Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy
Abstract
1. Introduction
2. Results
2.1. The Vps21 Module Regulates Vps34 and PI(3)P Localizations under Nitrogen Starvation
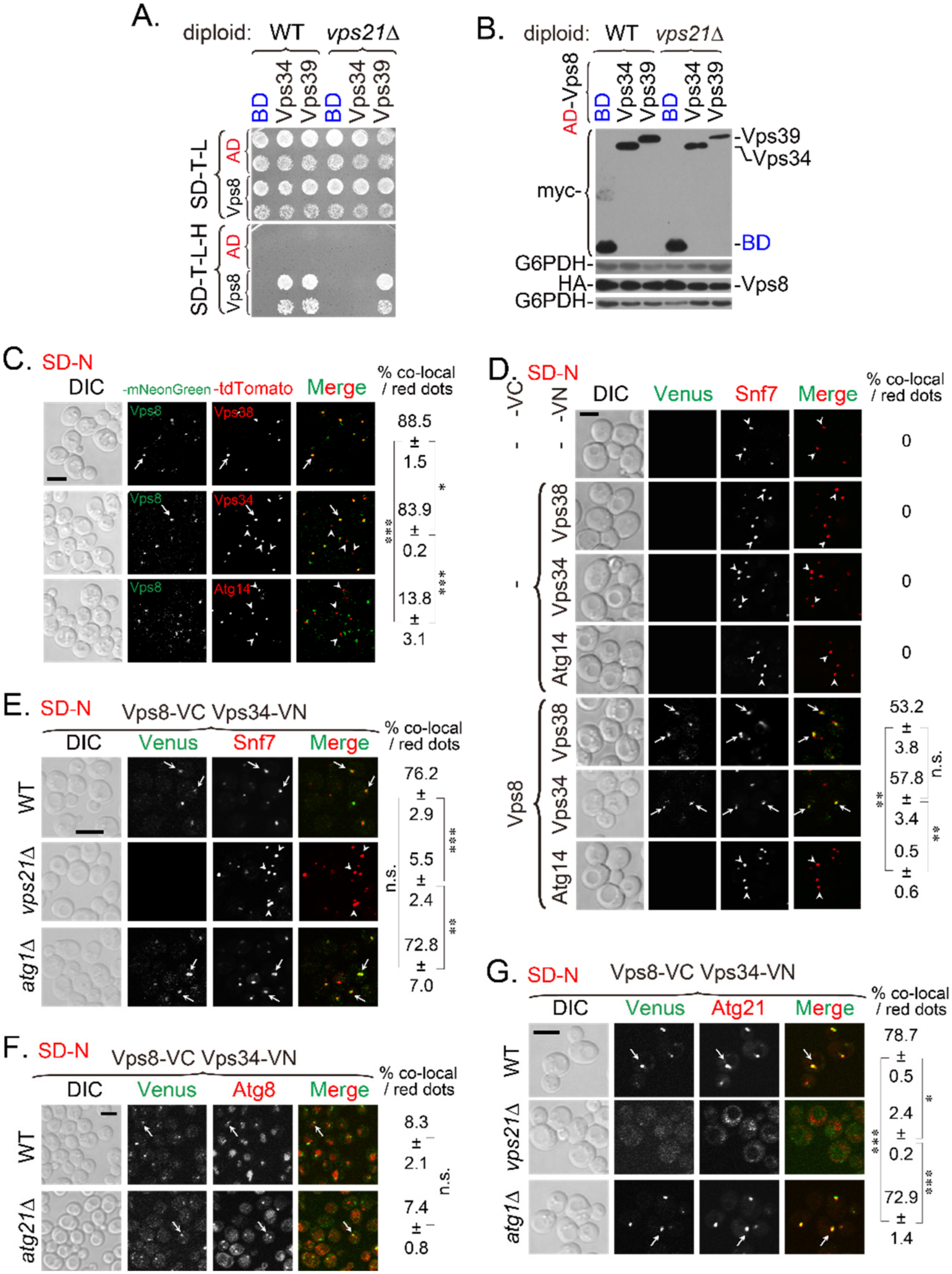
2.2. Vps8 Interacts with Vps34 on Endosomes and the PAS under Nitrogen Starvation, and This Interaction Depends on Vps21
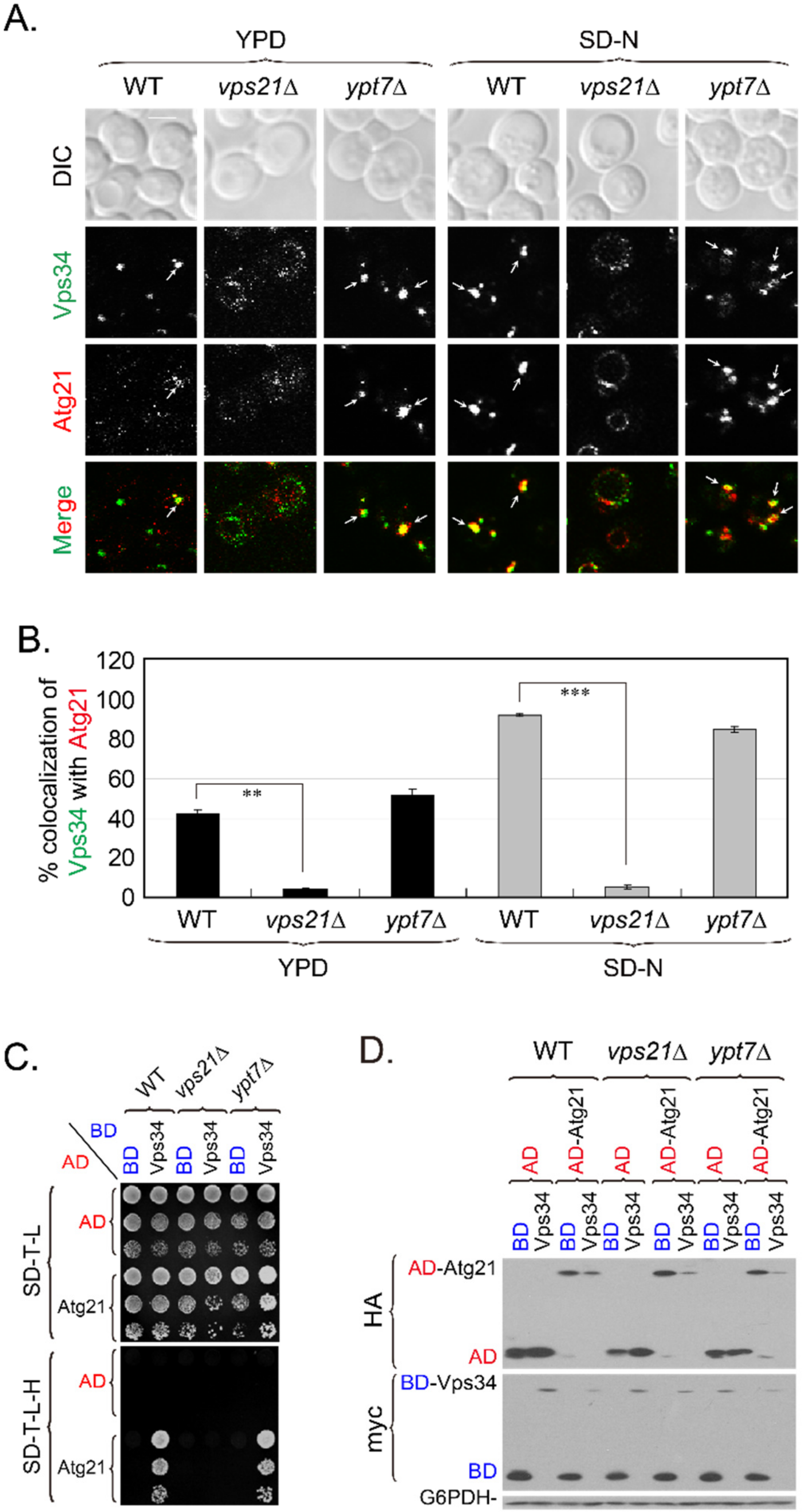
2.3. The Vps34-Atg21 Interaction on Endosomes Depends on Vps21
2.4. PI3K Complex II Subunits Mainly Localize to Endosomes and Partially Localize to the PAS, Depending on Vps21
2.5. Atg21 Mainly Localizes to Endosomes and Also to the PAS, in a Vps21-Dependent Manner
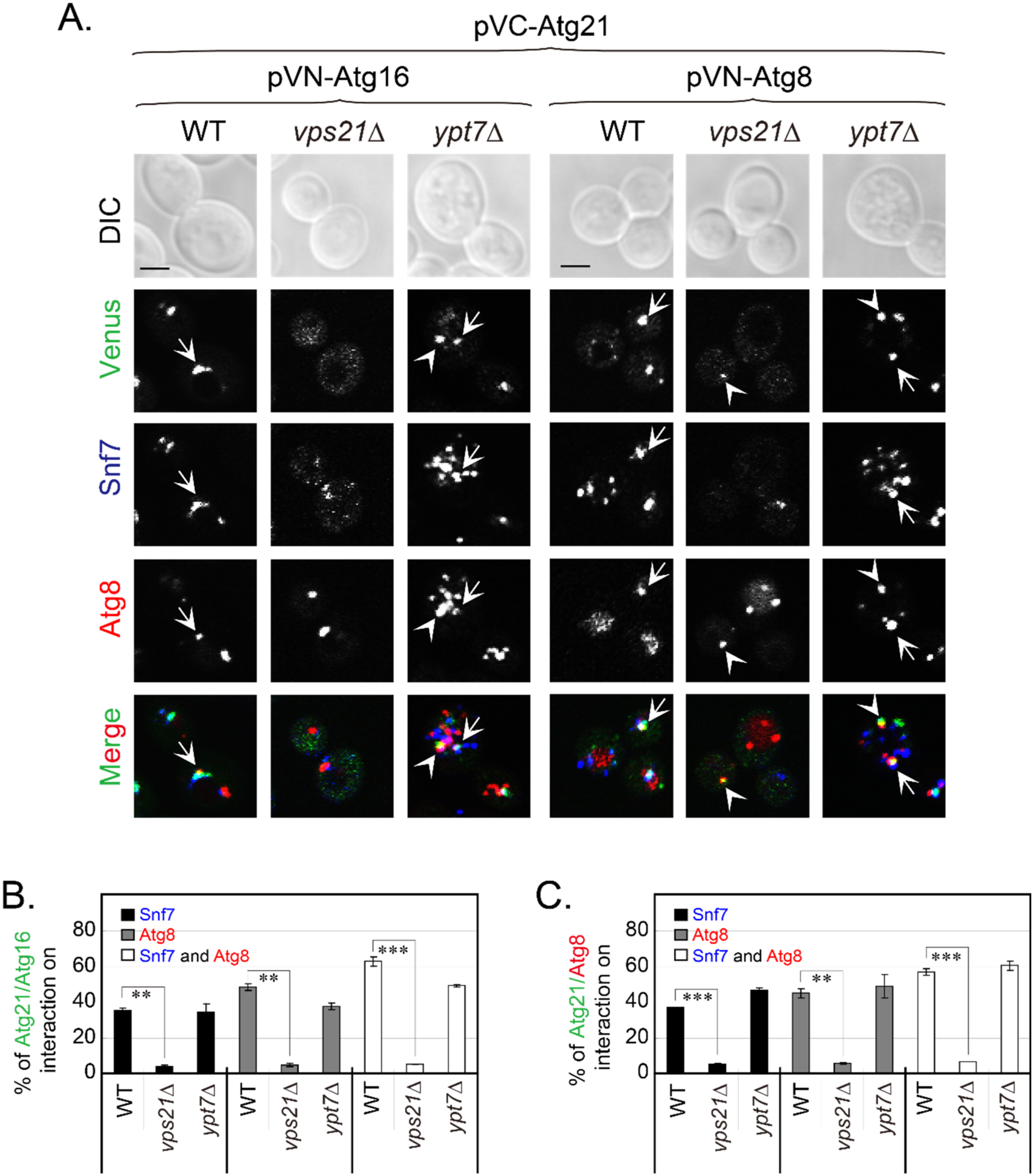
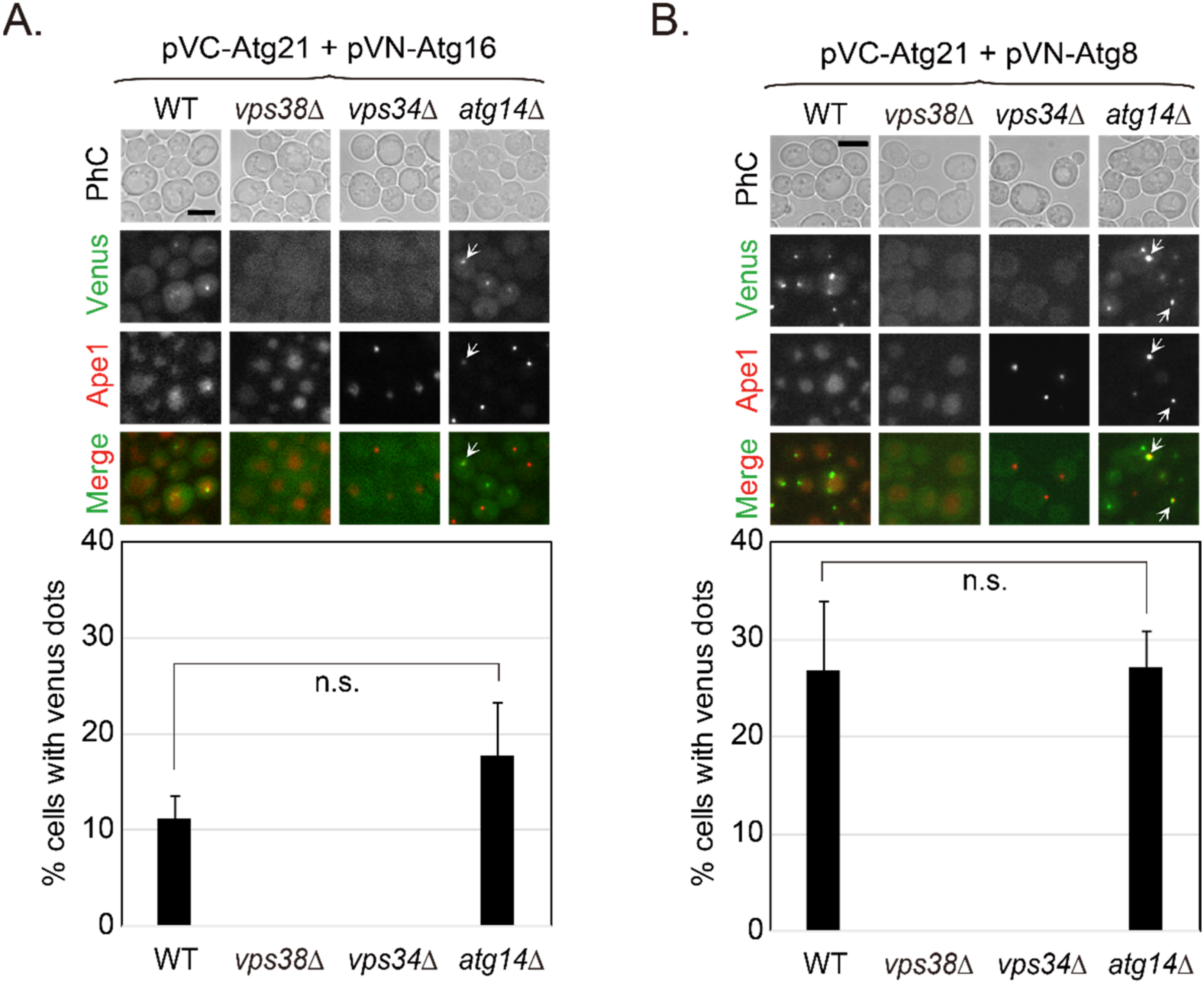
2.6. Interactions of Atg21 with Atg16 and Atg8 Depend on Vps21 and PI3K Complex II
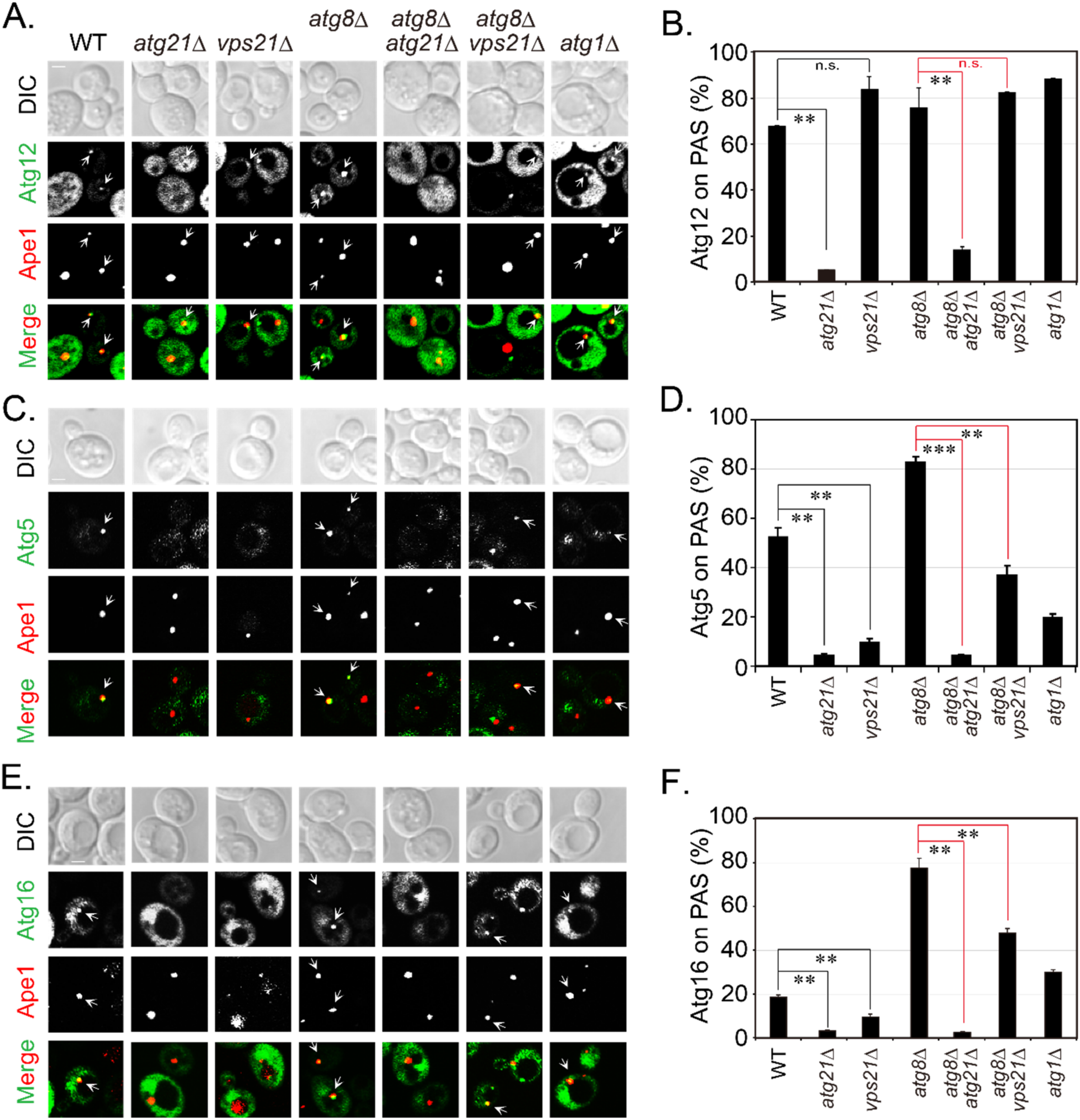
2.7. Vps21-Dependent Atg21 Localization Is Important for Atg5 and Atg16 Localization to the PAS
2.8. Vps21 and Atg21 Genetically Interact to Regulate Autophagy under Nitrogen Starvation
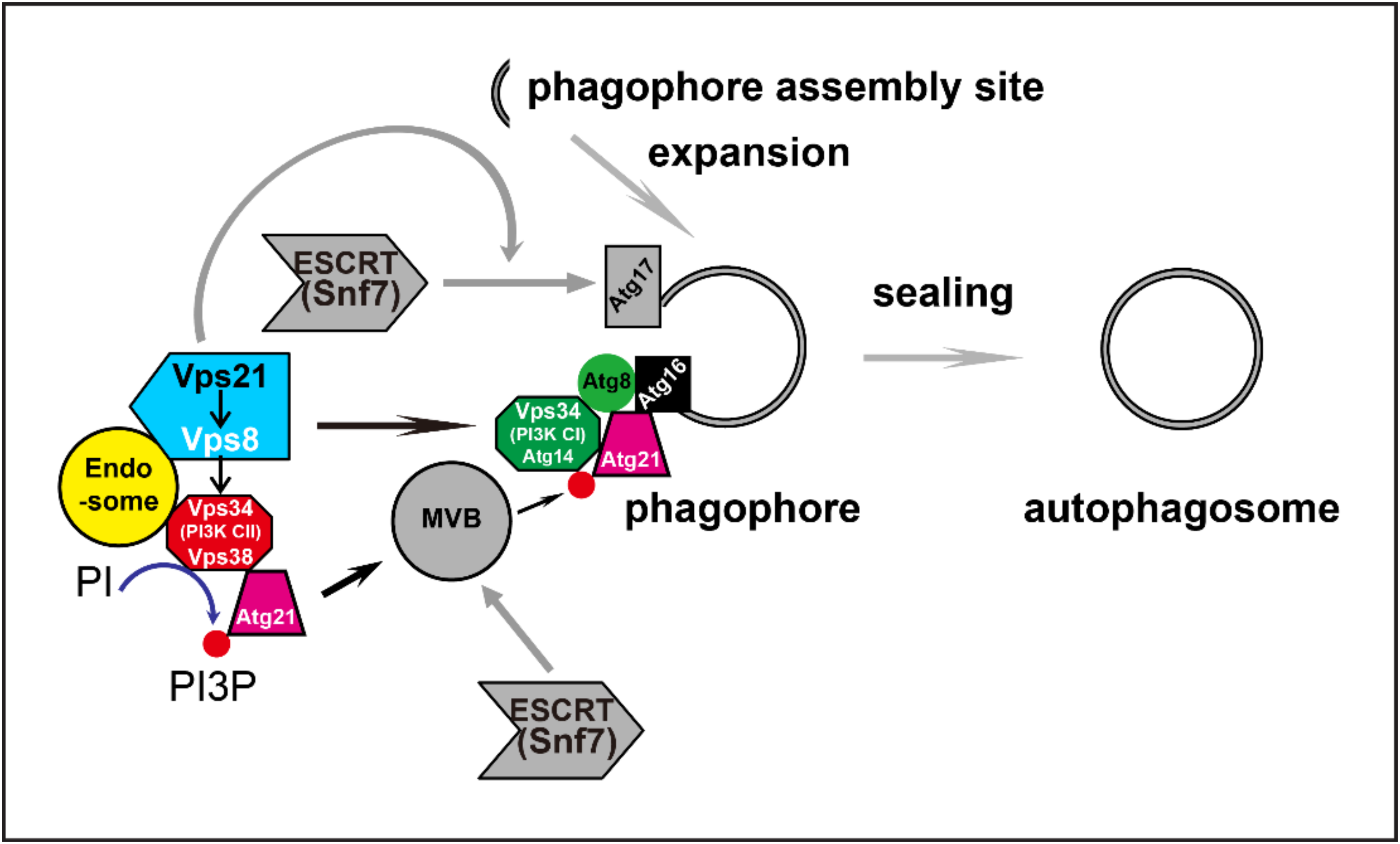
3. Discussion
4. Materials and Methods
4.1. Strains, Plasmids, and Reagents
4.2. Yeast Culture Conditions and Induction of Autophagy
4.3. Fluorescence Microscopy Observations and Quantifications
4.4. Y2H Assay
4.5. BiFC Assay
4.6. Immunoblotting Analysis
4.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef] [PubMed]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef] [PubMed]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Reggiori, F. Molecular regulation of autophagosome formation. Biochem. Soc. Trans. 2022, 50, 55–69. [Google Scholar] [CrossRef]
- Li, L.; Zhong, Q. Autophagosome-lysosome fusion: PIs to the rescue. EMBO J. 2016, 35, 1845–1847. [Google Scholar] [CrossRef]
- Vicinanza, M.; Rubinsztein, D.C. Mirror image phosphoinositides regulate autophagy. Mol. Cell. Oncol. 2016, 3, e1019974. [Google Scholar] [CrossRef][Green Version]
