Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells
Abstract
1. Introduction
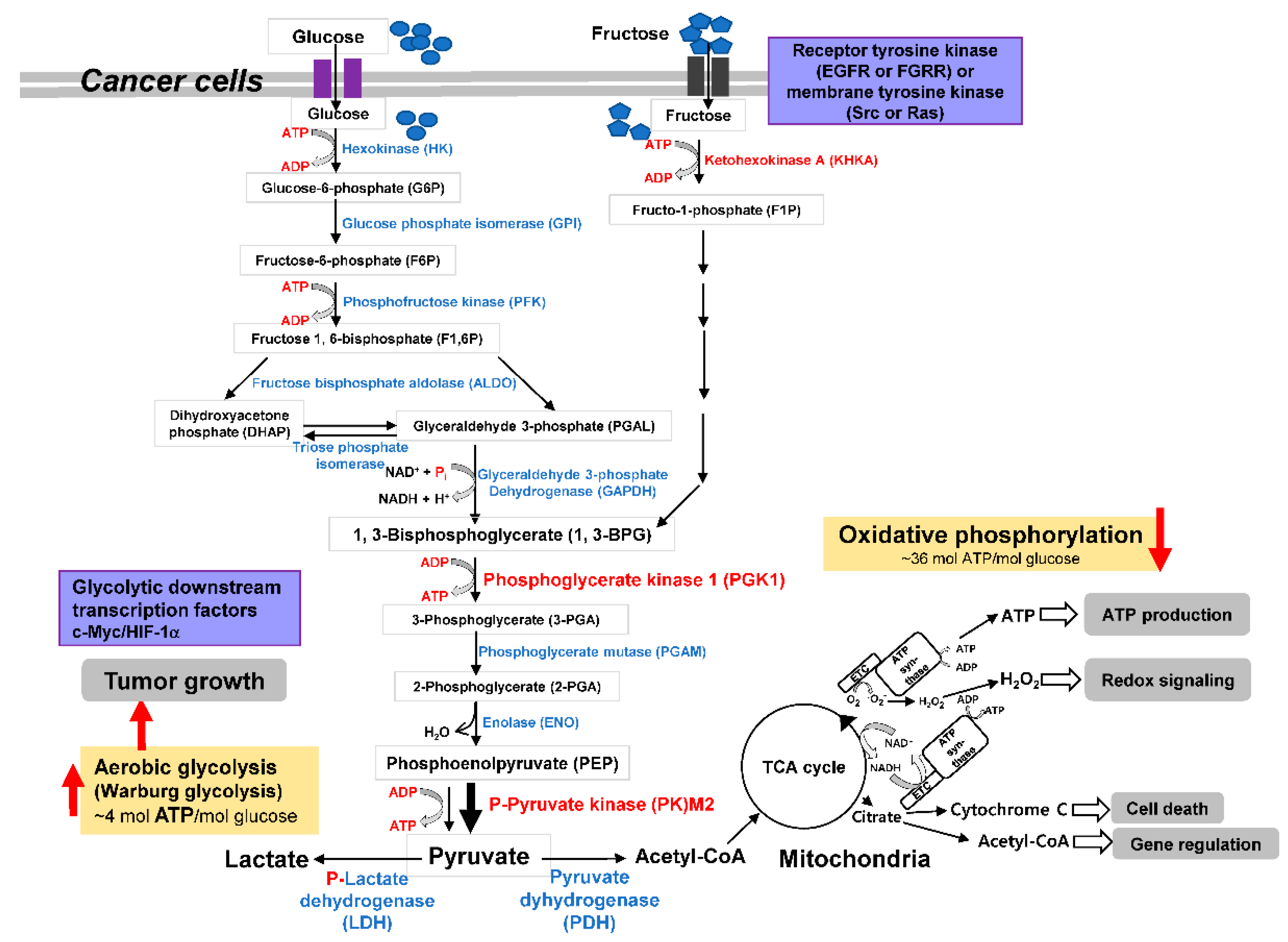
2. Carbohydrate Metabolic Reprogramming and Adaptation and Direction Decision of Tumor Cell Metabolism
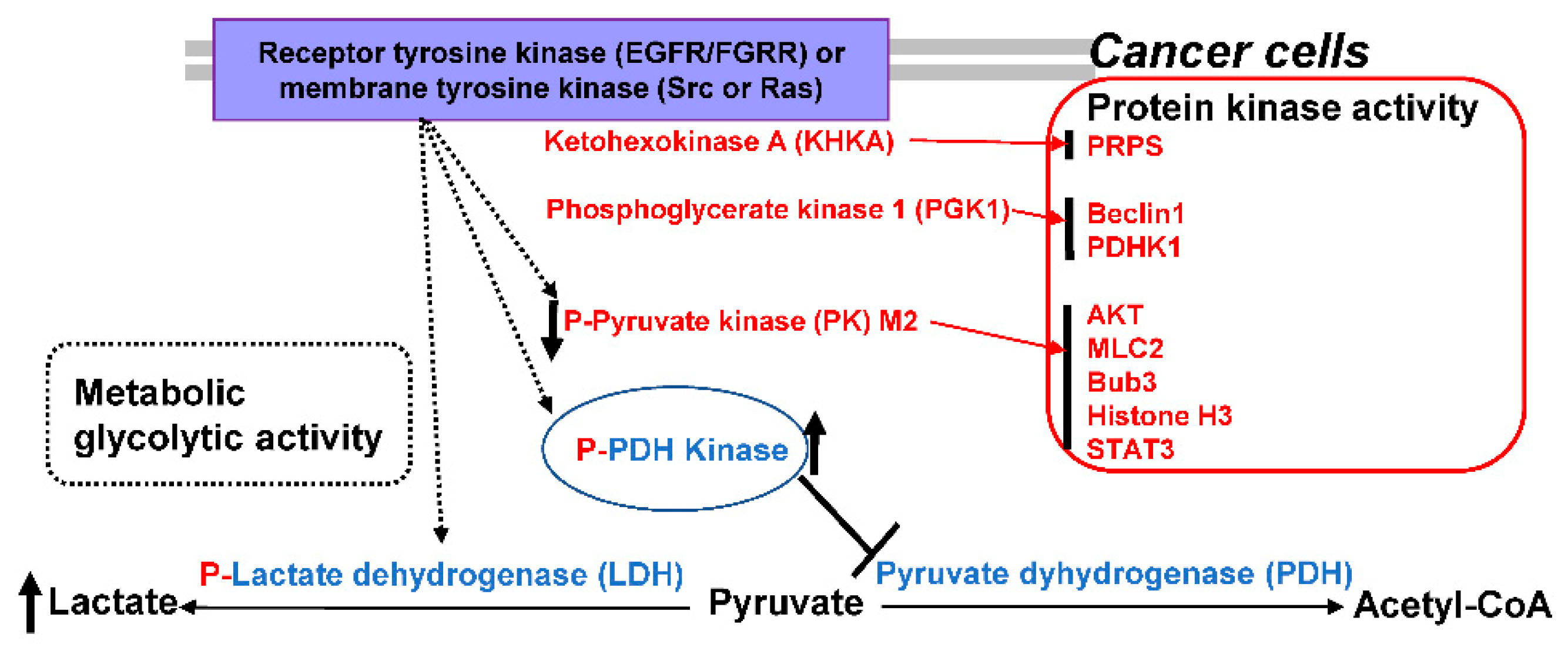
3. Revisiting the Warburg Effect where Metabolic Enzymes Suppress Tumor Behavior
4. Initial Step of Glc Metabolism in Tumor Metabolic Pathway
5. HK Inhibition in Cancer Cells
6. Keto-HK Inhibition in Cancer Cells
7. GPI Inhibition in Cancer Cells
8. PFK/PFKFB Inhibition in Cancer Cells
9. FBP and PKM2 Inhibition in Cancer Cells
10. Fructose-Bisphosphate Aldolase A (ALDOA) Inhibition in Cancer Cells
11. TPI Inhibition in Cancer Cells
12. GAPDH Inhibition in Cancer Cells
13. PGK Inhibition in Cancer Cells
14. PGAM Inhibition in Cancer Cells
15. ENO1 Inhibition in Cancer Cells
16. LDH Inhibition in Cancer Cells
17. PDHK Inhibition in Cancer Cells
18. Critical View of Anti-Cancer Therapeutics Based on Understanding the Warburg Effect
19. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.C.; Pohlmann, P.R.; Clarke, R.; Sengupta, S. Treatment against glucose-dependent cancers through metabolic PFKFB3 targeting of glycolytic flux. Cancer Metastasis Rev. 2022, 41, 447–458. [Google Scholar] [CrossRef]
- Brand, A.; Singer, K.; Koehl, G.E.; Kolitzus, M.; Schoenhammer, G.; Thiel, A.; Matos, C.; Bruss, C.; Klobuch, S.; Peter, K.; et al. LDHA-Associated Lactic Acid Production Blunts Tumor Immunosurveillance by T and NK Cells. Cell Metab. 2016, 24, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Marchiq, I.; Le Floch, R.; Roux, D.; Simon, M.P.; Pouyssegur, J. Genetic disruption of lactate/H+ symporters (MCTs) and their subunit CD147/BASIGIN sensitizes glycolytic tumor cells to phenformin. Cancer Res. 2015, 75, 171–180. [Google Scholar] [CrossRef]
- de Padua, M.C.; Delodi, G.; Vučetić, M.; Durivault, J.; Vial, V.; Bayer, P.; Noleto, G.R.; Mazure, N.M.; Ždralević, M.; Pouysségur, J. Disrupting glucose-6-phosphate isomerase fully suppresses the “Warburg effect” and activates OXPHOS with minimal impact on tumor growth except in hypoxia. Oncotarget 2017, 8, 87623–87637. [Google Scholar] [CrossRef]
- Nielson, T.C.; Le, H.V. Inhibition of Glycolysis and Glutaminolysis: An Emerging Drug Discovery Approach to Combat Cancer. Curr. Top. Med. Chem. 2018, 18, 494–504. [Google Scholar] [CrossRef]
- Available online: https://clinicaltrials.gov (accessed on 15 June 2019).
- Zhelev, Z.; Aoki, I.; Lazarova, D.; Vlaykova, T.; Higashi, T.; Bakalova, R. A “Weird” Mitochondrial Fatty Acid Oxidation as a Metabolic “Secret” of Cancer. Oxidative Med. Cell. Longev. 2022, 2022, 2339584. [Google Scholar] [CrossRef]
- Hönigova, K.; Navratil, J.; Peltanova, B.; Polanska, H.H.; Raudenska, M.; Masarik, M. Metabolic tricks of cancer cells. Biochim. Biophys. Acta 2022, 1877, 188705. [Google Scholar] [CrossRef]
- Li, T.; Tian, Y.; Wang, Y.; Cui, Z.; He, Z.; Wu, X.; Zhang, Y.; Jiang, H. Kiss1 Inhibits the Proliferation of Nasopharyngeal Carcinoma Cells Via Activation of the LKB1/AMPK Pathway. Front. Oncol. 2022, 11, 724251. [Google Scholar] [CrossRef]
- Vousden, K.H.; Ryan, K.M. p53 and metabolism. Nat. Rev. Cancer 2009, 9, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sinha, S.; Shrivastava, N. Therapeutic Targeting Hypoxia-Inducible Factor (HIF-1) in Cancer: Cutting Gordian Knot of Cancer Cell Metabolism. Front. Genet. 2022, 13, 849040. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Van Lith, S.; Navis, A.C.; Lenting, K.; Verrijp, K.; Schepens, J.T.G.; Hendriks, W.J.A.J.; Schubert, N.A.; Venselaar, H.; Wevers, R.A.; Van Rooij, A.; et al. Identification of a novel inactivating mutation in Isocitrate Dehydrogenase 1 (IDH1-R314C) in a high grade astrocytoma. Sci. Rep. 2016, 6, 30486. [Google Scholar] [CrossRef]
- Christofk, H.R.; Vander Heiden, M.G.; Harris, M.H.; Ramanathan, A.; Gerszten, R.E.; Wei, R.; Fleming, M.D.; Schreiber, S.L.; Cantley, L.C. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 2008, 452, 230–233. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S.; Yu, D. Protein kinase function of pyruvate kinase M2 and cancer. Cancer Cell Int. 2020, 20, 523. [Google Scholar] [CrossRef]
- Hitosugi, T.; Kang, S.; Heiden, M.G.V.; Chung, T.-W.; Elf, S.; Lythgoe, K.; Dong, S.; Lonial, S.; Wang, X.; Chen, G.Z.; et al. Tyrosine Phosphorylation Inhibits PKM2 to Promote the Warburg Effect and Tumor Growth. Sci. Signal. 2009, 2, ra73. [Google Scholar] [CrossRef]
- Bensard, C.L.; Wisidagama, D.R.; Olson, K.A.; Berg, J.A.; Krah, N.M.; Schell, J.C.; Nowinski, S.M.; Fogarty, S.; Bott, A.J.; Wei, P.; et al. Regulation of Tumor Initiation by the Mitochondrial Pyruvate Carrier. Cell Metab. 2020, 31, 284–300.e7. [Google Scholar] [CrossRef]
