Comprehensive Analysis of Subcellular Localization, Immune Function and Role in Bacterial wilt Disease Resistance of Solanum lycopersicum Linn. ROP Family Small GTPases
Abstract
:1. Introduction
2. Results
2.1. Identification and Phylogenetic Analysis of SlRops
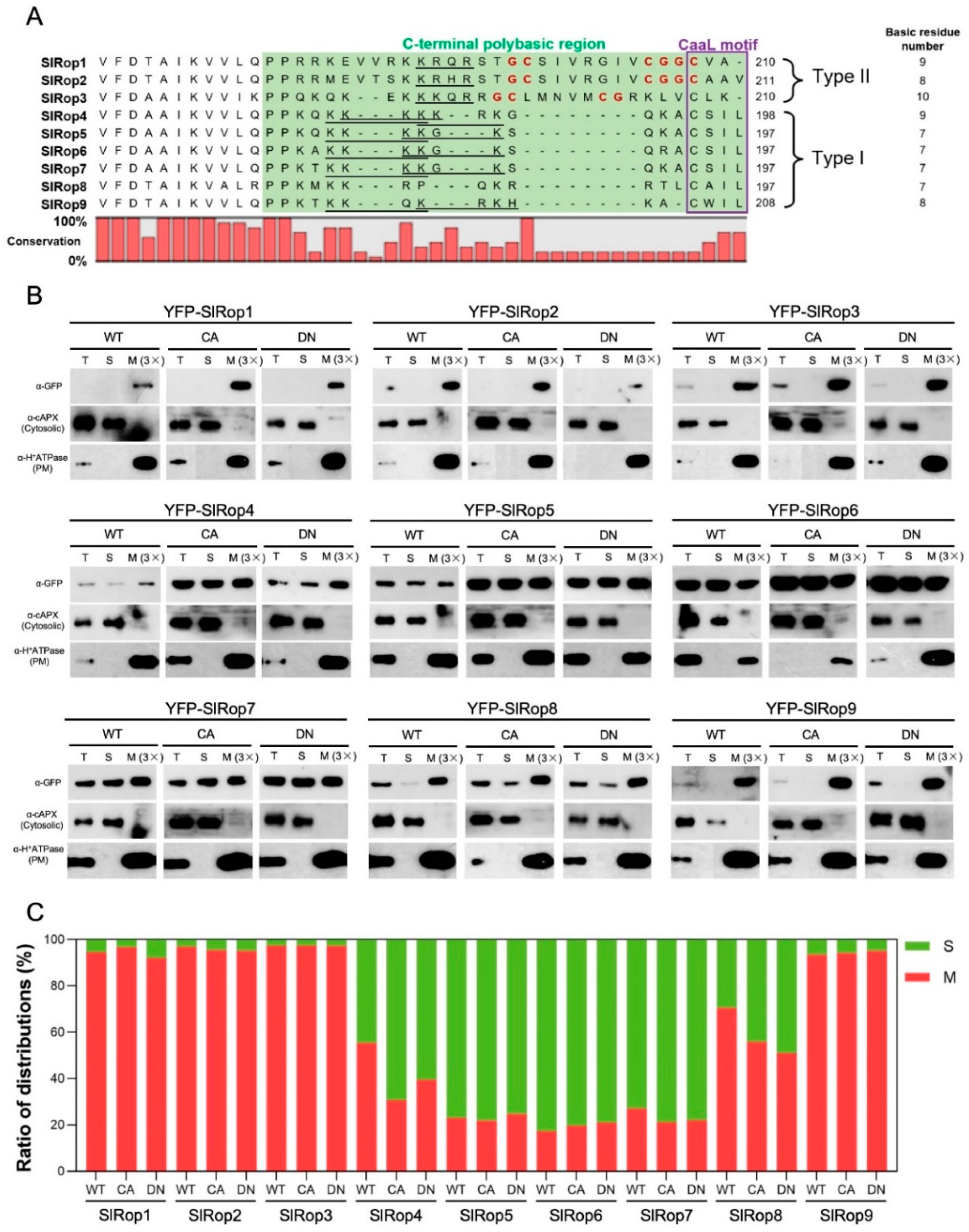
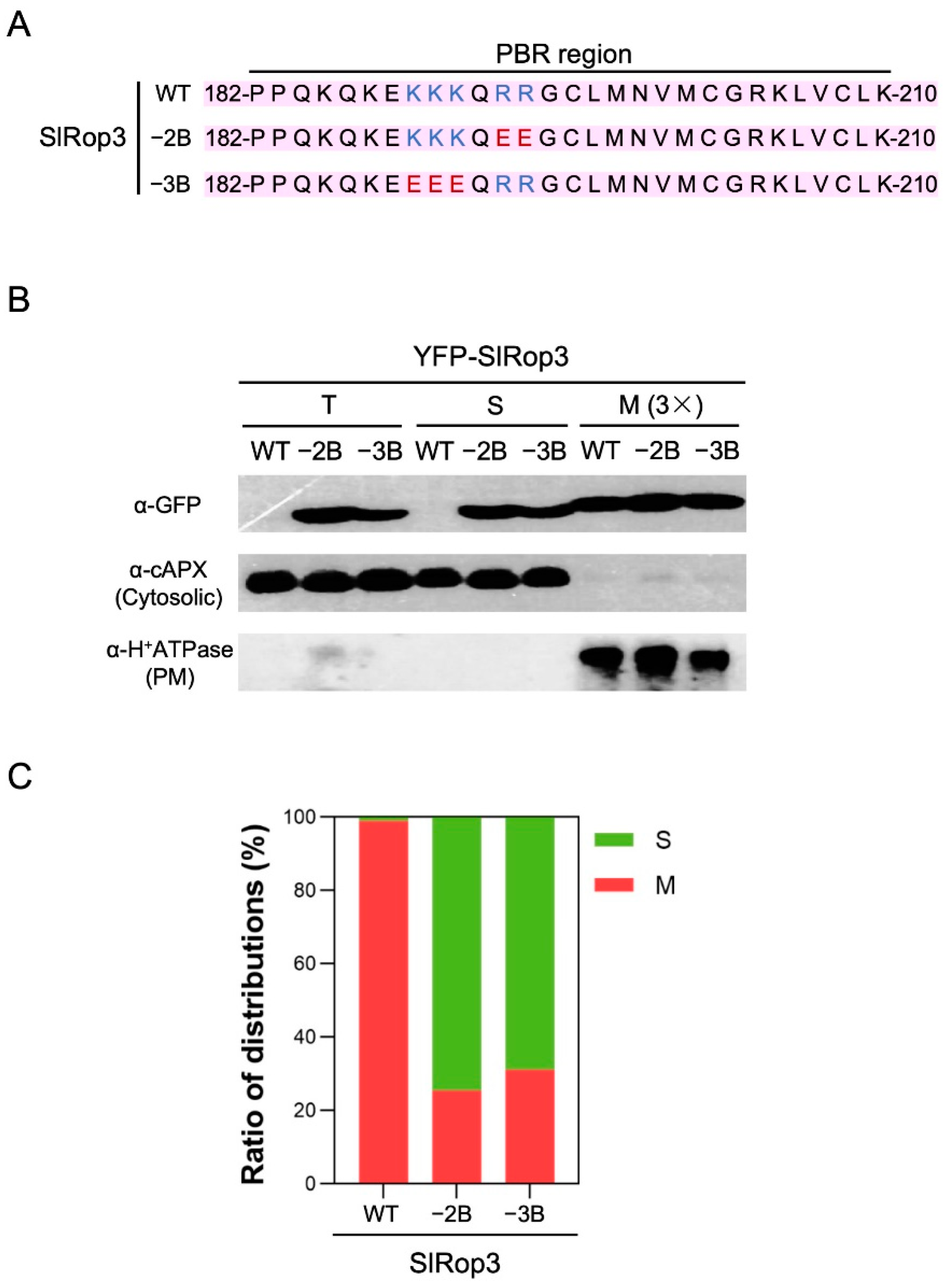
2.2. Relationship between the Polybasic Region and Subcellular Localization of SlRops
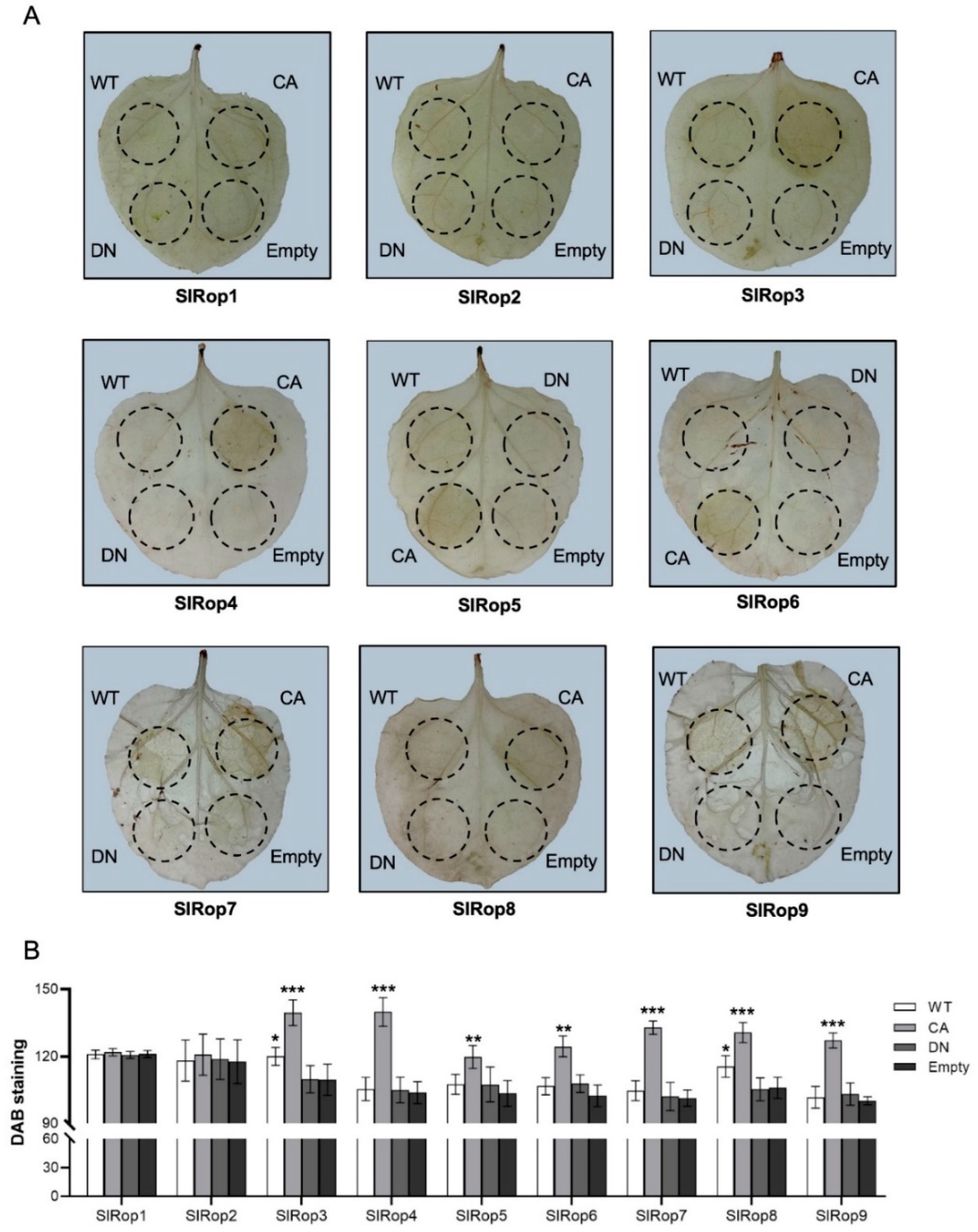
2.3. Involvement of SlRops in Hypersensitive Response (HR) Induction
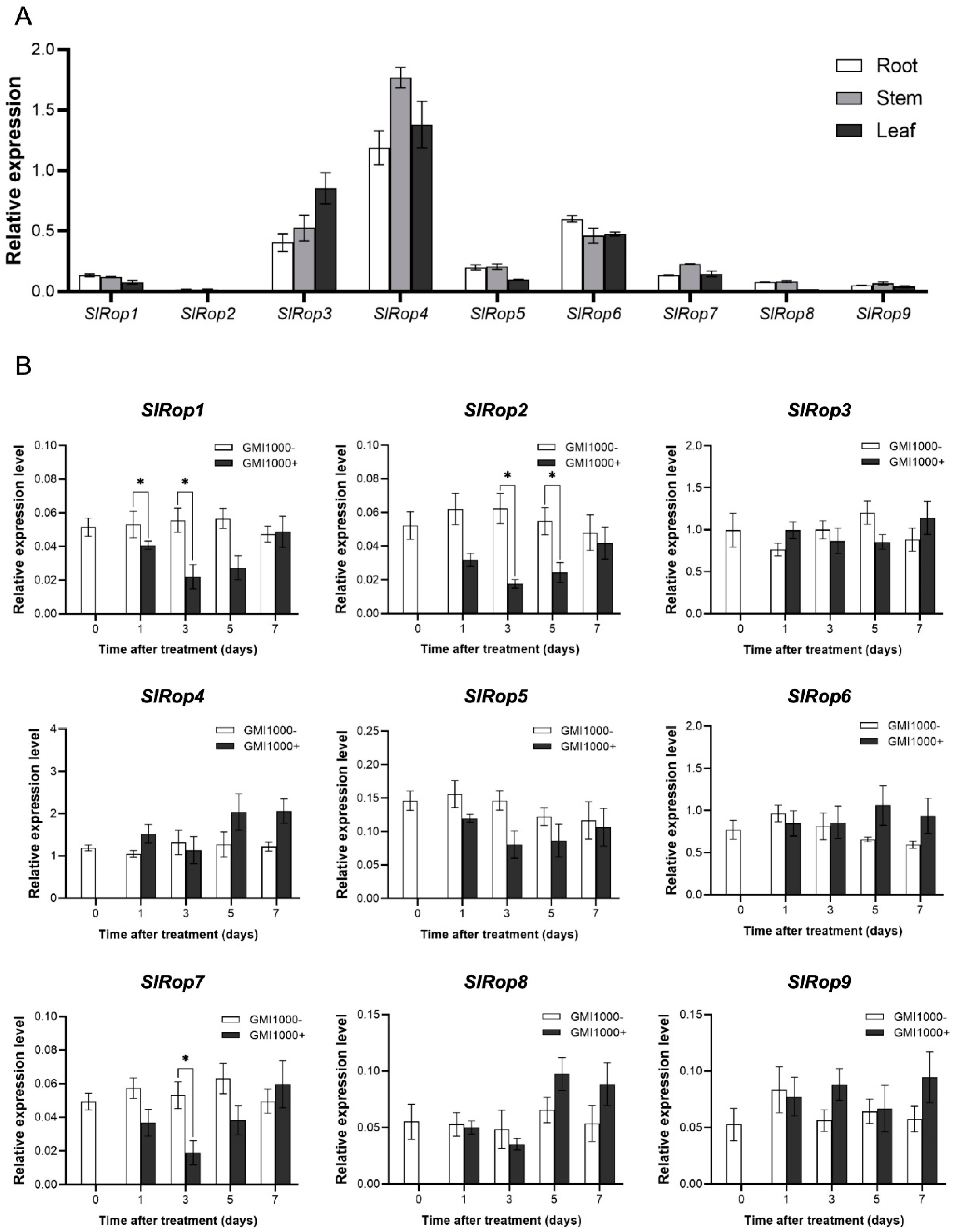
2.4. Tissue-Specific Expression Patterns of SlRops
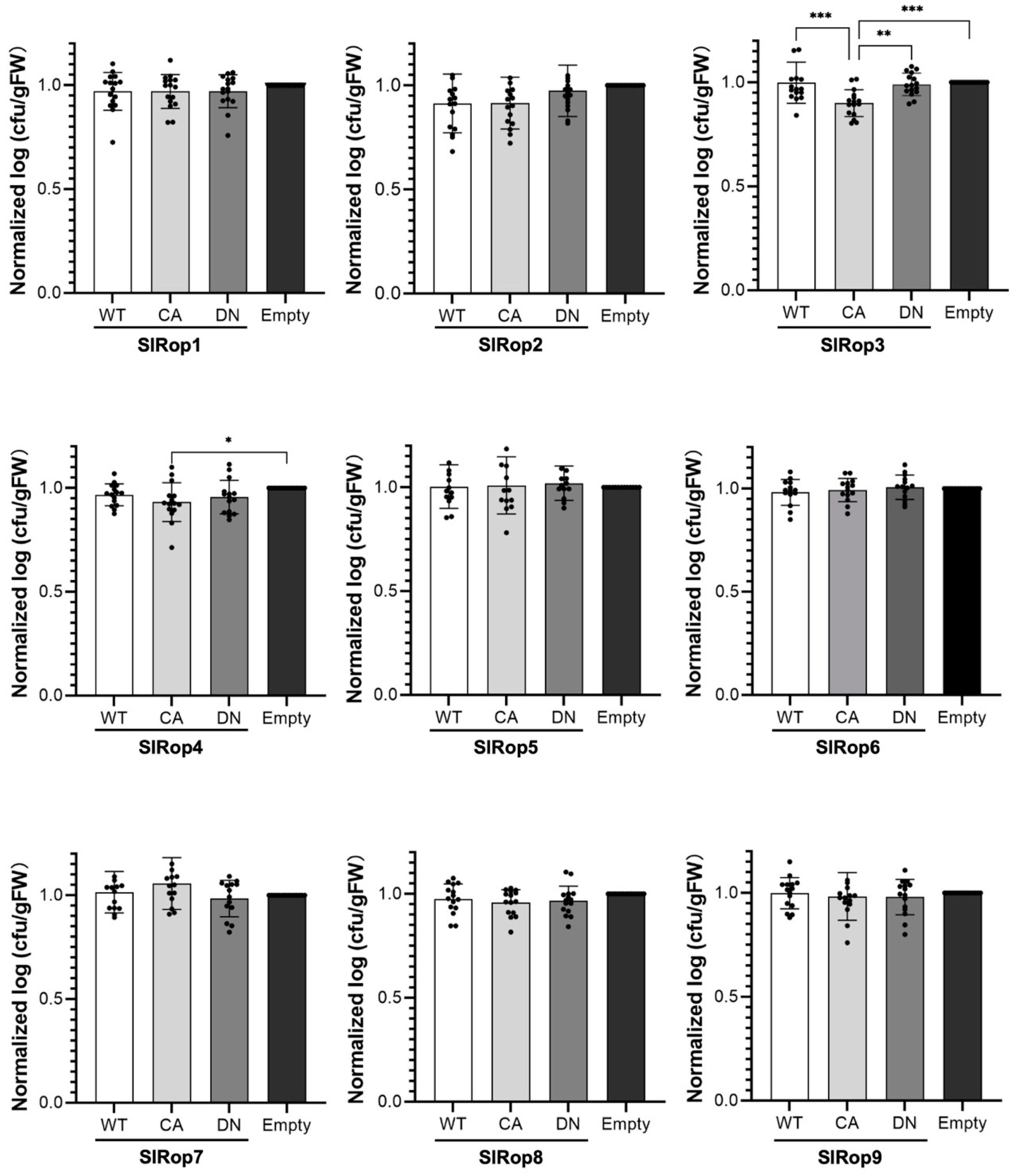
2.5. Roles of SlRops in Disease Resistance to Tomato Bacterial Wilt
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth
4.2. RNA Isolation and qRT-PCR
4.3. Plasmid Construction
4.4. Transient Expression in N. benthamiana
4.5. Subcellular Localization
4.6. Cell Fractionation and Quantification
4.7. HR Detection and Quantification
4.8. R. solanacearum Strains, Growth, and Infection
4.9. Western Blotting
4.10. Sequence Alignment and Phylogenetic Analysis
4.11. Analysis of Chromosomal Mapping, Conserved Motif, Gene Structure, and Collinearity
4.12. Statistical Analysis
4.13. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Xue, H.; Lozano-Duran, R.; Macho, A.P. Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum. Plants 2020, 9, 516. [Google Scholar] [CrossRef] [PubMed]
- Lowe-Power, T.M.; Khokhani, D.; Allen, C. How Ralstonia solanacearum Exploits and Thrives in the Flowing Plant Xylem Environment. Trends Microbiol. 2018, 26, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Genin, S. Molecular traits controlling host range and adaptation to plants in Ralstonia solanacearum. New Phytol. 2010, 187, 920–928. [Google Scholar] [CrossRef]
