M.; Chowdary, P.D.; Santoro, F.; Marks, J.R.; Grassart, A.; Drubin, D.G.; Cui, Y.; et al. Nanoscale Manipulation of Membrane Curvature for Probing Endocytosis in Live Cells. Nat. Nanotechnol. 2017, 12, 750–756. [Google Scholar] [CrossRef]
- Hu, X.; Margadant, F.M.; Yao, M.; Sheetz, M.P. Molecular Stretching Modulates Mechanosensing Pathways. Protein Sci. Publ. Protein Soc. 2017, 26, 1337–1351. [Google Scholar] [CrossRef]
- Cheng, K.; Xia, P.; Lin, Q.; Shen, S.; Gao, M.; Ren, S.; Li, X. Effects of Low-Intensity Pulsed Ultrasound on Integrin-FAK-PI3K/Akt Mechanochemical Transduction in Rabbit Osteoarthritis Chondrocytes. Ultrasound Med. Biol. 2014, 40, 1609–1618. [Google Scholar] [CrossRef]
- Mittag, U.; Kriechbaumer, A.; Bartsch, M.; Rittweger, J. Form Follows Function: A Computational Simulation Exercise on Bone Shape Forming and Conservation. J. Musculoskelet. Neuronal Interact. 2015, 15, 215–226. [Google Scholar] [PubMed]
- Grounds, M.D.; Sorokin, L.; White, J. Strength at the Extracellular Matrix-Muscle Interface. Scand. J. Med. Sci. Sports 2005, 15, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a Distance: Mechanically Coupling the Extracellular Matrix with the Nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Delalande, A.; Leduc, C.; Midoux, P.; Postema, M.; Pichon, C. Efficient Gene Delivery by Sonoporation Is Associated with Microbubble Entry into Cells and the Clathrin-Dependent Endocytosis Pathway. Ultrasound Med. Biol. 2015, 41, 1913–1926. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T. Role of Extracellular Matrix in Development of Skeletal Muscle and Postmortem Aging of Meat. Meat Sci. 2015, 109, 48–55. [Google Scholar] [CrossRef]
- Zuidema, A.; Wang, W.; Sonnenberg, A. Crosstalk between Cell Adhesion Complexes in Regulation of Mechanotransduction. BioEssays 2020, 42, 2000119. [Google Scholar] [CrossRef]
- Liu, H.; Hu, J.; Zheng, Q.; Feng, X.; Zhan, F.; Wang, X.; Xu, G.; Hua, F. Piezo1 Channels as Force Sensors in Mechanical Force-Related Chronic Inflammation. Front. Immunol. 2022, 13, 816149. [Google Scholar] [CrossRef]
- Poole, K.; Moroni, M.; Lewin, G.R. Sensory Mechanotransduction at Membrane-Matrix Interfaces. Pflugers Arch. 2015, 467, 121–132. [Google Scholar] [CrossRef][Green Version]
- Pardo, J.V.; Siliciano, J.D.; Craig, S.W. A Vinculin-Containing Cortical Lattice in Skeletal Muscle: Transverse Lattice Elements (“costameres”) Mark Sites of Attachment between Myofibrils and Sarcolemma. Proc. Natl. Acad. Sci. USA 1983, 80, 1008–1012. [Google Scholar] [CrossRef]
- Randazzo, D.; Blaauw, B.; Paolini, C.; Pierantozzi, E.; Spinozzi, S.; Lange, S.; Chen, J.; Protasi, F.; Reggiani, C.; Sorrentino, V. Exercise-Induced Alterations and Loss of Sarcomeric M-Line Organization in the Diaphragm Muscle of Obscurin Knockout Mice. Am. J. Physiol.-Cell Physiol. 2017, 312, C16–C28. [Google Scholar] [CrossRef]
- Jaka, O.; Casas-Fraile, L.; López de Munain, A.; Sáenz, A. Costamere Proteins and Their Involvement in Myopathic Processes. Expert Rev. Mol. Med. 2015, 17, e12. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Guo, S.S.; Fässler, R. Integrin-Mediated Mechanotransduction. J. Cell Biol. 2016, 215, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Garbincius, J.F.; Michele, D.E. Dystrophin–Glycoprotein Complex Regulates Muscle Nitric Oxide Production through Mechanoregulation of AMPK Signaling. Proc. Natl. Acad. Sci. USA 2015, 112, 13663–13668. [Google Scholar] [CrossRef] [PubMed]