- Mathupala, S.P.; Ko, Y.H.; Pedersen, P.L. Hexokinase-2 bound to mitochondria: Cancer‘s stygian link to the “Warburg effect” and a pivotal target for effective therapy. Semin. Cancer Biol. 2009, 19, 17–24. [Google Scholar] [CrossRef]
- Adams, V.; Griffin, L.D.; Gelb, B.D.; McCabe, E.R. Protein kinase activity of rat brain hexokinase. Biochem. Biophys. Res. Commun. 1991, 177, 1101–1106. [Google Scholar] [CrossRef]
- Kim, J.-W.; Gao, P.; Liu, Y.-C.; Semenza, G.L.; Dang, C.V. Hypoxia-Inducible Factor 1 and Dysregulated c-Myc Cooperatively Induce Vascular Endothelial Growth Factor and Metabolic Switches Hexokinase 2 and Pyruvate Dehydrogenase Kinase 1. Mol. Cell. Biol. 2007, 27, 7381–7393. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Wu, C.; Yang, K.; Yang, Y.; Liu, Y.; Gao, S.; Wang, Q.; Li, C.; Chen, L.; Li, H. Novel selective hexokinase 2 inhibitor Benitrobenrazide blocks cancer cells growth by targeting glycolysis. Pharmacol. Res. 2020, 164, 105367. [Google Scholar] [CrossRef] [PubMed]
- Gitenay, D.; Wiel, C.; Lallet-Daher, H.; Vindrieux, D.; Aubert, S.; Payen, L.; Simonnet, H.; Bernard, D. Glucose metabolism and hexosamine pathway regulate oncogene-induced senescence. Cell Death Dis. 2014, 5, e1089. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Onodera, Y.; Nam, J.-M.; Bissell, M.J. Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J. Clin. Investig. 2014, 124, 367–384. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, G.; Liu, Z.; Liu, S.; Cai, Z.; You, P.; Ke, Y.; Lai, L.; Huang, Y.; Gao, H.; et al. Aberrant FGFR Tyrosine Kinase Signaling Enhances the Warburg Effect by Reprogramming LDH Isoform Expression and Activity in Prostate Cancer. Cancer Res. 2018, 78, 4459–4470. [Google Scholar] [CrossRef]
- Brown, N.; Higham, S.E.; Perunovic, B.; Arafa, M.; Balasubramanian, S.; Rehman, I. Lactate Dehydrogenase-B Is Silenced by Promoter Methylation in a High Frequency of Human Breast Cancers. PLoS ONE 2013, 8, e57697. [Google Scholar] [CrossRef]
- Li, X.; Qian, X.; Lu, Z. Fructokinase A acts as a protein kinase to promote nucleotide synthesis. Cell Cycle 2016, 15, 2689–2690. [Google Scholar] [CrossRef]
- Asipu, A.; Hayward, B.E.; O’Reilly, J.; Bonthron, D.T. Properties of Normal and Mutant Recombinant Human Ketohexokinases and Implications for the Pathogenesis of Essential Fructosuria. Diabetes 2003, 52, 2426–2432. [Google Scholar] [CrossRef]
- Kim, J.-W.; Zeller, K.I.; Wang, Y.; Jegga, A.G.; Aronow, B.J.; O‘Donnell, K.A.; Dang, C.V. Evaluation of Myc E-Box Phylogenetic Footprints in Glycolytic Genes by Chromatin Immunoprecipitation Assays. Mol. Cell. Biol. 2004, 24, 5923–5936. [Google Scholar] [CrossRef]
- Fairbank, M.; St-Pierre, P.; Nabi, I.R. The complex biology of autocrine motility factor/phosphoglucose isomerase (AMF/PGI) and its receptor, the gp78/AMFR E3 ubiquitin ligase. Mol. BioSyst. 2009, 5, 793–801. [Google Scholar] [CrossRef]
- Boudreau, A.; Purkey, H.E.; Hitz, A.; Robarge, K.; Peterson, D.; Labadie, S.; Kwong, M.; Hong, R.; Gao, M.; Del Nagro, C.; et al. Metabolic plasticity underpins innate and acquired resistance to LDHA inhibition. Nat. Chem. Biol. 2016, 12, 779–786. [Google Scholar] [CrossRef] [PubMed]
- El-Maghrabi, M.R.; Noto, F.; Wu, N.; Manes, N. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase: Suiting structure to need, in a family of tissue-specific enzymes. Curr. Opin. Clin. Nutr. Metab. Care 2001, 4, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Leite, T.C.; Da Silva, D.; Coelho, R.G.; Zancan, P.; Sola-Penna, M. Lactate favours the dissociation of skeletal muscle 6-phosphofructo-1-kinase tetramers down-regulating the enzyme and muscle glycolysis. Biochem. J. 2007, 408, 123–130. [Google Scholar] [CrossRef]
- Usenik, A.; Legiša, M. Evolution of Allosteric Citrate Binding Sites on 6-phosphofructo-1-kinase. PLoS ONE 2010, 5, e15447. [Google Scholar] [CrossRef]
- Bartrons, R.; Rodríguez-García, A.; Simon-Molas, H.; Castaño, E.; Manzano, A.; Navarro-Sabaté, À. The potential utility of PFKFB3 as a therapeutic target. Expert Opin. Ther. Targets 2018, 22, 659–674. [Google Scholar] [CrossRef]
- Li, X.; Liu, J.; Qian, L.; Ke, H.; Yao, C.; Tian, W.; Liu, Y.; Zhang, J. Expression of PFKFB3 and Ki67 in lung adenocarcinomas and targeting PFKFB3 as a therapeutic strategy. Mol. Cell. Biochem. 2018, 445, 123–134. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, T.; Goldhardt, T.; Edelmann, M.; Rogge, T.; Rauch, K.; Kyuchukov, N.; Menck, K.; Bleckmann, A.; Kalucka, J.; Khan, S.; et al. Effects of the Novel PFKFB3 Inhibitor KAN0438757 on Colorectal Cancer Cells and Its Systemic Toxicity Evaluation In Vivo. Cancers 2021, 13, 1011. [Google Scholar] [CrossRef]
- Shi, L.; Pan, H.; Liu, Z.; Xie, J.; Han, W. Roles of PFKFB3 in cancer. Signal Transduct. Target. Ther. 2017, 2, 17044. [Google Scholar] [CrossRef]