- Li, N.; Wu, X.; Zhuang, W.; Xia, L.; Chen, Y.; Wu, C.; Rao, Z.; Du, L.; Zhao, R.; Yi, M.; et al. Tomato and lycopene and multiple health outcomes: Umbrella review. Food Chem. 2021, 343, 128396. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, E. The Small GTPase Superfamily in Plants: A Conserved Regulatory Module with Novel Functions. Annu. Rev. Plant Biol. 2020, 71, 247–272. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef]
- Vernoud, V.; Horton, A.C.; Yang, Z.; Nielsen, E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003, 131, 1191–1208. [Google Scholar] [CrossRef]
- Craddock, C.; Lavagi, I.; Yang, Z. New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 2012, 22, 492–501. [Google Scholar] [CrossRef]
- Kawano, Y.; Kaneko-Kawano, T.; Shimamoto, K. Rho family GTPase-dependent immunity in plants and animals. Front. Plant Sci. 2014, 5, 522. [Google Scholar] [CrossRef] [Green Version]
- Engelhardt, S.; Trutzenberg, A.; Huckelhoven, R. Regulation and Functions of ROP GTPases in Plant-Microbe Interactions. Cells 2020, 9, 2016. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, A.; Wong, H.L.; Fujiwara, M.; Okuda, J.; Nishide, K.; Uno, K.; Imai, K.; Umemura, K.; Kawasaki, T.; Kawano, Y.; et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 2013, 13, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, A.; Fujiwara, M.; Hamada, S.; Wakabayashi, M.; Yao, A.; Wang, Q.; Kosami, K.I.; Dang, T.T.; Kaneko-Kawano, T.; Fukada, F.; et al. The Small GTPase OsRac1 Forms Two Distinct Immune Receptor Complexes Containing the PRR OsCERK1 and the NLR Pit. Plant Cell Physiol. 