Effects of Two Distinct Psychoactive Microbes, Lacticaseibacillus rhamnosus JB-1 and Limosilactobacillus reuteri 6475, on Circulating and Hippocampal mRNA in Male Mice
Abstract
1. Introduction
2. Results
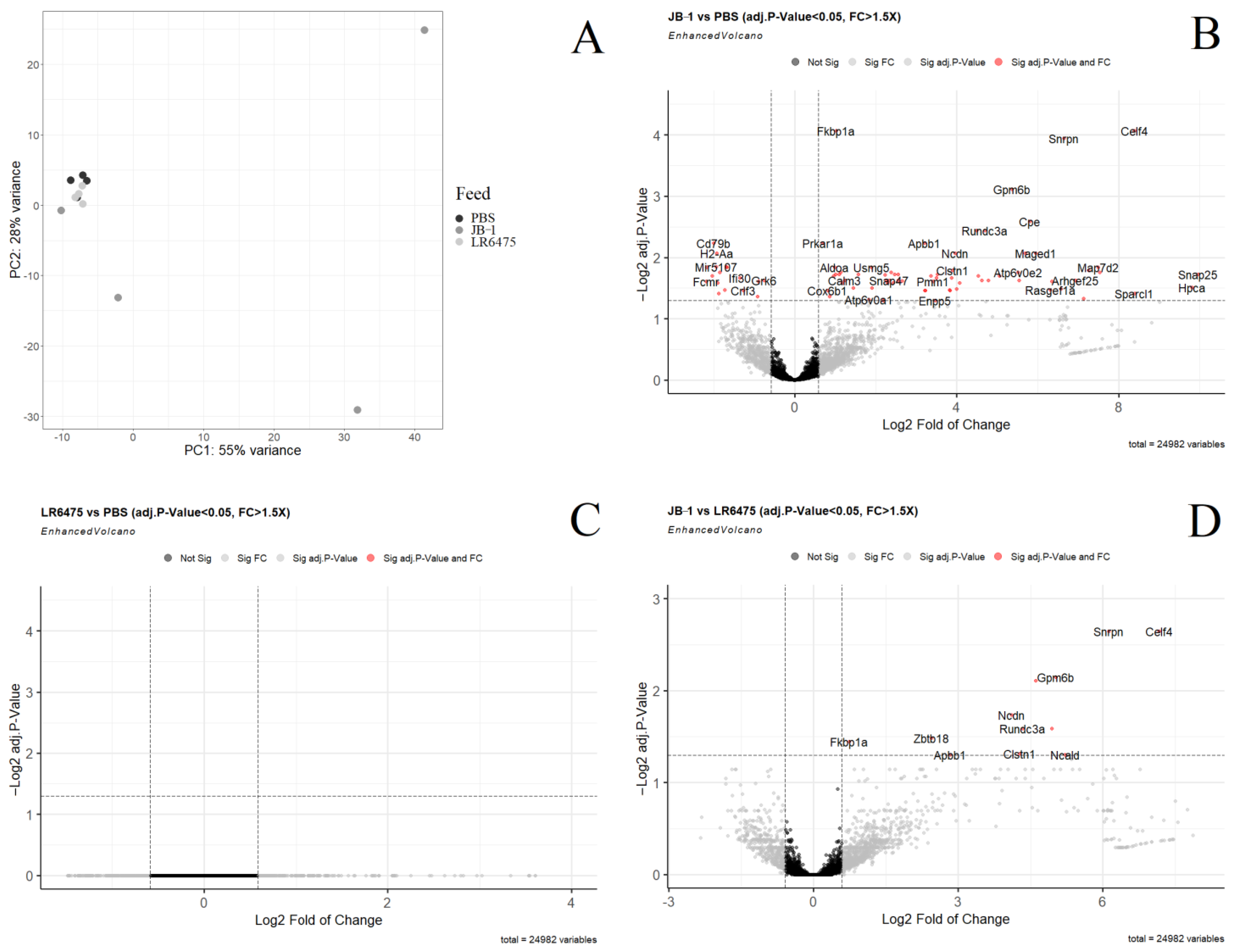
2.1. Many mRNAs and Gene Sets Are Altered in the Blood of JB-1-Fed Mice, but Not LR6475-Fed
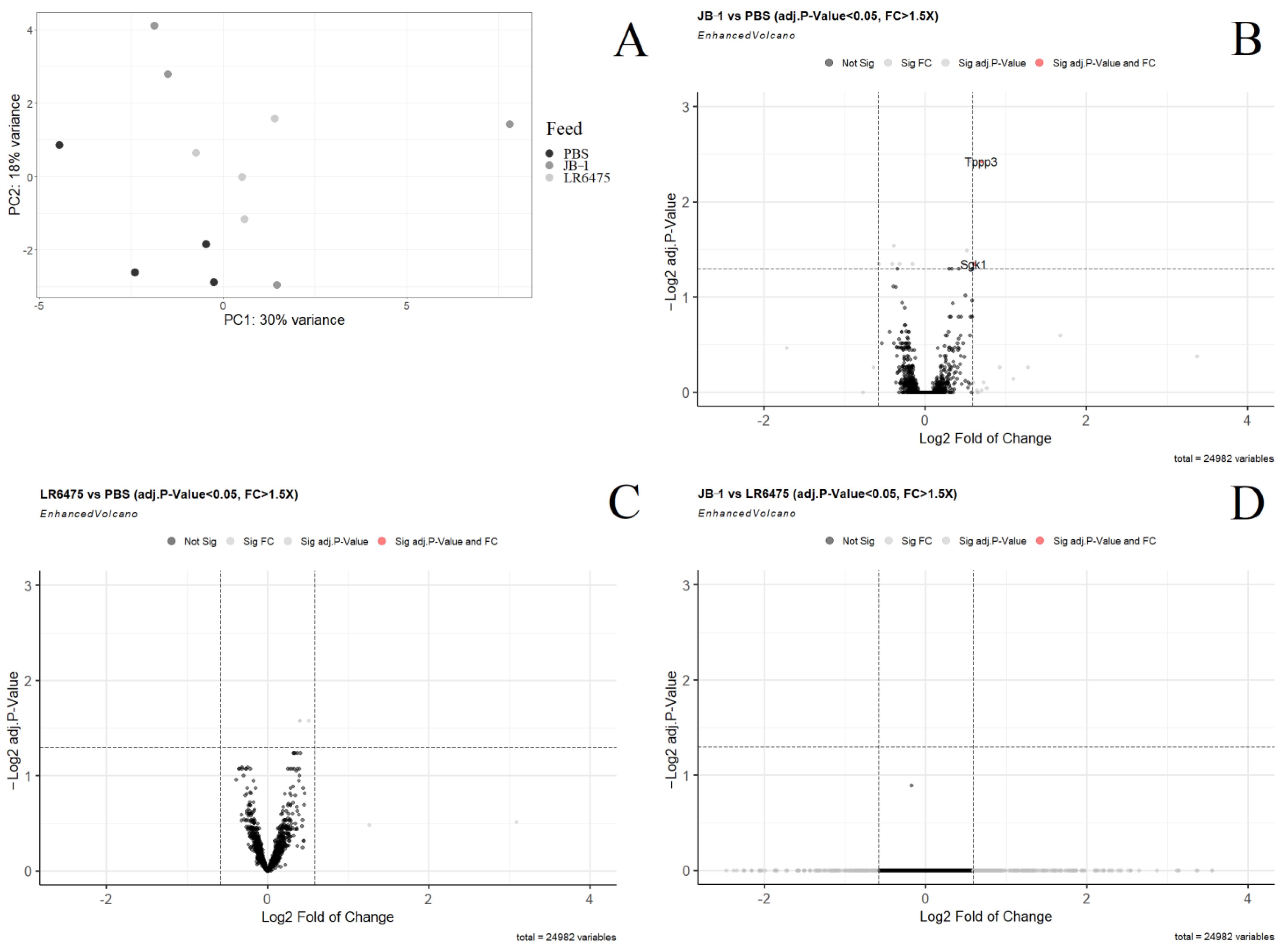
2.2. Few mRNAs and Gene Sets Are Altered in the Hippocampus of Psychobiotic-Fed Mice
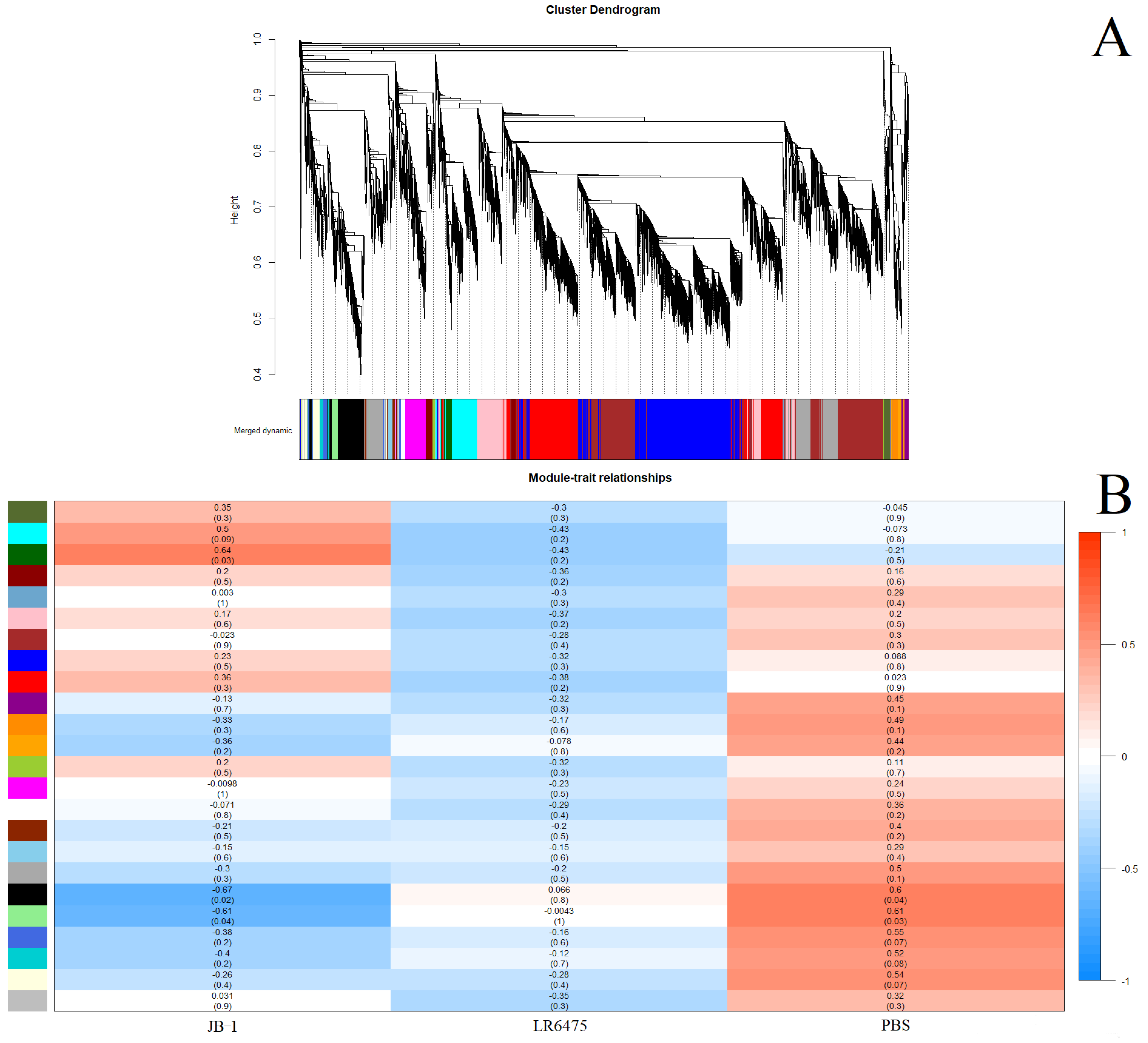