- Samarel, A.M.; Koshman, Y.; Swanson, E.R.; Russell, B. Biophysical Forces Modulate the Costamere and Z-Disc for Sarcomere Remodeling in Heart Failure. In Biophysics of the Failing Heart: Physics and Biology of Heart Muscle; Solaro, R.J., Tardiff, J.C., Eds.; Biological and Medical Physics, Biomedical Engineering; Springer: New York, NY, USA, 2013; pp. 141–174. [Google Scholar] [CrossRef]
- Henderson, C.A.; Gomez, C.G.; Novak, S.M.; Mi-Mi, L.; Gregorio, C.C. Overview of the Muscle Cytoskeleton. Compr. Physiol. 2017, 7, 891–924. [Google Scholar] [CrossRef] [PubMed]
- McNally, E.M.; Pytel, P. Muscle Diseases: The Muscular Dystrophies. Annu. Rev. Pathol. 2007, 2, 87–109. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.J.; Weihl, C.C.; Spencer, M.J. Molecular and Cellular Basis of Genetically Inherited Skeletal Muscle Disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 713–732. [Google Scholar] [CrossRef]
- Cazorla, O.; Vassort, G.; Garnier, D.; Le Guennec, J.Y. Length Modulation of Active Force in Rat Cardiac Myocytes: Is Titin the Sensor? J. Mol. Cell. Cardiol. 1999, 31, 1215–1227. [Google Scholar] [CrossRef]
- Knöll, R.; Hoshijima, M.; Chien, K.R. Muscle LIM Protein in Heart Failure. Exp. Clin. Cardiol. 2002, 7, 104–105. [Google Scholar]
- Bos, J.M.; Poley, R.N.; Ny, M.; Tester, D.J.; Xu, X.; Vatta, M.; Towbin, J.A.; Gersh, B.J.; Ommen, S.R.; Ackerman, M.J. Genotype—Phenotype Relationships Involving Hypertrophic Cardiomyopathy-Associated Mutations in Titin, Muscle LIM Protein, and Telethonin. Mol. Genet. Metab. 2006, 88, 78–85. [Google Scholar] [CrossRef]
- Galkin, V.E.; Orlova, A.; Egelman, E.H. Actin Filaments as Tension Sensors. Curr. Biol. 2012, 22, R96–R101. [Google Scholar] [CrossRef]
- Krüger, M.; Kötter, S. Titin, a Central Mediator for Hypertrophic Signaling, Exercise-Induced Mechanosignaling and Skeletal Muscle Remodeling. Front. Physiol. 2016, 7, 76. [Google Scholar] [CrossRef]
- Peter, A.K.; Cheng, H.; Ross, R.S.; Knowlton, K.U.; Chen, J. The Costamere Bridges Sarcomeres to the Sarcolemma in Striated Muscle. Prog. Pediatr. Cardiol. 2011, 31, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Qadota, H.; Benian, G.M. Molecular Structure of Sarcomere-to-Membrane Attachment at M-Lines in C. Elegans Muscle. J. Biomed. Biotechnol. 2010, 2010, 864749. [Google Scholar] [CrossRef] [PubMed]
- Gokhin, D.S.; Fowler, V.M. Tropomodulin Capping of Actin Filaments in Striated Muscle Development and Physiology. J. Biomed. Biotechnol. 2011, 2011, 103069. [Google Scholar] [CrossRef] [PubMed]
- Guilluy, C.; Burridge, K. Nuclear Mechanotransduction: Forcing the Nucleus to Respond. Nucleus 2015, 6, 19–22. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, M.L.; Lammerding, J. Keeping the LINC: The Importance of Nucleo-Cytoskeletal Coupling in Intracellular Force Transmission and Cellular Function. Biochem. Soc. Trans. 2011, 39, 1729–1734. [Google Scholar] [CrossRef]
- Fedorchak, G.R.; Kaminski, A.; Lammerding, J. Cellular Mechanosensing: Getting to the Nucleus of It All. Prog. Biophys. Mol. Biol. 2014, 115, 76–92. [Google Scholar] [CrossRef]
- Radke, M.H.; Polack, C.; Methawasin, M.; Fink, C.; Granzier, H.L.; Gotthardt, M. Deleting Full Length Titin versus the Titin M-Band Region Leads to Differential Mechanosignaling and Cardiac Phenotypes. Circulation 2019, 139, 1813. [Google Scholar] [CrossRef]