- Nascimbeni, A.C.; Codogno, P.; Morel, E. Phosphatidylinositol-3-phosphate in the regulation of autophagy membrane dynamics. FEBS J. 2017, 284, 1267–1278. [Google Scholar] [CrossRef]
- Schu, P.V.; Takegawa, K.; Fry, M.J.; Stack, J.H.; Waterfield, M.D.; Emr, S.D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 1993, 260, 88–91. [Google Scholar] [CrossRef]
- Hiles, I.D.; Otsu, M.; Volinia, S.; Fry, M.J.; Gout, I.; Dhand, R.; Panayotou, G.; Ruiz-Larrea, F.; Thompson, A.; Totty, N.F.; et al. Phosphatidylinositol 3-kinase: Structure and expression of the 110 kd catalytic subunit. Cell 1992, 70, 419–429. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef]
- Petiot, A.; Ogier-Denis, E.; Blommaart, E.F.; Meijer, A.J.; Codogno, P. Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000, 275, 992–998. [Google Scholar] [CrossRef]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Dooley, H.C.; Wilson, M.I.; Tooze, S.A. WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy 2015, 11, 190–191. [Google Scholar]
- Zerial, M.; McBride, H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001, 2, 107–117. [Google Scholar] [CrossRef]
- Bean, B.D.; Davey, M.; Snider, J.; Jessulat, M.; Deineko, V.; Tinney, M.; Stagljar, I.; Babu, M.; Conibear, E. Rab5-family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes. Mol. Biol. Cell 2015, 26, 1119–1128. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Takacs, Z.; Donnes, P.; Kohlbacher, O. WIPI proteins: Essential PtdIns3P effectors at the nascent autophagosome. J. Cell Sci. 2015, 128, 207–217. [Google Scholar] [CrossRef]
- Juris, L.; Montino, M.; Rube, P.; Schlotterhose, P.; Thumm, M.; Krick, R. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015, 34, 955–973. [Google Scholar] [CrossRef]
- Peplowska, K.; Markgraf, D.F.; Ostrowicz, C.W.; Bange, G.; Ungermann, C. The CORVET tethering complex interacts with the yeast Rab5 homolog Vps21 and is involved in endo-lysosomal biogenesis. Dev. Cell 2007, 12, 739–750. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, F.; Zou, S.; Yu, S.; Li, S.; Li, D.; Song, J.; Li, H.; He, Z.; Hu, B.; et al. A Vps21 endocytic module regulates autophagy. Mol. Biol. Cell 2014, 25, 3166–3177. [Google Scholar] [CrossRef]