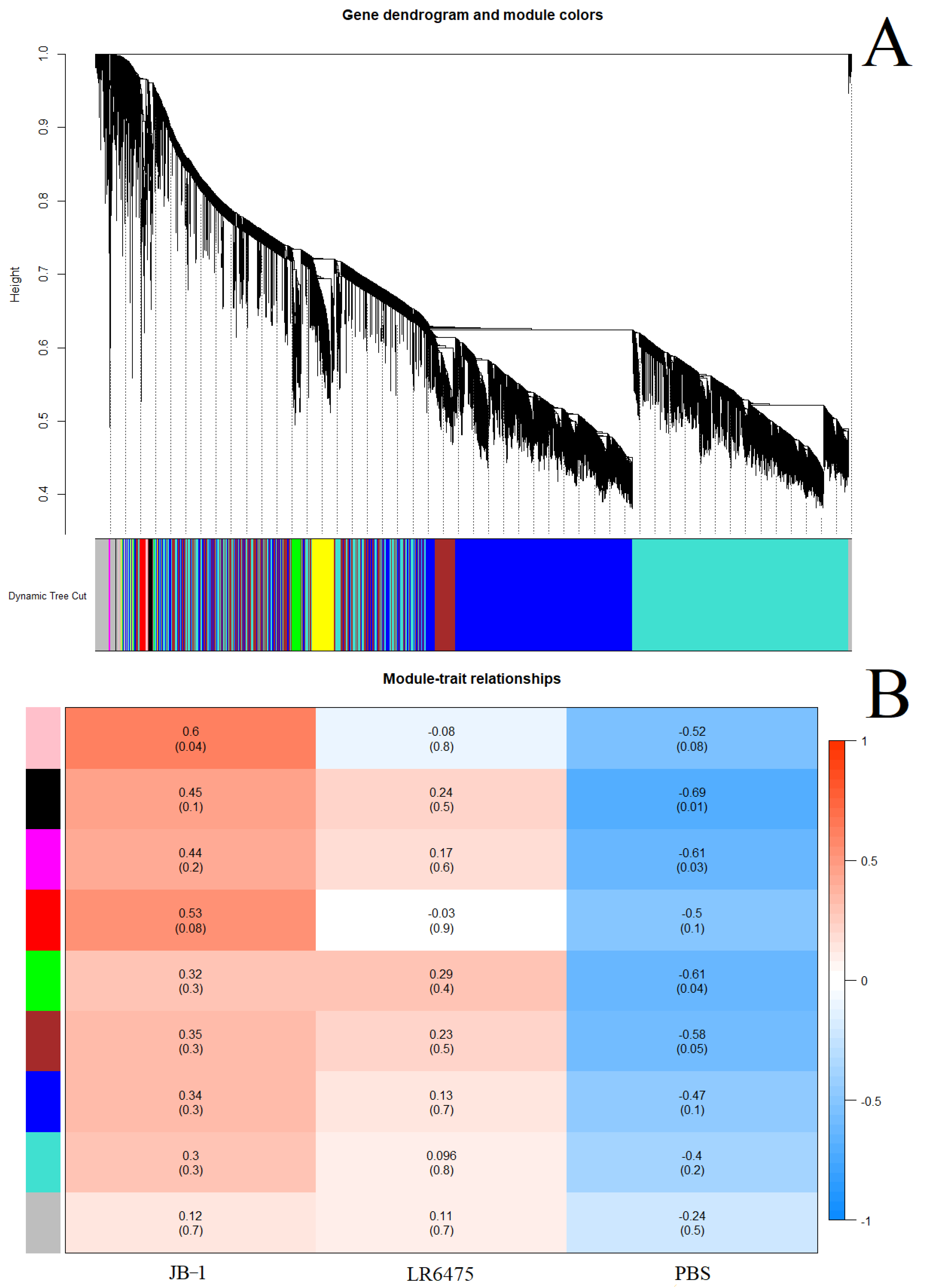
2.3. Weighted Correlation Network Analysis Confirms Differences in Blood Expression between Feeding Groups
- As a second line of evidence that JB-1, but not LR6475 has a unique impact on blood and hippocampal mRNA expression, we performed weighted correlation network analysis (WGCNA) which relies on unsupervised clustering of genes to construct a network with modules of commonly co-expressed genes.
- Using a power of 13 for blood and merging at a threshold of 0.05, 24 eigengenes were identified and mapped to the cluster dendogram (Figure 3A). There were several strong relationships identified when comparing these gene modules to the feed groups (traits) including statistically significant differences between JB-1 and PBS-fed mouse blood in the ‘black’ and ‘light green’ groups (Figure 3B). None of the modules had a significant relationship with LR6475-fed mice, making it statistically indiscernible from PBS, which is consistent with the PCA (Figure 1A).