2021, 62, 1662–1675. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Akamatsu, A.; Hayashi, K.; Housen, Y.; Okuda, J.; Yao, A.; Nakashima, A.; Takahashi, H.; Yoshida, H.; Wong, H.L.; et al. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 2010, 7, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shiotani, K.; Togashi, T.; Miki, D.; Aoyama, M.; Wong, H.L.; Kawasaki, T.; Shimamoto, K. Analysis of the Rac/Rop small GTPase family in rice: Expression, subcellular localization and role in disease resistance. Plant Cell Physiol. 2010, 51, 585–595. [Google Scholar] [CrossRef]
- Zhou, Z.; Pang, Z.; Zhao, S.; Zhang, L.; Lv, Q.; Yin, D.; Li, D.; Liu, X.; Zhao, X.; Li, X.; et al. Importance of OsRac1 and RAI1 in signalling of nucleotide-binding site leucine-rich repeat protein-mediated resistance to rice blast disease. New Phytol. 2019, 223, 828–838. [Google Scholar] [CrossRef]
- Kawano, Y.; Shimamoto, K. Early signaling network in rice PRR-mediated and R-mediated immunity. Curr. Opin. Plant Biol. 2013, 16, 496–504. [Google Scholar] [CrossRef]
- Wong, H.L.; Sakamoto, T.; Kawasaki, T.; Umemura, K.; Shimamoto, K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004, 135, 1447–1456. [Google Scholar] [CrossRef]
- Wong, H.L.; Pinontoan, R.; Hayashi, K.; Tabata, R.; Yaeno, T.; Hasegawa, K.; Kojima, C.; Yoshioka, H.; Iba, K.; Kawasaki, T.; et al. Regulation of rice NADPH oxidase by binding of Rac GTPase to its N-terminal extension. Plant Cell 2007, 19, 4022–4034. [Google Scholar] [CrossRef]