- Zhou, F.; Zou, S.; Chen, Y.; Lipatova, Z.; Sun, D.; Zhu, X.; Li, R.; Wu, Z.; You, W.; Cong, X.; et al. A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 2017, 13, e1007020. [Google Scholar] [CrossRef]
- Zhou, F.; Wu, Z.; Zhao, M.; Murtazina, R.; Cai, J.; Zhang, A.; Li, R.; Sun, D.; Li, W.; Zhao, L.; et al. Rab5-dependent autophagosome closure by ESCRT. J. Cell Biol. 2019, 218, 1908–1927. [Google Scholar] [CrossRef]
- Zhu, L.; Jorgensen, J.R.; Li, M.; Chuang, Y.S.; Emr, S.D. ESCRTs function directly on the lysosome membrane to downregulate ubiquitinated lysosomal membrane proteins. eLife 2017, 6, e26403. [Google Scholar] [CrossRef]
- Shin, M.E.; Ogburn, K.D.; Varban, O.A.; Gilbert, P.M.; Burd, C.G. FYVE domain targets Pib1p ubiquitin ligase to endosome and vacuolar membranes. J. Biol. Chem. 2001, 276, 41388–41393. [Google Scholar] [CrossRef]
- Seals, D.F.; Eitzen, G.; Margolis, N.; Wickner, W.T.; Price, A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc. Natl. Acad. Sci. USA 2000, 97, 9402–9407. [Google Scholar] [CrossRef]
- Krick, R.; Henke, S.; Tolstrup, J.; Thumm, M. Dissecting the localization and function of Atg18, Atg21 and Ygr223c. Autophagy 2008, 4, 896–910. [Google Scholar] [CrossRef]
- Cabrera, M.; Arlt, H.; Epp, N.; Lachmann, J.; Griffith, J.; Perz, A.; Reggiori, F.; Ungermann, C. Functional separation of endosomal fusion factors and the class C core vacuole/endosome tethering (CORVET) complex in endosome biogenesis. J. Biol. Chem. 2013, 288, 5166–5175. [Google Scholar] [CrossRef]
- Horazdovsky, B.F.; Cowles, C.R.; Mustol, P.; Holmes, M.; Emr, S.D. A novel RING finger protein, Vps8p, functionally interacts with the small GTPase, Vps21p, to facilitate soluble vacuolar protein localization. J. Biol. Chem. 1996, 271, 33607–33615. [Google Scholar] [CrossRef]