- Bershadsky, A.D.; Balaban, N.Q.; Geiger, B. Adhesion-Dependent Cell Mechanosensitivity. Annu. Rev. Cell Dev. Biol. 2003, 19, 677–695. [Google Scholar] [CrossRef]
- Jaalouk, D.E.; Lammerding, J. Mechanotransduction Gone Awry. Nat. Rev. Mol. Cell Biol. 2009, 10, 63–73. [Google Scholar] [CrossRef]
- Kolahi, K.S.; Mofrad, M.R.K. Mechanotransduction: A Major Regulator of Homeostasis and Development. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Pathological Mechanosensing. Nat. Biomed. Eng. 2021, 5, 1405–1406. [CrossRef] [PubMed]
- Wang, N.; Butler, J.P.; Ingber, D.E. Mechanotransduction across the Cell Surface and through the Cytoskeleton. Science 1993, 260, 1124–1127. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Sheetz, M. Local Force and Geometry Sensing Regulate Cell Functions. Nat. Rev. Mol. Cell Biol. 2006, 7, 265–275. [Google Scholar] [CrossRef]
- Koenderink, G.H.; Dogic, Z.; Nakamura, F.; Bendix, P.M.; MacKintosh, F.C.; Hartwig, J.H.; Stossel, T.P.; Weitz, D.A. An Active Biopolymer Network Controlled by Molecular Motors. Proc. Natl. Acad. Sci. USA 2009, 106, 15192–15197. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Kovar, D.R.; Gardel, M.L.; Winkelman, J.D. LIM Domain Proteins in Cell Mechanobiology. Cytoskeleton 2021, 78, 303–311. [Google Scholar] [CrossRef]
- Cassandri, M.; Smirnov, A.; Novelli, F.; Pitolli, C.; Agostini, M.; Malewicz, M.; Melino, G.; Raschellà, G. Zinc-Finger Proteins in Health and Disease. Cell Death Discov. 2017, 3, 17071. [Google Scholar] [CrossRef]
- Barash, I.A.; Mathew, L.; Lahey, M.; Greaser, M.L.; Lieber, R.L. Muscle LIM Protein Plays Both Structural and Functional Roles in Skeletal Muscle. Am. J. Physiol. Cell Physiol. 2005, 289, C1312–C1320. [Google Scholar] [CrossRef]
- Hoffmann, C.; Moreau, F.; Moes, M.; Luthold, C.; Dieterle, M.; Goretti, E.; Neumann, K.; Steinmetz, A.; Thomas, C. Human Muscle LIM Protein Dimerizes along the Actin Cytoskeleton and Cross-Links Actin Filaments. Mol. Cell. Biol. 2014, 34, 3053–3065. [Google Scholar] [CrossRef]
- Arber, S.; Halder, G.; Caroni, P. Muscle LIM Protein, a Novel Essential Regulator of Myogenesis, Promotes Myogenic Differentiation. Cell 1992, 79, 221–231. [Google Scholar] [CrossRef]
- Arber, S.; Hunter, J.J.; Ross, J.; Hongo, M.; Sansig, G.; Borg, J.; Perriard, J.C.; Chien, K.R.; Caroni, P. MLP-Deficient Mice Exhibit a Disruption of Cardiac Cytoarchitectural Organization, Dilated Cardiomyopathy, and Heart Failure. Cell 1997, 88, 393–403. [Google Scholar] [CrossRef]
- Wang, X.; Li, Q.; Adhikari, N.; Hall, J.L. A Role for Muscle LIM Protein (MLP) in Vascular Remodeling. J. Mol. Cell. Cardiol. 2006, 40, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Levin, E.; Leibinger, M.; Andreadaki, A.; Fischer, D. Neuronal Expression of Muscle LIM Protein in Postnatal Retinae of Rodents. PLoS ONE 2014, 9, e100756. [Google Scholar] [CrossRef]
- Levin, E.; Andreadaki, A.; Gobrecht, P.; Bosse, F.; Fischer, D. Nociceptive DRG Neurons Express Muscle Lim Protein upon Axonal Injury. Sci. Rep. 2017, 7, 643. [Google Scholar] [CrossRef] [PubMed]
- Gunkel, S.; Heineke, J.; Hilfiker-Kleiner, D.; Knöll, R. MLP: A Stress Sensor Goes Nuclear. J. Mol. Cell. Cardiol. 2009, 47, 423–425. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Papalouka, V.; Terzis, G.; Roumeliotis, T.I.; Spengos, K.; Garbis, S.D.; Manta, P.; Kranias, E.G.; Sanoudou, D. Muscle Lim Protein Isoform Negatively Regulates Striated Muscle Actin Dynamics and Differentiation. FEBS J. 2014, 281, 3261–3279. [Google Scholar] [CrossRef]