3. Discussion
3.1. Inflammatory Response
3.2. Cerebral Cortical Signaling
3.3. JB-1-Modulated Genes in the Blood
4. Materials and Methods
4.1. Laboratory Methods
4.1.1. Feeding and Tissue Collection
4.1.2. RNA Isolation and Analysis
4.2. Bioinformatic and Statistical Analysis
4.2.1. Data Preprocessing and Differential Expression
4.2.2. PCA
4.2.3. Enrichment Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wittchen, H.U.; Jacobi, F.; Rehm, J.; Gustavsson, A.; Svensson, M.; Jönsson, B.; Olesen, J.; Allgulander, C.; Alonso, J.; Faravelli, C.; et al. The Size and Burden of Mental Disorders and Other Disorders of the Brain in Europe 2010. Eur. Neuropsychopharmacol. 2011, 21, 655–679. [Google Scholar] [CrossRef] [PubMed]
- Patalay, P.; Gage, S.H. Changes in Millennial Adolescent Mental Health and Health-Related Behaviours over 10 Years: A Population Cohort Comparison Study. Int. J. Epidemiol. 2019, 48, 1650–1664. [Google Scholar] [CrossRef] [PubMed]
- Abbott, A. COVID’s Mental-Health Toll: How Scientists Are Tracking a Surge in Depression. Nature 2021, 590, 194–195. [Google Scholar] [CrossRef] [PubMed]
- Long-Smith, C.; O’Riordan, K.J.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota-Gut-Brain Axis: New Therapeutic Opportunities. Annu. Rev. Pharmacol. Toxicol. 2020, 60, 477–502. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The Gut-Brain Axis: Interactions between Enteric Microbiota, Central and Enteric Nervous Systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Sarkar, A.; Lehto, S.M.; Harty, S.; Dinan, T.G.; Cryan, J.F.; Burnet, P.W.J. Psychobiotics and the Manipulation of Bacteria–Gut–Brain Signals. Trends Neurosci. 2016, 39, 763–781. [Google Scholar] [CrossRef]
- Wallace, C.J.K.; Milev, R. The Effects of Probiotics on Depressive Symptoms in Humans: A Systematic Review. Ann. Gen. Psychiatry 2017, 16, 14. [Google Scholar] [CrossRef]
- Bharwani, A.; Mian, M.F.; Surette, M.G.; Bienenstock, J.; Forsythe, P. Oral Treatment with Lactobacillus Rhamnosus Attenuates Behavioural Deficits and Immune Changes in Chronic Social Stress. BMC Med. 2017, 15, 7. [Google Scholar] [CrossRef]
- Bravo, J.A.; Forsythe, P.; Chew, M.V.; Escaravage, E.; Savignac, H.M.; Dinan, T.G.; Bienenstock, J.; Cryan, J.F. Ingestion of Lactobacillus Strain Regulates Emotional Behavior and Central GABA Receptor Expression in a Mouse via the Vagus Nerve. Proc. Natl. Acad. Sci. USA 2011, 108, 16050–16055. [Google Scholar] [CrossRef]
- McVey Neufeld, K.-A.; Kay, S.; Bienenstock, J. Mouse Strain Affects Behavioral and Neuroendocrine Stress Responses Following Administration of Probiotic Lactobacillus Rhamnosus JB-1 or Traditional Antidepressant Fluoxetine. Front. Neurosci. 2018, 12, 294. [Google Scholar] [CrossRef]
- Liu, Y.; Mian, M.F.; McVey Neufeld, K.-A.; Forsythe, P. CD4+CD25+ T Cells Are Essential for Behavioral Effects of Lactobacillus Rhamnosus JB-1 in Male BALB/c Mice. Brain. Behav. Immun. 2020, 88, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Dinan, T.G.; Stanton, C.; Cryan, J.F. Psychobiotics: A Novel Class of Psychotropic. Biol. Psychiatry 2013, 74, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Kim, Y.-K.; Han, P.-L. Extracellular Vesicles Derived from Lactobacillus Plantarum Increase BDNF Expression in Cultured Hippocampal Neurons and Produce Antidepressant-like Effects in Mice. Exp. Neurobiol. 2019, 28, 158–171. [Google Scholar] [CrossRef] [PubMed]
- Haas-Neill, S.; Forsythe, P. A Budding Relationship: Bacterial Extracellular Vesicles in the Microbiota-Gut-Brain Axis. Int. J. Mol. Sci. 2020, 21, 8899. [Google Scholar] [CrossRef]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259. [Google Scholar] [CrossRef]
- Buffington, S.A.; Di Prisco, G.V.; Auchtung, T.A.; Ajami, N.J.; Petrosino, J.F.; Costa-Mattioli, M. Microbial Reconstitution Reverses Maternal Diet-Induced Social and Synaptic Deficits in Offspring. Cell 2016, 165, 1762–1775. [Google Scholar] [CrossRef] [PubMed]
- Poutahidis, T.; Kleinewietfeld, M.; Smillie, C.; Levkovich, T.; Perrotta, A.; Bhela, S.; Varian, B.J.; Ibrahim, Y.M.; Lakritz, J.R.; Kearney, S.M.; et al. Microbial Reprogramming Inhibits Western Diet-Associated Obesity. PLoS ONE 2013, 8, e68596. [Google Scholar] [CrossRef]
- Medicine, N.L.L. Reuteri DSM 17938 and L. Reuteri ATCC PTA 6475 in Moderate to Severe Irritable Bowel in Adults (Reuteri-IBS). 2019. Available online: https://clinicaltrials.gov/ct2/show/NCT04037826 (accessed on 28 February 2022).