- Wang, Y.; Qu, C.; Liu, T.; Wang, C. PFKFB3 inhibitors as potential anticancer agents: Mechanisms of action, current developments, and structure-activity relationships. Eur. J. Med. Chem. 2020, 203, 112612. [Google Scholar] [CrossRef]
- Al Hasawi, N.; Alkandari, M.F.; Luqmani, Y.A. Phosphofructokinase: A mediator of glycolytic flux in cancer progression. Crit. Rev. Oncol. 2014, 92, 312–321. [Google Scholar] [CrossRef]
- Wang, B.; Yuan, Y.; Zou, Y.; Qi, Z.; Huang, G.; Liu, Y.; Xia, S.; Huang, Y.; Huang, Z. Fructose-1,6-bisphosphatase 2 represses cervical cancer progression via inhibiting aerobic glycolysis through promoting pyruvate kinase isozyme type M2 ubiquitination. Anti-Cancer Drugs 2022, 33, e198–e206. [Google Scholar] [CrossRef] [PubMed]
- Huangyang, P.; Li, F.; Lee, P.; Nissim, I.; Weljie, A.M.; Mancuso, A.; Li, B.; Keith, B.; Yoon, S.S.; Simon, M.C. Fructose-1,6-bisphosphatase 2 inhibits sarcoma progression by restraining mitochondrial biogenesis. Cell Metab. 2020, 31, 1032. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, J.; Li, N.; Qian, Z.; Zhu, M.; Li, Q.; Zheng, J.; Wang, X.; Shi, G. Promoter Hypermethylation Mediated Downregulation of FBP1 in Human Hepatocellular Carcinoma and Colon Cancer. PLoS ONE 2011, 6, e25564. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O‘Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Lu, Z.; Hunter, T. Metabolic Kinases Moonlighting as Protein Kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef]
- Desai, S.; Ding, M.; Wang, B.; Lu, Z.; Zhao, Q.; Shaw, K.; Yung, W.A.; Weinstein, J.N.; Tan, M.; Yao, J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 2014, 5, 8202–8210. [Google Scholar] [CrossRef]
- Wang, H.-J.; Hsieh, Y.-J.; Cheng, W.-C.; Lin, C.-P.; Lin, Y.-S.; Yang, S.-F.; Chen, C.-C.; Izumiya, Y.; Yu, J.-S.; Kung, H.-J.; et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α–mediated glucose metabolism. Proc. Natl. Acad. Sci. USA 2014, 111, 279–284. [Google Scholar] [CrossRef]
- Lv, L.; Xu, Y.-P.; Zhao, D.; Li, F.-L.; Wang, W.; Sasaki, N.; Jiang, Y.; Zhou, X.; Li, T.-T.; Guan, K.-L.; et al. Mitogenic and Oncogenic Stimulation of K433 Acetylation Promotes PKM2 Protein Kinase Activity and Nuclear Localization. Mol. Cell 2013, 52, 340–352. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.K.; Han, Y.-M.; Kim, J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int. J. Biochem. Cell Biol. 2008, 40, 1043–1054. [Google Scholar] [CrossRef]
- Mikawa, T.; Lleonart, M.E.; Takaori-Kondo, A.; Inagaki, N.; Yokode, M.; Kondoh, H. Dysregulated glycolysis as an oncogenic event. Cell. Mol. Life Sci. 2015, 72, 1881–1892. [Google Scholar] [CrossRef]
- Yang, W.; Xia, Y.; Hawke, D.; Li, X.; Liang, J.; Xing, D.; Aldape, K.; Hunter, T.; Yung, W.K.A.; Lu, Z. PKM2 Phosphorylates Histone H3 and Promotes Gene Transcription and Tumorigenesis. Cell 2012, 150, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Xia, Y.; Ji, H.; Zheng, Y.; Liang, J.; Huang, W.; Gao, X.; Aldape, K.; Lu, Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature 2011, 480, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Son, J.Y.; Yoon, S.; Tae, I.H.; Park, Y.J.; De, U.; Jeon, Y.; Park, Y.J.; Rhyu, I.J.; Lee, B.M.; Chung, K.-H.; et al. Novel therapeutic roles of MC-4 in combination with everolimus against advanced renal cell carcinoma by dual targeting of Akt/pyruvate kinase muscle isozyme M2 and mechanistic target of rapamycin complex 1 pathways. Cancer Med. 2018, 7, 5083–5095. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, J.; Zhang, W.; Luo, H.; Shen, Z.; Cheng, H.; Zhu, X. PKM2 enhances chemosensitivity to cisplatin through interaction with the mTOR pathway in cervical cancer. Sci. Rep. 2016, 6, 30788. [Google Scholar] [CrossRef]
- Suzuki, A.; Puri, S.; Leland, P.; Puri, A.; Moudgil, T.; Fox, B.A.; Puri, R.K.; Joshi, B.H. Subcellular compartmentalization of PKM2 identifies anti-PKM2 therapy response in vitro and in vivo mouse model of human non-small-cell lung cancer. PLoS ONE 2019, 14, e0217131. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z. Pyruvate kinase M2 at a glance. J. Cell Sci. 2015, 128, 1655–1660. [Google Scholar] [CrossRef]
- Jiang, Y.; Li, X.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; Wei, C.; Guo, F.; Chen, Y.; et al. PKM2 Regulates Chromosome Segregation and Mitosis Progression of Tumor Cells. Mol. Cell 2014, 53, 75–87. [Google Scholar] [CrossRef]
- He, C.-L.; Bian, Y.-Y.; Xue, Y.; Liu, Z.-X.; Zhou, K.-Q.; Yao, C.-F.; Lin, Y.; Zou, H.-F.; Luo, F.-X.; Qu, Y.-Y.; et al. Pyruvate Kinase M2 Activates mTORC1 by Phosphorylating AKT1S1. Sci. Rep. 2016, 6, 21524. [Google Scholar] [CrossRef]
- Liang, J.; Cao, R.; Wang, X.; Zhang, Y.; Wang, P.; Gao, H.; Li, C.; Yang, F.; Zeng, R.; Wei, P.; et al. Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2. Cell Res. 2016, 27, 329–351. [Google Scholar] [CrossRef]