- Kuma, A.; Mizushima, N.; Ishihara, N.; Ohsumi, Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002, 277, 18619–18625. [Google Scholar] [CrossRef]
- Nair, U.; Cao, Y.; Xie, Z.; Klionsky, D.J. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 2010, 285, 11476–11488. [Google Scholar] [CrossRef] [PubMed]
- Stromhaug, P.E.; Reggiori, F.; Guan, J.; Wang, C.W.; Klionsky, D.J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 2004, 15, 3553–3566. [Google Scholar] [CrossRef] [PubMed]
- Meiling-Wesse, K.; Barth, H.; Voss, C.; Eskelinen, E.L.; Epple, U.D.; Thumm, M. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J. Biol. Chem. 2004, 279, 37741–37750. [Google Scholar] [CrossRef] [PubMed]
- Munzel, L.; Neumann, P.; Otto, F.B.; Krick, R.; Metje-Sprink, J.; Kroppen, B.; Karedla, N.; Enderlein, J.; Meinecke, M.; Ficner, R.; et al. Atg21 organizes Atg8 lipidation at the contact of the vacuole with the phagophore. Autophagy 2021, 17, 1458–1478. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Wu, Z.; Zhao, M.; Segev, N.; Liang, Y. Autophagosome closure by ESCRT: Vps21/RAB5-regulated ESCRT recruitment via an Atg17-Snf7 interaction. Autophagy 2019, 15, 1653–1654. [Google Scholar] [CrossRef]
- Araki, Y.; Ku, W.C.; Akioka, M.; May, A.I.; Hayashi, Y.; Arisaka, F.; Ishihama, Y.; Ohsumi, Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J. Cell Biol. 2013, 203, 299–313. [Google Scholar] [CrossRef]
- Ohashi, Y.; Soler, N.; Garcia Ortegon, M.; Zhang, L.; Kirsten, M.L.; Perisic, O.; Masson, G.R.; Burke, J.E.; Jakobi, A.J.; Apostolakis, A.A.; et al. Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex. Autophagy 2016, 12, 2129–2144. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef]
- Eisenberg-Lerner, A.; Kimchi, A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012, 19, 788–797. [Google Scholar] [CrossRef]
- Su, H.; Yang, F.; Wang, Q.; Shen, Q.; Huang, J.; Peng, C.; Zhang, Y.; Wan, W.; Wong, C.C.L.; Sun, Q.; et al. VPS34 acetylation controls its lipid kinase activity and the initiation of canonical and non-canonical autophagy. Mol. Cell 2017, 67, 907–921.e7. [Google Scholar] [CrossRef]