- Collins, N.; Han, S.-J.; Enamorado, M.; Link, V.M.; Huang, B.; Moseman, E.A.; Kishton, R.J.; Shannon, J.P.; Dixit, D.; Schwab, S.R.; et al. The Bone Marrow Protects and Optimizes Immunological Memory during Dietary Restriction. Cell 2019, 178, 1088–1101. [Google Scholar] [CrossRef]
- West, C.L.; McVey Neufeld, K.-A.; Mao, Y.-K.; Stanisz, A.M.; Forsythe, P.; Bienenstock, J.; Barbut, D.; Zasloff, M.; Kunze, W.A. Identification of SSRI-Evoked Antidepressant Sensory Signals by Decoding Vagus Nerve Activity. Sci. Rep. 2021, 11, 21130. [Google Scholar] [CrossRef]
- Campbell, S.; Macqueen, G. The Role of the Hippocampus in the Pathophysiology of Major Depression. J. Psychiatry Neurosci. 2004, 29, 417–426. [Google Scholar] [PubMed]
- MacQueen, G.; Frodl, T. The Hippocampus in Major Depression: Evidence for the Convergence of the Bench and Bedside in Psychiatric Research? Mol. Psychiatry 2011, 16, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Roddy, D.W.; Farrell, C.; Doolin, K.; Roman, E.; Tozzi, L.; Frodl, T.; O’Keane, V.; O’Hanlon, E. The Hippocampus in Depression: More Than the Sum of Its Parts? Advanced Hippocampal Substructure Segmentation in Depression. Biol. Psychiatry 2019, 85, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Knapp, B.; Omasits, U.; Schreiner, W.; Epstein, M.M. A Comparative Approach Linking Molecular Dynamics of Altered Peptide Ligands and MHC with In Vivo Immune Responses. PLoS ONE 2010, 5, e11653. [Google Scholar] [CrossRef] [PubMed]
- Maric, M.; Arunachalam, B.; Phan, U.T.; Dong, C.; Garrett, W.S.; Cannon, K.S.; Alfonso, C.; Karlsson, L.; Flavell, R.A.; Cresswell, P. Defective Antigen Processing in GILT-Free Mice. Science 2001, 294, 1361–1365. [Google Scholar] [CrossRef]
- Singh, R.; Cresswell, P. Defective Cross-Presentation of Viral Antigens in GILT-Free Mice. Science 2010, 328, 1394–1398. [Google Scholar] [CrossRef]
- Fuentes-Pananá, E.M.; Bannish, G.; van der Voort, D.; King, L.B.; Monroe, J.G. Igα/Igβ Complexes Generate Signals for B Cell Development Independent of Selective Plasma Membrane Compartmentalization. J. Immunol. 2005, 174, 1245–1252. [Google Scholar] [CrossRef]
- Siemasko, K.; Eisfelder, B.J.; Stebbins, C.; Kabak, S.; Sant, A.J.; Song, W.; Clark, M.R. Ig Alpha and Ig Beta Are Required for Efficient Trafficking to Late Endosomes and to Enhance Antigen Presentation. J. Immunol. 1999, 162, 6518–6525. [Google Scholar]
- Liu, Y.; Steinhausen, K.; Bharwani, A.; Mian, M.F.; McVey Neufeld, K.-A.; Forsythe, P. Increased Persistence of Avoidance Behaviour and Social Deficits with L.Rhamnosus JB-1 or Selective Serotonin Reuptake Inhibitor Treatment Following Social Defeat. Sci. Rep. 2020, 10, 13485. [Google Scholar] [CrossRef]
- Branco-de-Almeida, L.S.; Kajiya, M.; Cardoso, C.R.; Silva, M.J.B.; Ohta, K.; Rosalen, P.L.; Franco, G.C.N.; Han, X.; Taubman, M.A.; Kawai, T. Selective Serotonin Reuptake Inhibitors Attenuate the Antigen Presentation from Dendritic Cells to Effector T Lymphocytes. FEMS Immunol. Med. Microbiol. 2011, 62, 283–294. [Google Scholar] [CrossRef]
- Pinto, F.E.; Andrade, C. Interferon-Related Depression: A Primer on Mechanisms, Treatment, and Prevention of a Common Clinical Problem. Curr. Neuropharmacol. 2016, 14, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Inserra, A.; Mastronardi, C.A.; Rogers, G.; Licinio, J.; Wong, M.-L. Neuroimmunomodulation in Major Depressive Disorder: Focus on Caspase 1, Inducible Nitric Oxide Synthase, and Interferon-Gamma. Mol. Neurobiol. 2019, 56, 4288–4305. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Scharpé, S.; Meltzer, H.Y.; Okayli, G.; Bosmans, E.; D’Hondt, P.; Vanden Bossche, B.; Cosyns, P. Increased Neopterin and Interferon-Gamma Secretion and Lower Availability of L-Tryptophan in Major Depression: Further Evidence for an Immune Response. Psychiatry Res. 1994, 54, 143–160. [Google Scholar] [CrossRef]
- Maes, M. Negative Immunoregulatory Effects of Antidepressants Inhibition of Interferon-γ and Stimulation of Interleukin-10 Secretion. Neuropsychopharmacology 1999, 20, 370–379. [Google Scholar] [CrossRef]
- Karimi, K.; Kandiah, N.; Chau, J.; Bienenstock, J.; Forsythe, P. A Lactobacillus Rhamnosus Strain Induces a Heme Oxygenase Dependent Increase in Foxp3+ Regulatory T Cells. PLoS ONE 2012, 7, e47556. [Google Scholar] [CrossRef]
- Forsythe, P.; Wang, B.; Khambati, I.; Kunze, W.A. Systemic Effects of Ingested Lactobacillus Rhamnosus: Inhibition of Mast Cell Membrane Potassium (IKCa) Current and Degranulation. PLoS ONE 2012, 7, e41234. [Google Scholar] [CrossRef]
- Maes, M.; Lambrechts, J.; Bosmans, E.; Jacobs, J.; Suy, E.; Vandervorst, C.; De Jonckheere, C.; Minner, B.; Raus, J. Evidence for a Systemic Immune Activation during Depression: Results of Leukocyte Enumeration by Flow Cytometry in Conjunction with Monoclonal Antibody Staining. Psychol. Med. 1992, 22, 45–53. [Google Scholar] [CrossRef]