- Tochio, T.; Tanaka, H.; Nakata, S.; Hosoya, H. Fructose-1,6-bisphosphate aldolase A is involved in HaCaT cell migration by inducing lamellipodia formation. J. Dermatol. Sci. 2010, 58, 123–129. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Li, Q.-G.; Xue, J.-J.; Wang, Z.; Yuan, X.; Tong, J.-D.; Xu, L.-C. Downregulation of FBP1 Promotes Tumor Metastasis and Indicates Poor Prognosis in Gastric Cancer via Regulating Epithelial-Mesenchymal Transition. PLoS ONE 2016, 11, e0167857. [Google Scholar] [CrossRef] [PubMed]
- Li, X.R.; Zhou, K.Q.; Yin, Z.; Gao, Y.L.; Yang, X. Knockdown of FBP1 enhances radiosensitivity in prostate cancer cells by activating autophagy. Neoplasma 2020, 67, 982–991. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, L.; Pei, J.; Wang, Z.; Zhang, J.; Wang, B. CELF6 modulates triple-negative breast cancer progression by regulating the stability of FBP1 mRNA. Breast Cancer Res. Treat. 2020, 183, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Guan, Z.; Hao, L.; Song, Y.; Wang, L.; Gong, L.; Liu, L.; Qi, X.; Hou, Z.; Shao, S. Fructose-Bisphosphate Aldolase A Is a Potential Metastasis-Associated Marker of Lung Squamous Cell Carcinoma and Promotes Lung Cell Tumorigenesis and Migration. PLoS ONE 2014, 9, e85804. [Google Scholar] [CrossRef]
- Kawai, K.; Uemura, M.; Munakata, K.; Takahashi, H.; Haraguchi, N.; Nishimura, J.; Hata, T.; Matsuda, C.; Ikenaga, M.; Murata, K.; et al. Fructose-bisphosphate aldolase A is a key regulator of hypoxic adaptation in colorectal cancer cells and involved in treatment resistance and poor prognosis. Int. J. Oncol. 2017, 50, 525–534. [Google Scholar] [CrossRef]
- Chang, Y.-C.; Chiou, J.; Yang, Y.-F.; Su, C.-Y.; Lin, Y.-F.; Yang, C.-N.; Lu, P.-J.; Huang, M.-S.; Yang, C.-J.; Hsiao, M. Therapeutic Targeting of Aldolase A Interactions Inhibits Lung Cancer Metastasis and Prolongs Survival. Cancer Res. 2018, 79, 4754–4766. [Google Scholar] [CrossRef]
- Zhang, C.; Zhao, L.-M.; Wu, H.; Tian, G.; Dai, S.-L.; Zhao, R.-Y.; Shan, B.-E. C/D-Box Snord105b Promotes Tumorigenesis in Gastric Cancer via ALDOA/C-Myc Pathway. Cell. Physiol. Biochem. 2018, 45, 2471–2482. [Google Scholar] [CrossRef]
- Li, C.-H.; Chan, M.-H.; Chang, Y.-C. The role of fructose 1,6-bisphosphate-mediated glycolysis/gluconeogenesis genes in cancer prognosis. Aging 2022, 14, 3233–3258. [Google Scholar] [CrossRef]
- Lew, C.R.; Tolan, D.R. Targeting of Several Glycolytic Enzymes Using RNA Interference Reveals Aldolase Affects Cancer Cell Proliferation through a Non-glycolytic Mechanism. J. Biol. Chem. 2012, 287, 42554–42563. [Google Scholar] [CrossRef]
- Pekel, G.; Ari, F. Therapeutic Targeting of Cancer Metabolism with Triosephosphate Isomerase. Chem. Biodivers. 2020, 17, e2000012. [Google Scholar] [CrossRef]
- Chen, T.; Huang, Z.; Tian, Y.; Wang, H.; Ouyang, P.; Chen, H.; Wu, L.; Lin, B.; He, R. Role of triosephosphate isomerase and downstream functional genes on gastric cancer. Oncol. Rep. 2017, 38, 1822–1832. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Maqbool, R.; Parray, F.Q.; Hussain, M. Triose-phosphate isomerase is a novel target of miR-22 and miR-28, with implications in tumorigenesis. J. Cell Physiol. 2018, 233, 8919–8929. [Google Scholar] [CrossRef] [PubMed]
- Scatena, R.; Bottoni, P.; Pontoglio, A.; Mastrototaro, L.; Giardina, B. Glycolytic enzyme inhibitors in cancer treatment. Expert Opin Investig Drugs 2008, 17, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, P.; Lin, B.; Du, J.; Pan, H.; Yu, H.; He, R.; Huang, Z. Global gene expression analysis of knockdown Triosephosphate isomerase (TPI) gene in human gastric cancer cell line MGC-803. Gene 2018, 647, 61–72. [Google Scholar] [CrossRef]
- Jiang, H.; Ma, N.; Shang, Y.; Zhou, W.; Chen, T.; Guan, D.; Li, J.; Wang, J.; Zhang, E.; Feng, Y.; et al. Triosephosphate isomerase 1 suppresses growth, migration and invasion of hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2017, 482, 1048–1053. [Google Scholar] [CrossRef]
- Rodriguez-Bolaños, M.; Perez-Montfort, R. Medical and Veterinary Importance of the Moonlighting Functions of Triosephosphate Isomerase. Curr. Protein Pept. Sci. 2019, 20, 304–315. [Google Scholar] [CrossRef]
- Enríquez-Flores, S.; Flores-López, L.A.; García-Torres, I.; Mora, I.D.L.M.-D.L.; Cabrera, N.; Gutiérrez-Castrellón, P.; Martínez-Pérez, Y.; López-Velázquez, G. Deamidated Human Triosephosphate Isomerase is a Promising Druggable Target. Biomolecules 2020, 10, 1050. [Google Scholar] [CrossRef]
- Ahmed, N.; Battah, S.; Karachalias, N.; Babaei-Jadidi, R.; Horányi, M.; Baróti, K.; Hollan, S.; Thornalley, P.J. Increased formation of methylglyoxal and protein glycation, oxidation and nitrosation in triosephosphate isomerase deficiency. Biochim. Biophys. Acta (BBA) -Mol. Basis Dis. 2003, 1639, 121–132. [Google Scholar] [CrossRef]