- Lieberherr, D.; Thao, N.P.; Nakashima, A.; Umemura, K.; Kawasaki, T.; Shimamoto, K. A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice 1 [w]. Plant Physiol. 2005, 138, 1644–1652. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Oikawa, T.; Kyozuka, J.; Wong, H.L.; Umemura, K.; Kishi-Kaboshi, M.; Takahashi, A.; Kawano, Y.; Kawasaki, T.; Shimamoto, K. The bHLH Rac Immunity1 (RAI1) Is Activated by OsRac1 via OsMAPK3 and OsMAPK6 in Rice Immunity. Plant Cell Physiol. 2012, 53, 740–754. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Henmi, K.; Ono, E.; Hatakeyama, S.; Iwano, M.; Satoh, H.; Shimamoto, K. The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 10922–10926. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Koita, H.; Nakatsubo, T.; Hasegawa, K.; Wakabayashi, K.; Takahashi, H.; Umemura, K.; Umezawa, T.; Shimamoto, K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Thao, N.P.; Chen, L.; Nakashima, A.; Hara, S.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell 2007, 19, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Y.; Ishikawa, K.; Kosami, K.I.; Uno, K.; Nagawa, S.; Tan, L.; Du, J.; Shimamoto, K.; Kawano, Y. Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. USA 2018, 115, E11551–E11560. [Google Scholar] [CrossRef]
- Yu, M.; Zhou, Z.; Liu, X.; Yin, D.; Li, D.; Zhao, X.; Li, X.; Li, S.; Chen, R.; Lu, L.; et al. The OsSPK1-OsRac1-RAI1 defense signaling pathway is shared by two distantly related NLR proteins in rice blast resistance. Plant Physiol. 2021, 187, 2852–2864. [Google Scholar] [CrossRef]
- Moeder, W.; Yoshioka, K.; Klessig, D.F. Involvement of the small GTPase Rac in the defense responses of tobacco to pathogens. Mol. Plant Microbe Interact. 2005, 18, 116–124. [Google Scholar] [CrossRef]