- Wilding, J.R.; Schneider, J.E.; Sang, A.E.; Davies, K.E.; Neubauer, S.; Clarke, K. Dystrophin- and MLP-Deficient Mouse Hearts: Marked Differences in Morphology and Function, but Similar Accumulation of Cytoskeletal Proteins. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2005, 19, 79–81. [Google Scholar] [CrossRef]
- Pasqualini, F.S.; Nesmith, A.P.; Horton, R.E.; Sheehy, S.P.; Parker, K.K. Mechanotransduction and Metabolism in Cardiomyocyte Microdomains. BioMed Res. Int. 2016, 2016, 4081638. [Google Scholar] [CrossRef]
- Vafiadaki, E.; Arvanitis, D.A.; Sanoudou, D. Muscle LIM Protein: Master Regulator of Cardiac and Skeletal Muscle Functions. Gene 2015, 566, 1–7. [Google Scholar] [CrossRef]
- Boateng, S.Y.; Belin, R.J.; Geenen, D.L.; Margulies, K.B.; Martin, J.L.; Hoshijima, M.; de Tombe, P.P.; Russell, B. Cardiac Dysfunction and Heart Failure Are Associated with Abnormalities in the Subcellular Distribution and Amounts of Oligomeric Muscle LIM Protein. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H259–H269. [Google Scholar] [CrossRef]
- Buyandelger, B.; Mansfield, C.; Knöll, R. Mechano-Signaling in Heart Failure. Pflugers Arch. 2014, 466, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Paudyal, A.; Dewan, S.; Ikie, C.; Whalley, B.J.; de Tombe, P.P.; Boateng, S.Y. Nuclear Accumulation of Myocyte Muscle LIM Protein Is Regulated by Heme Oxygenase 1 and Correlates with Cardiac Function in the Transition to Failure. J. Physiol. 2016, 592, 3287–3305. [Google Scholar] [CrossRef] [PubMed]
- Koskinen, S.O.A.; Kyröläinen, H.; Flink, R.; Selänne, H.P.; Gagnon, S.S.; Ahtiainen, J.P.; Nindl, B.C.; Lehti, M. Human Skeletal Muscle Type 1 Fibre Distribution and Response of Stress-Sensing Proteins along the Titin Molecule after Submaximal Exhaustive Exercise. Histochem. Cell Biol. 2017, 148, 545–555. [Google Scholar] [CrossRef]
- Blom, N.; Gammeltoft, S.; Brunak, S. Sequence and Structure-Based Prediction of Eukaryotic Protein Phosphorylation Sites. J. Mol. Biol. 1999, 292, 1351–1362. [Google Scholar] [CrossRef]
- Available online: https://www.phosphosite.org/proteinAction.action?id=18990&showAllSites=true (accessed on 1 December 2021).
- Postel, R.; Vakeel, P.; Topczewski, J.; Knöll, R.; Bakkers, J. Zebrafish Integrin-Linked Kinase Is Required in Skeletal Muscles for Strengthening the Integrin–ECM Adhesion Complex. Dev. Biol. 2008, 318, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Gorza, L.; Sorge, M.; Seclì, L.; Brancaccio, M. Master Regulators of Muscle Atrophy: Role of Costamere Components. Cells 2021, 10, 61. [Google Scholar] [CrossRef]
- Riaz, M.; Park, J.; Sewanan, L.R.; Ren, Y.; Schwan, J.; Das, S.K.; Pomianowski, P.T.; Huang, Y.; Ellis, M.W.; Luo, J.; et al. Muscle LIM Protein Force-Sensing Mediates Sarcomeric Biomechanical Signaling in Human Familial Hypertrophic Cardiomyopathy. Circulation 2022, 145, 1238–1253. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Tian, L.; Zhao, Y.; Yan, Y.; Li, S.; Li, S.; Tong, H. Vitamin C Promotes Muscle Development Mediated by the Interaction of CSRP3 with MyoD and MyoG. J. Agric. Food Chem. 2022, 70, 7158–7169. [Google Scholar] [CrossRef]
- Kong, Y.; Flick, M.J.; Kudla, A.J.; Konieczny, S.F. Muscle LIM Protein Promotes Myogenesis by Enhancing the Activity of MyoD. Mol. Cell. Biol. 1997, 17, 4750–4760. [Google Scholar] [CrossRef]