- Enríquez-Flores, S.; Flores-López, L.A.; Mora, I.D.L.M.-D.L.; García-Torres, I.; Gracia-Mora, I.; Gutiérrez-Castrellón, P.; Fernández-Lainez, C.; Martínez-Pérez, Y.; Olaya-Vargas, A.; de Vos, P.; et al. Naturally occurring deamidated triosephosphate isomerase is a promising target for cell-selective therapy in cancer. Sci. Rep. 2022, 12, 4028. [Google Scholar] [CrossRef]
- Marcus, Y.; Tal, N.; Ronen, M.; Carmieli, R.; Gurevitz, M. The drug ornidazole inhibits photosynthesis in a different mechanism described for protozoa and anaerobic bacteria. Biochem. J. 2016, 473, 4413–4426. [Google Scholar] [CrossRef]
- Liu, P.; Sun, S.-J.; Ai, Y.-J.; Feng, X.; Zheng, Y.-M.; Gao, Y.; Zhang, J.-Y.; Zhang, L.; Sun, Y.-P.; Xiong, Y.; et al. Elevated nuclear localization of glycolytic enzyme TPI1 promotes lung adenocarcinoma and enhances chemoresistance. Cell Death Dis. 2022, 13, 205. [Google Scholar] [CrossRef]
- Zhu, X.; Jin, C.; Pan, Q.; Hu, X. Determining the quantitative relationship between glycolysis and GAPDH in cancer cells exhibiting the Warburg effect. J. Biol. Chem. 2021, 296, 100369. [Google Scholar] [CrossRef]
- Lazarev, V.F.; Mikhaylova, E.R.; Dutysheva, E.A.; Suezov, R.V.; Guzhova, I.V.; Margulis, B.A. A hydrocortisone derivative binds to GAPDH and reduces the toxicity of extracellular polyglutamine-containing aggregates. Biochem. Biophys. Res. Commun. 2017, 487, 723–727. [Google Scholar] [CrossRef]
- Li, T.; Tan, X.; Yang, R.; Miao, Y.; Zhang, M.; Xi, Y.; Guo, R.; Zheng, M.; Li, B. Discovery of novel glyceraldehyde-3-phosphate dehydrogenase inhibitor via docking-based virtual screening. Bioorg. Chem. 2020, 96, 103620. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Mullarky, E.; Lu, C.; Bosch, K.N.; Kavalier, A.; Rivera, K.; Roper, J.; Chio, I.I.C.; Giannopoulou, E.G.; Rago, C.; et al. Faculty Opinions recommendation of Vitamin C selectively kills KRAS and BRAF mutant colorectal cancer cells by targeting GAPDH. Science 2015, 350, 1391–1396. [Google Scholar] [CrossRef] [PubMed]
- Rahier, N.J.; Molinier, N.; Long, C.; Deshmukh, S.K.; Kate, A.S.; Ranadive, P.; Verekar, S.A.; Jiotode, M.; Lavhale, R.R.; Tokdar, P.; et al. Anticancer activity of koningic acid and semisynthetic derivatives. Bioorg. Med. Chem. 2015, 23, 3712–3721. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, M.D.; Bhargava, P.; Kim, P.M.; Putluri, V.; Snowman, A.M.; Putluri, N.; Calabresi, P.A.; Snyder, S.H. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 2018, 360, 449–453. [Google Scholar] [CrossRef]
- Vanle, B.C.; Florang, V.R.; Murry, D.J.; Aguirre, A.L.; Doorn, J.A. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by the dopamine metabolite, 3,4-dihydroxyphenylacetaldehyde. Biochem. Biophys. Res. Commun. 2017, 492, 275–281. [Google Scholar] [CrossRef]
- Liao, S.-T.; Han, C.; Xu, D.-Q.; Fu, X.-W.; Wang, J.-S.; Kong, L.-Y. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 2019, 10, 5091. [Google Scholar] [CrossRef]
- Carneiro, A.S.; Lameira, J.; Alves, C.N. A theoretical study of the molecular mechanism of the GAPDH Trypanosoma cruzi enzyme involving iodoacetate inhibitor. Chem. Phys. Lett. 2011, 514, 336–340. [Google Scholar] [CrossRef]
- Sakai, K.; Hasumi, K.; Endo, A. Inactivation of rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by koningic acid. Biochim. et Biophys. Acta (BBA) -Protein Struct. Mol. Enzym. 1988, 952, 297–303. [Google Scholar] [CrossRef]
- Li, W.; Liao, L.-P.; Song, N.; Liu, Y.-J.; Ding, Y.-L.; Zhang, Y.-Y.; Zhou, X.-R.; Sun, Z.-Y.; Xiao, S.-H.; Wang, H.-B.; et al. Natural product 1,2,3,4,6-penta-O-galloyl-β-D-glucopyranose is a reversible inhibitor of glyceraldehyde 3-phosphate dehydrogenase. Acta Pharmacol. Sin. 2022, 43, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Chiarelli, L.R.; Morera, S.M.; Bianchi, P.; Fermo, E.; Zanella, A.; Galizzi, A.; Valentini, G. Molecular Insights on Pathogenic Effects of Mutations Causing Phosphoglycerate Kinase Deficiency. PLoS ONE 2012, 7, e32065. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, Y.; Lu, Z. PGK1 is a new member of the protein kinome. Cell Cycle 2016, 15, 1803–1804. [Google Scholar] [CrossRef] [PubMed]
- Gründker, C.; Wokoun, U.; Hellriegel, M.; Emons, G. Inhibition of aerobic glycolysis enhances the anti-tumor efficacy of Zoptarelin Doxorubicin in triple-negative breast cancer cells. J. Obstet. Gynaecol. Res. 2019, 45, 1334–1342. [Google Scholar] [CrossRef]
- Xie, H.; Tong, G.; Zhang, Y.; Liang, S.; Tang, K.; Yang, Q. PGK1 Drives Hepatocellular Carcinoma Metastasis by Enhancing Metabolic Process. Int. J. Mol. Sci. 2017, 18, 1630. [Google Scholar] [CrossRef]
- Shashni, B.; Sakharkar, K.R.; Nagasaki, Y.; Sakharkar, M.K. Glycolytic enzymes PGK1 and PKM2 as novel transcriptional targets of PPARγ in breast cancer pathophysiology. J. Drug Target. 2013, 21, 161–174. [Google Scholar] [CrossRef]