- McKnight, N.C.; Zhenyu, Y. Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr. Pathobiol. Rep. 2013, 1, 231–238. [Google Scholar] [CrossRef]
- Hill, S.M.; Wrobel, L.; Ashkenazi, A.; Fernandez-Estevez, M.; Tan, K.; Burli, R.W.; Rubinsztein, D.C. VCP/p97 regulates Beclin-1-dependent autophagy initiation. Nat. Chem. Biol. 2021, 17, 448–455. [Google Scholar] [CrossRef]
- Vergne, I.; Roberts, E.; Elmaoued, R.A.; Tosch, V.; Delgado, M.A.; Proikas-Cezanne, T.; Laporte, J.; Deretic, V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009, 28, 2244–2258. [Google Scholar] [CrossRef]
- Taguchi-Atarashi, N.; Hamasaki, M.; Matsunaga, K.; Omori, H.; Ktistakis, N.T.; Yoshimori, T.; Noda, T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 2010, 11, 468–478. [Google Scholar] [CrossRef]
- Cebollero, E.; van der Vaart, A.; Zhao, M.; Rieter, E.; Klionsky, D.J.; Helms, J.B.; Reggiori, F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 2012, 22, 1545–1553. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, S.; Zhao, H.; Zou, W.; Yoshina, S.; Mitani, S.; Zhang, H.; Wang, X. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 2014, 15, 973–981. [Google Scholar] [CrossRef]
- Polson, H.E.; de Lartigue, J.; Rigden, D.J.; Reedijk, M.; Urbe, S.; Clague, M.J.; Tooze, S.A. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010, 6, 506–522. [Google Scholar] [CrossRef]
- Proikas-Cezanne, T.; Pfisterer, S.G. Assessing mammalian autophagy by WIPI-1/Atg18 puncta formation. Methods Enzymol. 2009, 452, 247–260. [Google Scholar]
- Krick, R.; Busse, R.A.; Scacioc, A.; Stephan, M.; Janshoff, A.; Thumm, M.; Kuhnel, K. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc. Natl. Acad. Sci. USA 2012, 109, E2042–E2049. [Google Scholar] [CrossRef]
- Baskaran, S.; Ragusa, M.J.; Boura, E.; Hurley, J.H. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol. Cell 2012, 47, 339–348. [Google Scholar] [CrossRef]