- Kong, X.; Zhang, Z.; Fu, T.; Ji, J.; Yang, J.; Gu, Z. TNF-α Regulates Microglial Activation via the NF-ΚB Signaling Pathway in Systemic Lupus Erythematosus with Depression. Int. J. Biol. Macromol. 2019, 125, 892–900. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, J.; Liu, Y.; Li, Z.; Li, X. TLR4-NF-κ B Signal Involved in Depressive-Like Behaviors and Cytokine Expression of Frontal Cortex and Hippocampus in Stressed C57BL/6 and Ob/Ob Mice. Neural Plast. 2018, 2018, 7254016. [Google Scholar] [CrossRef]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR Kinase Structure, Mechanism and Regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef]
- Hu, L.; Chen, F.; Wu, C.; Wang, J.; Chen, S.; Li, X.; Wang, J.; Wu, L.; Ding, J.; Wang, J.; et al. Rapamycin Recruits SIRT2 for FKBP12 Deacetylation during MTOR Activity Modulation in Innate Immunity. iScience 2021, 24, 103177. [Google Scholar] [CrossRef] [PubMed]
- Chi, H. Regulation and Function of MTOR Signalling in T Cell Fate Decisions. Nat. Rev. Immunol. 2012, 12, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Weichhart, T.; Hengstschläger, M.; Linke, M. Regulation of Innate Immune Cell Function by mTOR. Nat. Rev. Immunol. 2015, 15, 599–614. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. STAT Family of Transcription Factors in Cytokine-Mediated Biological Responses. Cytokine Growth Factor Rev. 2000, 11, 199–207. [Google Scholar] [CrossRef]
- Jankovic, D.; Trinchieri, G. IL-10 or Not IL-10: That Is the Question. Nat. Immunol. 2007, 8, 1281–1283. [Google Scholar] [CrossRef]
- Shariq, A.S.; Brietzke, E.; Rosenblat, J.D.; Pan, Z.; Rong, C.; Ragguett, R.-M.; Park, C.; McIntyre, R.S. Therapeutic Potential of JAK/STAT Pathway Modulation in Mood Disorders. Rev. Neurosci. 2018, 30, 1–7. [Google Scholar] [CrossRef]
- Al-Samhari, M.M.; Al-Rasheed, N.M.; Al-Rejaie, S.; Al-Rasheed, N.M.; Hasan, I.H.; Mahmoud, A.M.; Dzimiri, N. Possible Involvement of the JAK/STAT Signaling Pathway in N-Acetylcysteine-Mediated Antidepressant-like Effects. Exp. Biol. Med. 2016, 241, 509–518. [Google Scholar] [CrossRef]
- Vincze, O.; Tökési, N.; Oláh, J.; Hlavanda, E.; Zotter, Á.; Horváth, I.; Lehotzky, A.; Tirián, L.; Medzihradszky, K.F.; Kovács, J.; et al. Tubulin Polymerization Promoting Proteins (TPPPs): Members of a New Family with Distinct Structures and Functions. Biochemistry 2006, 45, 13818–13826. [Google Scholar] [CrossRef]
- Leong, M.L.L.; Maiyar, A.C.; Kim, B.; O’Keeffe, B.A.; Firestone, G.L. Expression of the Serum- and Glucocorticoid-Inducible Protein Kinase, Sgk, Is a Cell Survival Response to Multiple Types of Environmental Stress Stimuli in Mammary Epithelial Cells. J. Biol. Chem. 2003, 278, 5871–5882. [Google Scholar] [CrossRef]
- Yoo, D.; Kim, B.Y.; Campo, C.; Nance, L.; King, A.; Maouyo, D.; Welling, P.A. Cell Surface Expression of the ROMK (Kir 1.1) Channel Is Regulated by the Aldosterone-Induced Kinase, SGK-1, and Protein Kinase A. J. Biol. Chem. 2003, 278, 23066–23075. [Google Scholar] [CrossRef]
- Eylenstein, A.; Gehring, E.; Heise, N.; Shumilina, E.; Schmidt, S.; Szteyn, K.; Münzer, P.; Nurbaeva, M.K.; Eichenmüller, M.; Tyan, L.; et al. Stimulation of Ca2+-channel Orai1/STIM1 by Serum-and Glucocorticoid-inducible Kinase 1 (SGK1). FASEB J. 2011, 25, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.-S.; Ann, E.-J.; Yoon, J.-H.; Jung, J.; Choi, Y.-H.; Kim, H.-Y.; Ahn, J.-S.; Kim, S.-M.; Kim, M.-Y.; Hong, J.-A.; et al. Serum- and Glucocorticoid-Inducible Kinase 1 (SGK1) Controls Notch1 Signaling by Downregulation of Protein Stability through Fbw7 Ubiquitin Ligase. J. Cell Sci. 2011, 124, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Dattilo, V.; Amato, R.; Perrotti, N.; Gennarelli, M. The Emerging Role of SGK1 (Serum- and Glucocorticoid-Regulated Kinase 1) in Major Depressive Disorder: Hypothesis and Mechanisms. Front. Genet. 2020, 11, 826. [Google Scholar] [CrossRef] [PubMed]
- Anacker, C.; Cattaneo, A.; Musaelyan, K.; Zunszain, P.A.; Horowitz, M.; Molteni, R.; Luoni, A.; Calabrese, F.; Tansey, K.; Gennarelli, M.; et al. Role for the Kinase SGK1 in Stress, Depression, and Glucocorticoid Effects on Hippocampal Neurogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 8708–8713. [Google Scholar] [CrossRef] [PubMed]
- Licznerski, P.; Duric, V.; Banasr, M.; Alavian, K.N.; Ota, K.T.; Kang, H.J.; Jonas, E.A.; Ursano, R.; Krystal, J.H.; Duman, R.S. Decreased SGK1 Expression and Function Contributes to Behavioral Deficits Induced by Traumatic Stress. PLoS Biol. 2015, 13, e1002282. [Google Scholar] [CrossRef]