- Hassanain, H.H.; Sharma, Y.K.; Moldovan, L.; Khramtsov, V.; Berliner, L.J.; Duvick, J.P.; Goldschmidt-Clermont, P.J. Plant rac proteins induce superoxide production in mammalian cells. Biochem. Biophys. Res. Commun. 2000, 272, 783–788. [Google Scholar] [CrossRef]
- Yang, S.; Yan, N.; Bouwmeester, K.; Na, R.; Zhang, Z.; Zhao, J. Genome-wide identification of small G protein ROPs and their potential roles in Solanaceous family. Gene 2020, 753, 144809. [Google Scholar] [CrossRef]
- Schultheiss, H.; Dechert, C.; Kogel, K.H.; Huckelhoven, R. Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J. 2003, 36, 589–601. [Google Scholar] [CrossRef]
- Schultheiss, H.; Dechert, C.; Kogel, K.H.; Huckelhoven, R. A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol. 2002, 128, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yang, F.; Na, R.; Zhang, X.; Yang, S.; Gao, J.; Fan, M.; Zhao, Y.; Zhao, J. AtROP1 negatively regulates potato resistance to Phytophthora infestans via NADPH oxidase-mediated accumulation of H2O2. BMC Plant Biol. 2014, 14, 392. [Google Scholar] [CrossRef] [PubMed]
- Qiu, A.; Liu, Z.; Li, J.; Chen, Y.; Guan, D.; He, S. The Ectopic Expression of CaRop1 Modulates the Response of Tobacco Plants to Ralstonia solanacearum and Aphids. Front. Plant Sci. 2016, 7, 1177. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Wang, J.; Gao, H.; Yang, Q.; Wang, Y.; Day, B.; Ma, Q. The small GTP-binding protein TaRop10 interacts with TaTrxh9 and functions as a negative regulator of wheat resistance against the stripe rust. Plant Sci. 2021, 309, 110937. [Google Scholar] [CrossRef]