- Dooley, H.C.; Razi, M.; Polson, H.E.; Girardin, S.E.; Wilson, M.I.; Tooze, S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell 2014, 55, 238–252. [Google Scholar] [CrossRef]
- Fracchiolla, D.; Chang, C.; Hurley, J.H.; Martens, S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. J. Cell Biol. 2020, 219, e201912098. [Google Scholar] [CrossRef]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.; Lamark, T.; et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef]
- Deretic, V. A master conductor for aggregate clearance by autophagy. Dev. Cell 2010, 18, 694–696. [Google Scholar] [CrossRef][Green Version]
- Grotemeier, A.; Alers, S.; Pfisterer, S.G.; Paasch, F.; Daubrawa, M.; Dieterle, A.; Viollet, B.; Wesselborg, S.; Proikas-Cezanne, T.; Stork, B. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell. Signal. 2010, 22, 914–925. [Google Scholar] [CrossRef]
- Bhati, K.K.; Luong, A.M.; Batoko, H. VPS34 Complexes in Plants: Untangled Enough? Trends Plant Sci. 2021, 26, 303–305. [Google Scholar] [CrossRef]
- Tasnin, M.N.; Ito, K.; Katsuta, H.; Takuma, T.; Sharmin, T.; Ushimaru, T. The PI3 kinase complex II-PI3P-Vps27 axis on vacuolar membranes is critical for microautophagy induction and nutrient stress adaptation. J. Mol. Biol. 2022, 434, 167360. [Google Scholar] [CrossRef]
- Liu, F.; Hu, W.; Li, F.; Marshall, R.S.; Zarza, X.; Munnik, T.; Vierstra, R.D. AUTOPHAGY-RELATED14 and its associated phosphatidylinositol 3-kinase complex promote autophagy in Arabidopsis. Plant Cell 2020, 32, 3939–3960. [Google Scholar] [CrossRef]
- Vicinanza, M.; Korolchuk, V.I.; Ashkenazi, A.; Puri, C.; Menzies, F.M.; Clarke, J.H.; Rubinsztein, D.C. PI(5)P regulates autophagosome biogenesis. Mol. Cell 2015, 57, 219–234. [Google Scholar] [CrossRef]
- McEwan, D.G.; Ryan, K.M. ATG2 and VPS13 proteins: Molecular highways transporting lipids to drive membrane expansion and organelle communication. FEBS J. 2021. [Google Scholar] [CrossRef]