- Hu, H.; Zhu, W.; Qin, J.; Chen, M.; Gong, L.; Li, L.; Liu, X.; Tao, Y.; Yin, H.; Zhou, H.; et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology 2017, 65, 515–528. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, G.; Chu, H.; Wang, X.; Xiong, L.; Cai, G.; Liu, R.; Gao, H.; Tao, B.; Li, W.; et al. Macrophage-Associated PGK1 Phosphorylation Promotes Aerobic Glycolysis and Tumorigenesis. Mol. Cell 2018, 71, 201–215.e7. [Google Scholar] [CrossRef]
- Jin, C.; Zhu, X.; Wu, H.; Wang, Y.; Hu, X. Perturbation of phosphoglycerate kinase 1 (PGK1) only marginally affects glycolysis in cancer cells. J. Biol. Chem. 2020, 295, 6425–6446. [Google Scholar] [CrossRef]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, Y.C.; Fang, C.; Russell, R.; Kim, J.H.; Fan, W.; Liu, R.; Zhong, Q.; Guan, K.-L. Differential Regulation of Distinct Vps34 Complexes by AMPK in Nutrient Stress and Autophagy. Cell 2013, 152, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Li, X.; Cai, Q.; Zhang, C.; Yu, Q.; Jiang, Y.; Lee, J.-H.; Hawke, D.; Wang, Y.; Xia, Y.; et al. Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Mol. Cell 2017, 65, 917–931.e6. [Google Scholar] [CrossRef]
- Wang, W.-L.; Jiang, Z.-R.; Hu, C.; Chen, C.; Hu, Z.-Q.; Wang, A.-L.; Wang, L.; Liu, J.; Liu, Q.-S. Pharmacologically inhibiting phosphoglycerate kinase 1 for glioma with NG52. Acta Pharmacol. Sin. 2021, 42, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, L.; Yu, G.; Qi, X.; Zhang, A.; Lu, Z.; Li, D.; Li, J. Identification of a novel non-ATP-competitive protein kinase inhibitor of PGK1 from marine nature products. Biochem. Pharmacol. 2021, 183, 114343. [Google Scholar] [CrossRef]
- Mikawa, T.; Shibata, E.; Shimada, M.; Ito, K.; Ito, T.; Kanda, H.; Takubo, K.; Shimada, A.; Lleonart, M.E.; Inagaki, N.; et al. Characterization of genetically modified mice for phosphoglycerate mutase, a vitally-essential enzyme in glycolysis. PLoS ONE 2021, 16, e0250856. [Google Scholar] [CrossRef]
- Li, N.; Liu, X. Phosphoglycerate Mutase 1: Its Glycolytic and Non-Glycolytic Roles in Tumor Malignant Behaviors and Potential Therapeutic Significance. Onco Targets Ther. 2020, 13, 1787–1795. [Google Scholar] [CrossRef]
- Huang, K.; Liang, Q.; Zhou, Y.; Jiang, L.-L.; Gu, W.-M.; Luo, M.-Y.; Tang, Y.-B.; Wang, Y.; Lu, W.; Huang, M.; et al. A Novel Allosteric Inhibitor of Phosphoglycerate Mutase 1 Suppresses Growth and Metastasis of Non-Small-Cell Lung Cancer. Cell Metab. 2019, 30, 1107–1119.e8. [Google Scholar] [CrossRef]
- Qu, J.; Sun, W.; Zhong, J.; Lv, H.; Zhu, M.; Xu, J.; Jin, N.; Xie, Z.; Tan, M.; Lin, S.-H.; et al. Phosphoglycerate mutase 1 regulates dNTP pool and promotes homologous recombination repair in cancer cells. J. Cell Biol. 2017, 216, 409–424. [Google Scholar] [CrossRef]
- Zhang, D.; Jin, N.; Sun, W.; Li, X.; Liu, B.; Xie, Z.; Qu, J.; Xu, J.; Yang, X.; Su, Y.; et al. Phosphoglycerate mutase 1 promotes cancer cell migration independent of its metabolic activity. Oncogene 2017, 36, 2900–2909. [Google Scholar] [CrossRef]
- Rodwell, V.W.; Towne, J.C.; Grisolia, S. The Kinetic Properties of Yeast and Muscle Phosphoglyceric Acid Mutase. J. Biol. Chem. 1957, 228, 875–890. [Google Scholar] [CrossRef]
- Mikawa, T.; Maruyama, T.; Okamoto, K.; Nakagama, H.; Lleonart, M.E.; Tsusaka, T.; Hori, K.; Murakami, I.; Izumi, T.; Takaori-Kondo, A.; et al. Senescence-inducing stress promotes proteolysis of phosphoglycerate mutase via ubiquitin ligase Mdm2. J. Cell Biol. 2014, 204, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Hitosugi, T.; Zhou, L.; Elf, S.; Fan, J.; Kang, H.-B.; Seo, J.H.; Shan, C.; Dai, Q.; Zhang, L.; Xie, J.; et al. Phosphoglycerate Mutase 1 Coordinates Glycolysis and Biosynthesis to Promote Tumor Growth. Cancer Cell 2012, 22, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cai, W.-S.; Chen, L.; Wang, G. Molecular dynamics simulation reveals how phosphorylation of tyrosine 26 of phosphoglycerate mutase 1 upregulates glycolysis and promotes tumor growth. Oncotarget 2017, 8, 12093–12107. [Google Scholar] [CrossRef]
- Mikawa, T.; Shibata, E.; Shimada, M.; Ito, K.; Ito, T.; Kanda, H.; Takubo, K.; Lleonart, M.E.; Inagaki, N.; Yokode, M.; et al. Phosphoglycerate Mutase Cooperates with Chk1 Kinase to Regulate Glycolysis. iScience 2020, 23, 101306. [Google Scholar] [CrossRef]
- Saghatelian, M.J.; Sorensen, A.; Cravatt, B.F. Target discovery in small-molecule cell-based screens by in situ proteome reactivity profiling. Nat. Biotechnol. 2005, 23, 1303–1307. [Google Scholar]
- Li, X.; Tang, S.; Wang, Q.-Q.; Leung, E.L.-H.; Jin, H.; Huang, Y.; Liu, J.; Geng, M.; Huang, M.; Yuan, S.; et al. Identification of Epigallocatechin-3- Gallate as an Inhibitor of Phosphoglycerate Mutase 1. Front. Pharmacol. 2017, 8, 325. [Google Scholar] [CrossRef]