- Han, H.; Zou, J.; Zhou, J.; Zeng, M.; Zheng, D.; Yuan, X.; Xi, D. The small GTPase NtRHO1 negatively regulates tobacco defense response to tobacco mosaic virus by interacting with NtWRKY50. J. Exp. Bot. 2022, 73, 366–381. [Google Scholar] [CrossRef]
- Smokvarska, M.; Jaillais, Y.; Martiniere, A. Function of membrane domains in rho-of-plant signaling. Plant Physiol. 2021, 185, 663–681. [Google Scholar] [CrossRef]
- Lavy, M.; Yalovsky, S. Association of Arabidopsis type-II ROPs with the plasma membrane requires a conserved C-terminal sequence motif and a proximal polybasic domain. Plant J. 2006, 46, 934–947. [Google Scholar] [CrossRef]
- Sorek, N.; Henis, Y.I.; Yalovsky, S. How prenylation and S-acylation regulate subcellular targeting and function of ROP GTPases. Plant Signal Behav. 2011, 6, 1026–1029. [Google Scholar] [CrossRef]
- Ono, E.; Wong, H.L.; Kawasaki, T.; Hasegawa, M.; Kodama, O.; Shimamoto, K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA 2001, 98, 759–764. [Google Scholar] [CrossRef]
- Poraty-Gavra, L.; Zimmermann, P.; Haigis, S.; Bednarek, P.; Hazak, O.; Stelmakh, O.R.; Sadot, E.; Schulze-Lefert, P.; Gruissem, W.; Yalovsky, S. The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiol. 2013, 161, 1172–1188. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.L. The polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal 2003, 15, 1071–1080. [Google Scholar] [CrossRef]
- Kadota, Y.; Shirasu, K.; Zipfel, C. Regulation of the NADPH Oxidase RBOHD During Plant Immunity. Plant Cell Physiol. 2015, 56, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Macho, A.P. A Fast and Easy Method to Study Ralstonia solanacearum Virulence upon Transient Gene Expression or Gene Silencing in Nicotiana benthamiana Leaves. Bio-Protocol 2021, 11, e4116. [Google Scholar] [CrossRef] [PubMed]