- Bas, L.; Papinski, D.; Licheva, M.; Torggler, R.; Rohringer, S.; Schuschnig, M.; Kraft, C. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J. Cell Biol. 2018, 217, 3656–3669. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.; Jotwani, A.; Geng, J.; Gammoh, N.; Richerson, D.; Yen, W.L.; Griffith, J.; Nag, S.; Wang, K.; Moss, T.; et al. SNARE proteins are required for macroautophagy. Cell 2011, 146, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mao, K.; Yu, A.Y.; Omairi-Nasser, A.; Austin, J., 2nd; Glick, B.S.; Yip, C.K.; Klionsky, D.J. The Atg17-Atg31-Atg29 complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr. Biol. 2016, 26, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Morozova, N.; Tokarev, A.A.; Mulholland, J.W.; Segev, N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol. Biol. Cell 2007, 18, 2533–2541. [Google Scholar] [CrossRef]
- Morozova, N.; Liang, Y.; Tokarev, A.A.; Chen, S.H.; Cox, R.; Andrejic, J.; Lipatova, Z.; Sciorra, V.A.; Emr, S.D.; Segev, N. TRAPPII subunits are required for the specificity switch of a Ypt-Rab GEF. Nat. Cell Biol. 2006, 8, 1263–1269. [Google Scholar] [CrossRef]
- Shintani, T.; Reggiori, F. Fluorescence microscopy-based assays for monitoring yeast Atg protein trafficking. Methods Enzymol. 2008, 451, 43–56. [Google Scholar]
- Gong, T.; Liao, Y.; He, F.; Yang, Y.; Yang, D.D.; Chen, X.D.; Gao, X.D. Control of polarized growth by the Rho family GTPase Rho4 in budding yeast: Requirement of the N-terminal extension of Rho4 and regulation by the Rho GTPase-activating protein Bem2. Eukaryot. Cell 2013, 12, 368–377. [Google Scholar] [CrossRef]
- Tokarev, A.A.; Taussig, D.; Sundaram, G.; Lipatova, Z.; Liang, Y.; Mulholland, J.W.; Segev, N. TRAPP II complex assembly requires Trs33 or Trs65. Traffic 2009, 10, 1831–1844. [Google Scholar] [CrossRef]
- Zou, S.; Chen, Y.; Liu, Y.; Segev, N.; Yu, S.; Liu, Y.; Min, G.; Ye, M.; Zeng, Y.; Zhu, X.; et al. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic 2013, 14, 233–246. [Google Scholar] [CrossRef]