- Zhang, K.; Pan, X.; Wang, F.; Ma, J.; Su, G.; Dong, Y.; Yang, J.; Wu, C. Baicalin Promotes Hippocampal Neurogenesis via SGK1- and FKBP5-Mediated Glucocorticoid Receptor Phosphorylation in a Neuroendocrine Mouse Model of Anxiety/Depression. Sci. Rep. 2016, 6, 30951. [Google Scholar] [CrossRef]
- Drabek, K.; van de Peppel, J.; Eijken, M.; van Leeuwen, J.P. GPM6B Regulates Osteoblast Function and Induction of Mineralization by Controlling Cytoskeleton and Matrix Vesicle Release. J. Bone Miner. Res. 2011, 26, 2045–2051. [Google Scholar] [CrossRef]
- Fjorback, A.W.; Müller, H.K.; Wiborg, O. Membrane Glycoprotein M6B Interacts with the Human Serotonin Transporter. J. Mol. Neurosci. 2009, 37, 191–200. [Google Scholar] [CrossRef]
- Haase, J.; Grudzinska-Goebel, J.; Müller, H.K.; Münster-Wandowski, A.; Chow, E.; Wynne, K.; Farsi, Z.; Zander, J.-F.; Ahnert-Hilger, G. Serotonin Transporter Associated Protein Complexes Are Enriched in Synaptic Vesicle Proteins and Proteins Involved in Energy Metabolism and Ion Homeostasis. ACS Chem. Neurosci. 2017, 8, 1101–1116. [Google Scholar] [CrossRef]
- Fuchsova, B.; Alvarez Juliá, A.; Rizavi, H.S.; Frasch, A.C.; Pandey, G.N. Altered Expression of Neuroplasticity-Related Genes in the Brain of Depressed Suicides. Neuroscience 2015, 299, 1–17. [Google Scholar] [CrossRef]
- Mariani, N.; Cattane, N.; Pariante, C.; Cattaneo, A. Gene Expression Studies in Depression Development and Treatment: An Overview of the Underlying Molecular Mechanisms and Biological Processes to Identify Biomarkers. Transl. Psychiatry 2021, 11, 354. [Google Scholar] [CrossRef] [PubMed]
- Rodriguiz, R.M.; Wilkins, J.J.; Creson, T.K.; Biswas, R.; Berezniuk, I.; Fricker, A.D.; Fricker, L.D.; Wetsel, W.C. Emergence of Anxiety-like Behaviours in Depressive-like Cpefat/Fat Mice. Int. J. Neuropsychopharmacol. 2013, 16, 1623–1634. [Google Scholar] [CrossRef][Green Version]
- Cheng, Y.; Cawley, N.X.; Yanik, T.; Murthy, S.R.K.; Liu, C.; Kasikci, F.; Abebe, D.; Loh, Y.P. A Human Carboxypeptidase E/NF-A1 Gene Mutation in an Alzheimer’s Disease Patient Leads to Dementia and Depression in Mice. Transl. Psychiatry 2016, 6, e973. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Kawano, T.; Taru, H.; Saito, Y.; Wada, S.; Miyamoto, K.; Kobayashi, H.; Ishikawa, H.O.; Ohsugi, Y.; Yamamoto, T.; et al. The Novel Cargo Alcadein Induces Vesicle Association of Kinesin-1 Motor Components and Activates Axonal Transport. EMBO J. 2007, 26, 1475–1486. [Google Scholar] [CrossRef]
- Davies, M.N.; Krause, L.; Bell, J.T.; Gao, F.; Ward, K.J.; Wu, H.; Lu, H.; Liu, Y.; Tsai, P.-C.; Collier, D.A.; et al. Hypermethylation in the ZBTB20 Gene Is Associated with Major Depressive Disorder. Genome Biol. 2014, 15, R56. [Google Scholar] [CrossRef]
- Li, S.; Cao, W.; Zhou, S.; Ma, M.; Zhang, W.; Li, F.; Li, C. Expression of Cntn1 Is Regulated by Stress and Associated with Anxiety and Depression Phenotypes. Brain. Behav. Immun. 2021, 95, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Saunders, C.T.; Wong, W.S.W.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate Somatic Small-Variant Calling from Sequenced Tumor–Normal Sample Pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally Applicable Gene Set Enrichment for Pathway Analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor Package for Pathway-Based Data Integration and Visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Litteljohn, D.; Nelson, E.; Hayley, S. IFN-Υ Differentially Modulates Memory-Related Processes under Basal and Chronic Stressor Conditions. Front. Cell. Neurosci. 2014, 8, 391. [Google Scholar] [CrossRef] [PubMed]
- Lambert, G.; Johansson, M.; Ågren, H.; Friberg, P. Reduced Brain Norepinephrine and Dopamine Release in Treatment-Refractory Depressive Illness. Arch. Gen. Psychiatry 2000, 57, 787. [Google Scholar] [CrossRef] [PubMed]
- Gulbins, A.; Grassmé, H.; Hoehn, R.; Kohnen, M.; Edwards, M.J.; Kornhuber, J.; Gulbins, E. Role of Janus-Kinases in Major Depressive Disorder. Neurosignals 2016, 24, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Kubera, M.; Basta-Kaim, A.; Papp, M. The Effect of Chronic Treatment with Imipramine on the Immunoreactivity of Animals Subjected to a Chronic Mild Stress Model of Depression. Immunopharmacology 1995, 30, 225–230. [Google Scholar] [CrossRef]
- Fatima, A.; Hoeber, J.; Schuster, J.; Koshimizu, E.; Maya-Gonzalez, C.; Keren, B.; Mignot, C.; Akram, T.; Ali, Z.; Miyatake, S.; et al. Monoallelic and Bi-Allelic Variants in NCDN Cause Neurodevelopmental Delay, Intellectual Disability, and Epilepsy. Am. J. Hum. Genet. 2021, 108, 739–748. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, W.-Y.; Xia, X.; Kong, L.-D. Effects of Icariin on Hypothalamic-Pituitary-Adrenal Axis Action and Cytokine Levels in Stressed Sprague-Dawley Rats. Biol. Pharm. Bull. 2006, 29, 2399–2403. [Google Scholar] [CrossRef]