- Wang, P.; Jiang, L.; Cao, Y.; Zhang, X.; Chen, B.; Zhang, S.; Huang, K.; Ye, D.; Zhou, L. Xanthone derivatives as phosphoglycerate mutase 1 inhibitors: Design, synthesis, and biological evaluation. Bioorg. Med. Chem. 2018, 26, 1961–1970. [Google Scholar] [CrossRef]
- Novelli, F.; Cappello, P.; Principe, M.; Bulfamante, S. Alpha-Enolase i ENO1 i a potential target in novel immunotherapies. Front. Biosci. 2017, 22, 944–959. [Google Scholar] [CrossRef]
- Ceruti, P.; Principe, M.; Capello, M.; Cappello, P.; Novelli, F. Three are better than one: Plasminogen receptors as cancer theranostic targets. Exp. Hematol. Oncol. 2013, 2, 12. [Google Scholar] [CrossRef]
- Shih, N.-Y.; Lai, H.-L.; Chang, G.-C.; Lin, H.-C.; Wu, Y.-C.; Liu, J.M.; Liu, K.-J.; Tseng, S.-W. Anti- -enolase Autoantibodies Are Down-regulated in Advanced Cancer Patients. Jpn. J. Clin. Oncol. 2010, 40, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Capello, M.; Ferri-Borgogno, S.; Riganti, C.; Chattaragada, M.S.; Principe, M.; Roux, C.; Zhou, W.; Petricoin, E.F.; Cappello, P.; Novelli, F. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget 2016, 7, 5598–5612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Mao, Y.; Zhao, Y.; Cao, Y.; Chen, X. Role of multifaceted regulators in cancer glucose metabolism and their clinical significance. Oncotarget 2016, 7, 31572–31585. [Google Scholar] [CrossRef] [PubMed]
- Sanzey, M.; Rahim, S.A.A.; Oudin, A.; Dirkse, A.; Kaoma, T.; Vallar, L.; Herold-Mende, C.; Bjerkvig, R.; Golebiewska, A.; Niclou, S.P. Comprehensive Analysis of Glycolytic Enzymes as Therapeutic Targets in the Treatment of Glioblastoma. PLoS ONE 2015, 10, e0123544. [Google Scholar] [CrossRef]
- Tomaino, B.; Cappello, P.; Capello, M.; Fredolini, C.; Sperduti, I.; Migliorini, P.; Salacone, P.; Novarino, A.; Giacobino, A.; Ciuffreda, L.; et al. Circulating Autoantibodies to Phosphorylated α-Enolase are a Hallmark of Pancreatic Cancer. J. Proteome Res. 2011, 10, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Feo, S.; Arcuri, D.; Piddini, E.; Passantino, R.; Giallongo, A. ENO1 gene product binds to the c-myc promoter and acts as a transcriptional repressor: Relationship with Myc promoter-binding protein 1 (MBP-1). FEBS Lett. 2000, 473, 47–52. [Google Scholar] [CrossRef]
- Sonveaux, P.; Vegran, F.; Schroeder, T.; Wergin, M.C.; Verrax, J.; Rabbani, Z.N.; De Saedeleer, C.J.; Kennedy, K.M.; Diepart, C.; Jordan, B.F.; et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J. Clin. Investig. 2008, 118, 3930–3942. [Google Scholar] [CrossRef]
- Le, A.; Cooper, C.R.; Gouw, A.M.; Dinavahi, R.; Maitra, A.; Deck, L.M.; Royer, R.E.; Vander Jagt, D.L.; Semenza, G.L.; Dang, C.V. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proc. Natl. Acad. Sci. USA 2010, 107, 2037–2042. [Google Scholar] [CrossRef]
- Billiard, J.; Dennison, J.B.; Briand, J.; Annan, R.S.; Chai, D.; Colón, M.; Dodson, C.S.; A Gilbert, S.; Greshock, J.; Jing, J.; et al. Quinoline 3-sulfonamides inhibit lactate dehydrogenase A and reverse aerobic glycolysis in cancer cells. Cancer Metab. 2013, 1, 19. [Google Scholar] [CrossRef]
- Pusapati, R.V.; Daemen, A.; Wilson, C.; Sandoval, W.; Gao, M.; Haley, B.; Baudy, A.R.; Hatzivassiliou, G.; Evangelista, M.; Settleman, J. mTORC1-Dependent Metabolic Reprogramming Underlies Escape from Glycolysis Addiction in Cancer Cells. Cancer Cell 2016, 29, 548–562. [Google Scholar] [CrossRef]
- Tennant, D.A.; Durán, R.V.; Gottlieb, E. Targeting metabolic transformation for cancer therapy. Nat. Cancer 2010, 10, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, N.; Han, S.; Wang, N.; Mo, F.; Loo, T.Y.; Shen, J.; Huang, H.; Chen, J. Bioactivity-Guided Identification and Cell Signaling Technology to Delineate the Lactate Dehydrogenase A Inhibition Effects of Spatholobus suberectus on Breast Cancer. PLoS ONE 2013, 8, e56631. [Google Scholar] [CrossRef]
- Keijer, J.; van Dartel, D.A. Reprogrammed metabolism of cancer cells as a potential therapeutic target. Curr. Pharm. Des. 2014, 20, 2580–2594. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, L.; Zhu, L.T.; Wang, Y.; Pan, D.; Yao, J.; You, Q.D.; Guo, Q.L. Wogonin reverses hypoxia resistance of human colon cancer HCT116 cells via downregulation of HIF-1alpha and glycolysis, by inhibiting PI3K/Akt signaling pathway. Mol. Carcinog. 2014, 53 (Suppl. 1), E107–E118. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Korotchkina, L.G. Regulation of the pyruvate dehydrogenase complex. Biochem. Soc. Trans. 2006, 34 Pt 2, 217–222. [Google Scholar] [CrossRef]
- Feldman-Salit, A.; Hering, S.; Messiha, H.L.; Veith, N.; Cojocaru, V.; Sieg, A.; Westerhoff, H.; Kreikemeyer, B.; Wade, R.C.; Fiedler, T. Regulation of the Activity of Lactate Dehydrogenases from Four Lactic Acid Bacteria. J. Biol. Chem. 2013, 288, 21295–21306. [Google Scholar] [CrossRef]