- Comai, G.; Tajbakhsh, S. Molecular and Cellular Regulation of Skeletal Myogenesis. Curr. Top. Dev. Biol. 2014, 110, 1–73. [Google Scholar] [CrossRef]
- Conerly, M.L.; Yao, Z.; Zhong, J.W.; Groudine, M.; Tapscott, S.J. Distinct Activities of Myf5 and MyoD Indicate Separate Roles in Skeletal Muscle Lineage Specification and Differentiation. Dev. Cell 2016, 36, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Rigby, P.W.J. Gene Regulatory Networks and Transcriptional Mechanisms That Control Myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef]
- Hinterberger, T.J.; Sassoon, D.A.; Rhodes, S.J.; Konieczny, S.F. Expression of the Muscle Regulatory Factor MRF4 during Somite and Skeletal Myofiber Development. Dev. Biol. 1991, 147, 144–156. [Google Scholar] [CrossRef]
- Rudnicki, M.A.; Jaenisch, R. The MyoD Family of Transcription Factors and Skeletal Myogenesis. BioEssays News Rev. Mol. Cell. Dev. Biol. 1995, 17, 203–209. [Google Scholar] [CrossRef]
- Olson, E.N. MyoD Family: A Paradigm for Development? Genes Dev. 1990, 4, 1454–1461. [Google Scholar] [CrossRef] [PubMed]
- Ontell, M.; Ontell, M.P.; Sopper, M.M.; Mallonga, R.; Lyons, G.; Buckingham, M. Contractile Protein Gene Expression in Primary Myotubes of Embryonic Mouse Hindlimb Muscles. Dev. Camb. Engl. 1993, 117, 1435–1444. [Google Scholar]
- Groisman, R.; Masutani, H.; Leibovitch, M.P.; Robin, P.; Soudant, I.; Trouche, D.; Harel-Bellan, A. Physical Interaction between the Mitogen-Responsive Serum Response Factor and Myogenic Basic-Helix-Loop-Helix Proteins. J. Biol. Chem. 1996, 271, 5258–5264. [Google Scholar] [CrossRef]
- Cao, Y.; Yao, Z.; Sarkar, D.; Lawrence, M.; Sanchez, G.J.; Parker, M.H.; MacQuarrie, K.L.; Davison, J.; Morgan, M.T.; Ruzzo, W.L.; et al. Genome-Wide MyoD Binding in Skeletal Muscle Cells: A Potential for Broad Cellular Reprogramming. Dev. Cell 2010, 18, 662–674. [Google Scholar] [CrossRef]
- Olson, E.N.; Rosenthal, N. Homeobox Genes and Muscle Patterning. Cell 1992, 79, 9–12. [Google Scholar] [CrossRef]
- Weintraub, H. The MyoD Family and Myogenesis: Redundancy, Networks, and Thresholds. Cell 1993, 75, 1241–1244. [Google Scholar] [CrossRef]
- Shintaku, J.; Peterson, J.M.; Talbert, E.E.; Gu, J.-M.; Ladner, K.J.; Williams, D.R.; Mousavi, K.; Wang, R.; Sartorelli, V.; Guttridge, D.C. MyoD Regulates Skeletal Muscle Oxidative Metabolism Cooperatively with Alternative NF-κ B. Cell Rep. 2016, 17, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Ward, M.; Iskratsch, T. Mix and (Mis-)Match—The Mechanosensing Machinery in the Changing Environment of the Developing, Healthy Adult and Diseased Heart. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gao, Y. The Role of Transmembrane Proteins on Force Transmission in Skeletal Muscle. J. Biomech. 2014, 47, 3232–3236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Virgilio, K.M.; Martin, K.S.; Peirce, S.M.; Blemker, S.S. Multiscale Models of Skeletal Muscle Reveal the Complex Effects of Muscular Dystrophy on Tissue Mechanics and Damage Susceptibility. Interface Focus 2015, 5, 20140080. [Google Scholar] [CrossRef] [PubMed]
- Winkelman, J.D.; Anderson, C.A.; Suarez, C.; Kovar, D.R.; Gardel, M.L. Evolutionarily Diverse LIM Domain-Containing Proteins Bind Stressed Actin Filaments through a Conserved Mechanism. Proc. Natl. Acad. Sci. USA 2020, 117, 25532–25542. [Google Scholar] [CrossRef]