- Wei, K.; Xu, Y.; Zhao, Z.; Wu, X.; Du, Y.; Sun, J.; Yi, T.; Dong, J.; Liu, B. Icariin Alters the Expression of Glucocorticoid Receptor, FKBP5 and SGK1 in Rat Brains Following Exposure to Chronic Mild Stress. Int. J. Mol. Med. 2016, 38, 337–344. [Google Scholar] [CrossRef]
- Wesseling, H.; Rahmoune, H.; Tricklebank, M.; Guest, P.C.; Bahn, S. A Targeted Multiplexed Proteomic Investigation Identifies Ketamine-Induced Changes in Immune Markers in Rat Serum and Expression Changes in Protein Kinases/Phosphatases in Rat Brain. J. Proteome Res. 2015, 14, 411–421. [Google Scholar] [CrossRef] [PubMed]
| Enriched In | Pathway Name | Adj.p-Val (JvC, JvR) |
|---|---|---|
| JB-1 | HALLMARK_OXIDATIVE_PHOSPHORYLATION | 3.4 × 10−4, 3.4 × 10−4 |
| JB-1 | KEGG_OXIDATIVE_PHOSPHORYLATION | 3.7 × 10−3, 9.9 × 10−3 |
| JB-1 | KEGG_PARKINSONS_DISEASE | 1.3 × 10−2, 2.3 × 10−2 |
| JB-1 | KEGG_HUNTINGTONS_DISEASE | 1.3 × 10−2, 2.3 × 10−2 |
| JB-1 | KEGG_ALZHEIMERS_DISEASE | 2.1 × 10−2, 3.7 × 10−2 |
| PBS | HALLMARK_ALLOGRAFT_REJECTION | 1.5 × 10−6, 2.7 × 10−6 |
| PBS | HALLMARK_INTERFERON_GAMMA_RESPONSE | 5.4 × 10-6, 8.9 × 10-6 |
| PBS | HALLMARK_INFLAMMATORY_RESPONSE | 4.6 × 10−3, 9.4 × 10−3 |
| PBS | HALLMARK_INTERFERON_ALPHA_RESPONSE | 7.7 × 10−3, 8.3 × 10−3 |
| PBS | HALLMARK_TNFA_SIGNALLING_VIA_NFKB | 3.2 × 10−2, 3.1 × 10−2 |
| PBS | HALLMARK_IL6_JAK_STAT3_SIGNALLING | 3.2 × 10-2, 2.5 × 10-2 |
| PBS | KEGG_PRIMARY_IMMUNODEFICIENCY | 1.5 × 10−3, 3.6 × 10−3 |
| PBS | KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 4.9 × 10−3, 9.2 × 10−3 |
| PBS | KEGG_JAK_STAT_SIGNALLING_PATHWAY | 4.9 × 10−3, 8.0 × 10−3 |
| PBS | KEGG_CHEMOKINE_SIGNALLING_PATHWAY | 1.8 × 10−2, 1.0 × 10−2 |
| PBS | KEGG_HEMATOPOIETIC_CELL_LINEAGE | 1.9 × 10−2, 1.0 × 10−2 |
| PBS | KEGG_B_CELL_RECEPTOR_SIGNALLING_PATHWAY | 1.9 × 10−2, 2.8 × 10−2 |
| PBS | KEGG_RIBOSOME | 2.1 × 10−2, 6.7 × 10−3 |
| PBS | KEGG_T_CELL_RECEPTOR_SIGNALLING_PATHWAY | 2.2 × 10−2, 2.8 × 10−2 |
| PBS | KEGG_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION | 3.8 × 10−2 |
| PBS | KEGG_NATURAL_KILLER_CELL_MEDIATED_CYTOTOXICITY | 4.5 × 10−2 |
| Pathway | INF-γ/INF-α | JAK/STAT | TNF-α via NF-KB |
|---|---|---|---|
| Change in Depression | ↑ generally increased, but suboptimal expression has also been associated with depression [33,76,77] | ↑ activation in stress and depression in mice [78] | ↑ in prefrontal cortex and hippocampus of mice [40] |
| Change in MDD Treatment | ↓ after treatment with either sertraline, clomipramine, or trazodone in human blood [35] | ↓ phosphorylation (activation) of Jak-3 returned to normal in mice following Amitriptyline treatment [78] | ↓ SSRIs such as imipramine reduce TNF-α levels in rats [79] |
| Blood Change Following JB-1 | ↓ mRNA (5.4 × 10−6) (7.7 × 10−3) | ↓ mRNA (4.9 × 10−3) | ↓ mRNA (3.2 × 10−2) |
| Gene | SGK1 | GPM6B | NCDN | CLSTN1 |
|---|---|---|---|---|
| Change in Depression | ↓ in hippocampus of rats [56] | ↓ in the hippocampus of humans [60] | ↓ in CNS of mice [80] | ↓ in blood of humans [66], ↑ in hippocampus of rats and mice [67] |
| Change in Depression Treatment | ↑ mRNA in hippocampus and prefrontal cortex of rats following icariin or baicalin treatment [81,82] | N/A | ↓ in hippocampus after ketamine treatment in rats [83] | N/A |
| Change Following JB-1 | ↑ mRNA in hippocampus of mice (4.5 × 10−2) | ↑ mRNA in blood of mice (7.6 × 10−4) | ↑ mRNA in blood of mice (8.5 × 10−3) | ↑ mRNA in blood of mice (1.6 × 10−2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haas-Neill, S.; Iwashita, E.; Dvorkin-Gheva, A.; Forsythe, P. Effects of Two Distinct Psychoactive Microbes, Lacticaseibacillus rhamnosus JB-1 and Limosilactobacillus reuteri 6475, on Circulating and Hippocampal mRNA in Male Mice. Int. J. Mol. Sci. 2022, 23, 9653. https://doi.org/10.3390/ijms23179653
Haas-Neill S, Iwashita E, Dvorkin-Gheva A, Forsythe P. Effects of Two Distinct Psychoactive Microbes, Lacticaseibacillus rhamnosus JB-1 and Limosilactobacillus reuteri 6475, on Circulating and Hippocampal mRNA in Male Mice. International Journal of Molecular Sciences. 2022; 23(17):9653. https://doi.org/10.3390/ijms23179653
Chicago/Turabian StyleHaas-Neill, Sandor, Eiko Iwashita, Anna Dvorkin-Gheva, and Paul Forsythe. 2022. "Effects of Two Distinct Psychoactive Microbes, Lacticaseibacillus rhamnosus JB-1 and Limosilactobacillus reuteri 6475, on Circulating and Hippocampal mRNA in Male Mice" International Journal of Molecular Sciences 23, no. 17: 9653. https://doi.org/10.3390/ijms23179653
APA StyleHaas-Neill, S., Iwashita, E., Dvorkin-Gheva, A., & Forsythe, P. (2022). Effects of Two Distinct Psychoactive Microbes, Lacticaseibacillus rhamnosus JB-1 and Limosilactobacillus reuteri 6475, on Circulating and Hippocampal mRNA in Male Mice. International Journal of Molecular Sciences, 23(17), 9653. https://doi.org/10.3390/ijms23179653