- Pathuri, I.P.; Zellerhoff, N.; Schaffrath, U.; Hensel, G.; Kumlehn, J.; Kogel, K.H.; Eichmann, R.; Huckelhoven, R. Constitutively activated barley ROPs modulate epidermal cell size, defense reactions and interactions with fungal leaf pathogens. Plant Cell Rep. 2008, 27, 1877–1887. [Google Scholar] [CrossRef]
- Ma, Q.H.; Zhu, H.H.; Han, J.Q. Wheat ROP proteins modulate defense response through lignin metabolism. Plant Sci. 2017, 262, 32–38. [Google Scholar] [CrossRef]
- Oda, T.; Hashimoto, H.; Kuwabara, N.; Akashi, S.; Hayashi, K.; Kojima, C.; Wong, H.L.; Kawasaki, T.; Shimamoto, K.; Sato, M.; et al. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 2010, 285, 1435–1445. [Google Scholar] [CrossRef]
- El Kasmi, F.; Chung, E.H.; Anderson, R.G.; Li, J.; Wan, L.; Eitas, T.K.; Gao, Z.; Dangl, J.L. Signaling from the plasma-membrane localized plant immune receptor RPM1 requires self-association of the full-length protein. Proc. Natl. Acad. Sci. USA 2017, 114, E7385–E7394. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Q.; Jing, M.; Guo, B.; Wu, J.; Wang, H.; Wang, Y.; Lin, L.; Wang, Y.; Ye, W.; et al. Distinct regions of the Phytophthora essential effector Avh238 determine its function in cell death activation and plant immunity suppression. New Phytol. 2017, 214, 361–375. [Google Scholar] [CrossRef]
- Salanoubat, M.; Genin, S.; Artiguenave, F.; Gouzy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J.C.; Cattolico, L.; et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar] [CrossRef]