- Ehler, E.; Horowits, R.; Zuppinger, C.; Price, R.L.; Perriard, E.; Leu, M.; Caroni, P.; Sussman, M.; Eppenberger, H.M.; Perriard, J.C. Alterations at the Intercalated Disk Associated with the Absence of Muscle LIM Protein. J. Cell Biol. 2001, 153, 763–772. [Google Scholar] [PubMed]
- Nayak, A.; Amrute-Nayak, M. SUMO System—A Key Regulator in Sarcomere Organization. FEBS J. 2020, 287, 2176–2190. [Google Scholar] [CrossRef]
- Gupta, M.P.; Samant, S.A.; Smith, S.H.; Shroff, S.G. HDAC4 and PCAF Bind to Cardiac Sarcomeres and Play a Role in Regulating Myofilament Contractile Activity. J. Biol. Chem. 2008, 283, 10135–10146. [Google Scholar] [CrossRef]
- VanHecke, G.C.; Abeywardana, M.Y.; Ahn, Y.-H. Proteomic Identification of Protein Glutathionylation in Cardiomyocytes. J. Proteome Res. 2019, 18, 1806–1818. [Google Scholar] [CrossRef]
- Han, S.; Cui, C.; Wang, Y.; He, H.; Liu, Z.; Shen, X.; Chen, Y.; Li, D.; Zhu, Q.; Yin, H. Knockdown of CSRP3 Inhibits Differentiation of Chicken Satellite Cells by Promoting TGF-β/Smad3 Signaling. Gene 2019, 707, 36–43. [Google Scholar] [CrossRef]
- Chang, Y.; Geng, F.; Hu, Y.; Ding, Y.; Zhang, R. Zebrafish Cysteine and Glycine-Rich Protein 3 Is Essential for Mechanical Stability in Skeletal Muscles. Biochem. Biophys. Res. Commun. 2019, 511, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Offerhaus, J.A.; Tadros, R.; Bezzina, C.R. Minor Hypertrophic Cardiomyopathy Genes, Major Insights into the Genetics of Cardiomyopathies. Nat. Rev. Cardiol. 2022, 19, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.K.; Sowdhamini, R. LIM Domain-Wide Comprehensive Virtual Mutagenesis Provides Structural Rationale for Cardiomyopathy Mutations in CSRP3. Sci. Rep. 2022, 12, 3562. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Chen, Y.; Jin, J.; Du, R.; Tang, K.; Fan, L.; Xiang, R. CSRP3, p.Arg122*, Is Responsible for Hypertrophic Cardiomyopathy in a Chinese Family. J. Gene Med. 2022, 24, e3390. [Google Scholar] [CrossRef]
- Sun, L.; Li, J.; Li, E.; Niu, S.; Qin, Z.; Zhi, Q.; Zhao, J.; Xiong, H.; Li, Y.; Jian, L.; et al. CRISPR/Cas9 Mediated Establishment of a Human CSRP3 Compound Heterozygous Knockout HESC Line to Model Cardiomyopathy and Heart Failure. Stem Cell Res. 2020, 49, 102077. [Google Scholar] [CrossRef]
- Gehmlich, K.; Geier, C.; Milting, H.; Fürst, D.; Ehler, E. Back to Square One: What Do We Know about the Functions of Muscle LIM Protein in the Heart? J. Muscle Res. Cell Motil. 2008, 29, 155–158. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Germain, P.; Delalande, A.; Pichon, C. Role of Muscle LIM Protein in Mechanotransduction Process. Int. J. Mol. Sci. 2022, 23, 9785. https://doi.org/10.3390/ijms23179785
Germain P, Delalande A, Pichon C. Role of Muscle LIM Protein in Mechanotransduction Process. International Journal of Molecular Sciences. 2022; 23(17):9785. https://doi.org/10.3390/ijms23179785
Chicago/Turabian StyleGermain, Philippe, Anthony Delalande, and Chantal Pichon. 2022. "Role of Muscle LIM Protein in Mechanotransduction Process" International Journal of Molecular Sciences 23, no. 17: 9785. https://doi.org/10.3390/ijms23179785
APA StyleGermain, P., Delalande, A., & Pichon, C. (2022). Role of Muscle LIM Protein in Mechanotransduction Process. International Journal of Molecular Sciences, 23(17), 9785. https://doi.org/10.3390/ijms23179785