- Sung, M.K.; Huh, W.K. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast 2007, 24, 767–775. [Google Scholar] [CrossRef]
- Graef, M.; Friedman, J.R.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Z.T.; Tang, S.W.; Zhao, B.S.; Li, H.; Song, J.Z.; Li, D.; Xie, Z. A Validated Set of Fluorescent-Protein-Based Markers for Major Organelles in Yeast (Saccharomyces cerevisiae). mBio 2019, 10, e01691-19. [Google Scholar] [CrossRef]
- Katzmann, D.J.; Stefan, C.J.; Babst, M.; Emr, S.D. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 2003, 162, 413–423. [Google Scholar] [CrossRef]
- Markgraf, D.F.; Ahnert, F.; Arlt, H.; Mari, M.; Peplowska, K.; Epp, N.; Griffith, J.; Reggiori, F.; Ungermann, C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell 2009, 20, 5276–5289. [Google Scholar] [CrossRef]
- Li, D.; Song, J.Z.; Shan, M.H.; Li, S.P.; Liu, W.; Li, H.; Zhu, J.; Wang, Y.; Lin, J.; Xie, Z. A fluorescent tool set for yeast Atg proteins. Autophagy 2015, 11, 954–960. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; You, W.; Sun, D.; Xu, H.; You, X.; Xu, H.; Wu, Z.; Xie, Z.; Liang, Y. Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy. Int. J. Mol. Sci. 2022, 23, 9550. https://doi.org/10.3390/ijms23179550
Zhao L, You W, Sun D, Xu H, You X, Xu H, Wu Z, Xie Z, Liang Y. Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy. International Journal of Molecular Sciences. 2022; 23(17):9550. https://doi.org/10.3390/ijms23179550
Chicago/Turabian StyleZhao, Lei, Weiming You, Dan Sun, Hui Xu, Xia You, Haiqian Xu, Zulin Wu, Zhiping Xie, and Yongheng Liang. 2022. "Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy" International Journal of Molecular Sciences 23, no. 17: 9550. https://doi.org/10.3390/ijms23179550
APA StyleZhao, L., You, W., Sun, D., Xu, H., You, X., Xu, H., Wu, Z., Xie, Z., & Liang, Y. (2022). Vps21 Directs the PI3K-PI(3)P-Atg21-Atg16 Module to Phagophores via Vps8 for Autophagy. International Journal of Molecular Sciences, 23(17), 9550. https://doi.org/10.3390/ijms23179550