- Singh, N.; Phukan, T.; Sharma, P.L.; Kabyashree, K.; Barman, A.; Kumar, R.; Sonti, R.V.; Genin, S.; Ray, S.K. An Innovative Root Inoculation Method to Study Ralstonia solanacearum Pathogenicity in Tomato Seedlings. Phytopathology 2018, 108, 436–442. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Nystrom, S.L.; McKay, D.J. Memes: A motif analysis environment in R using tools from the MEME Suite. PLoS Comput. Biol. 2021, 17, e1008991. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhang, D.; Liu, C.; Li, Y.; Miao, Y. Comprehensive Analysis of Subcellular Localization, Immune Function and Role in Bacterial wilt Disease Resistance of Solanum lycopersicum Linn. ROP Family Small GTPases. Int. J. Mol. Sci. 2022, 23, 9727. https://doi.org/10.3390/ijms23179727
Wang Q, Zhang D, Liu C, Li Y, Miao Y. Comprehensive Analysis of Subcellular Localization, Immune Function and Role in Bacterial wilt Disease Resistance of Solanum lycopersicum Linn. ROP Family Small GTPases. International Journal of Molecular Sciences. 2022; 23(17):9727. https://doi.org/10.3390/ijms23179727
Chicago/Turabian StyleWang, Qiong, Dan Zhang, Chaochao Liu, Yuying Li, and Yanni Miao. 2022. "Comprehensive Analysis of Subcellular Localization, Immune Function and Role in Bacterial wilt Disease Resistance of Solanum lycopersicum Linn. ROP Family Small GTPases" International Journal of Molecular Sciences 23, no. 17: 9727. https://doi.org/10.3390/ijms23179727
APA StyleWang, Q., Zhang, D., Liu, C., Li, Y., & Miao, Y. (2022). Comprehensive Analysis of Subcellular Localization, Immune Function and Role in Bacterial wilt Disease Resistance of Solanum lycopersicum Linn. ROP Family Small GTPases. International Journal of Molecular Sciences, 23(17), 9727. https://doi.org/10.3390/ijms23179727