- I Koukourakis, M.; For the ‘Tumour and Angiogenesis Research Group‘; Giatromanolaki, A.; Sivridis, E.; Bougioukas, G.; Didilis, V.; Gatter, K.C.; Harris, A. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 2003, 89, 877–885. [Google Scholar] [CrossRef]
- Xie, H.; Hanai, J.-I.; Ren, J.-G.; Kats, L.; Burgess, K.; Bhargava, P.; Signoretti, S.; Billiard, J.; Duffy, K.J.; Grant, A.; et al. Targeting Lactate Dehydrogenase-A Inhibits Tumorigenesis and Tumor Progression in Mouse Models of Lung Cancer and Impacts Tumor-Initiating Cells. Cell Metab. 2014, 19, 795–809. [Google Scholar] [CrossRef]
- Sutendra, G.; Michelakis, E.D. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front. Oncol. 2013, 3, 38. [Google Scholar] [CrossRef]
- Jane, E.P.; Premkumar, D.R.; Rajasundaram, D.; Thambireddy, S.; Reslink, M.C.; Agnihotri, S.; Pollack, I.F. Reversing tozasertib resistance in glioma through inhibition of pyruvate dehydrogenase kinases. Mol. Oncol. 2022, 16, 219–249. [Google Scholar] [CrossRef]
- Sun, W.; Xie, Z.; Liu, Y.; Zhao, D.; Wu, Z.; Zhang, D.; Lv, H.; Tang, S.; Jin, N.; Jiang, H.; et al. JX06 Selectively Inhibits Pyruvate Dehydrogenase Kinase PDK1 by a Covalent Cysteine Modification. Cancer Res. 2015, 75, 4923–4936. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Guo, Y.; Tam, K.Y. Targeting glucose metabolism to develop anticancer treatments and therapeutic patents. Expert Opin. Ther. Patents 2022, 32, 441–453. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, K.; Morii, E. Metabolic Reprogramming of Cancer Cells during Tumor Progression and Metastasis. Metabolites 2021, 11, 28. [Google Scholar] [CrossRef]
- Bonnet, S.; Archer, S.L.; Allalunis-Turner, J.; Haromy, A.; Beaulieu, C.; Thompson, R.; Lee, C.T.; Lopaschuk, G.D.; Puttagunta, L.; Bonnet, S.; et al. A Mitochondria-K+ Channel Axis Is Suppressed in Cancer and Its Normalization Promotes Apoptosis and Inhibits Cancer Growth. Cancer Cell 2007, 11, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Tso, S.-C.; Qi, X.; Gui, W.-J.; Wu, C.-Y.; Chuang, J.L.; Wernstedt-Asterholm, I.; Morlock, L.K.; Owens, K.R.; Scherer, P.E.; Williams, N.S.; et al. Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket. J. Biol. Chem. 2014, 289, 4432–4443. [Google Scholar] [CrossRef]
- Zhou, X.; Yu, S.; Su, J.; Sun, L. Computational Study on New Natural Compound Inhibitors of Pyruvate Dehydrogenase Kinases. Int. J. Mol. Sci. 2016, 17, 340. [Google Scholar] [CrossRef]
- Stacpoole, P.W. Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. JNCI J. Natl. Cancer Inst. 2017, 109, djx071. [Google Scholar] [CrossRef]
- Kooshki, L.; Mahdavi, P.; Fakhri, S.; Akkol, E.K.; Khan, H. Targeting lactate metabolism and glycolytic pathways in the tumor microenvironment by natural products: A promising strategy in combating cancer. BioFactors 2022, 48, 359–383. [Google Scholar] [CrossRef]
- Gao, J.-L.; Chen, Y.-G. Natural Compounds Regulate Glycolysis in Hypoxic Tumor Microenvironment. BioMed Res. Int. 2015, 2015, 354143. [Google Scholar] [CrossRef]
- Pérez-Escalante, E.; Cariño-Cortés, R.; Fernández-Martínez, E.; Ortiz, M.I.; Muñoz-Pérez, V.M.; Sánchez-Crisóstomo, I.; Jiménez-Ángeles, L.; Emmanuel, P.-E.; Raquel, C.-C.; Eduardo, F.-M.; et al. Colorectal Cancer: Causes and Evidence of Chemopreventive Treatments. Curr. Pharm. Biotechnol. 2018, 19, 1135–1155. [Google Scholar] [CrossRef]
- Sheng, H.; Sun, H. Synthesis, biology and clinical significance of pentacyclic triterpenes: A multi-target approach to prevention and treatment of metabolic and vascular diseases. Nat. Prod. Rep. 2011, 28, 543–593. [Google Scholar] [CrossRef] [PubMed]
- Sheng, H.; Tang, W. Glycolysis Inhibitors for Anticancer Therapy: A Review of Recent Patents. Recent Patents Anti-Cancer Drug Discov. 2016, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukushi, A.; Kim, H.-D.; Chang, Y.-C.; Kim, C.-H. Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells. Int. J. Mol. Sci. 2022, 23, 10037. https://doi.org/10.3390/ijms231710037
Fukushi A, Kim H-D, Chang Y-C, Kim C-H. Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells. International Journal of Molecular Sciences. 2022; 23(17):10037. https://doi.org/10.3390/ijms231710037
Chicago/Turabian StyleFukushi, Abekura, Hee-Do Kim, Yu-Chan Chang, and Cheorl-Ho Kim. 2022. "Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells" International Journal of Molecular Sciences 23, no. 17: 10037. https://doi.org/10.3390/ijms231710037
APA StyleFukushi, A., Kim, H.-D., Chang, Y.-C., & Kim, C.-H. (2022). Revisited Metabolic Control and Reprogramming Cancers by Means of the Warburg Effect in Tumor Cells. International Journal of Molecular Sciences, 23(17), 10037. https://doi.org/10.3390/ijms231710037








